Significance
Selfing species wheat are bred as pure-line varieties with stagnating yield growths. In contrast, selection gain in maize is high, owing to massive investment sustained by hybrid seed sales, coupled with an efficient exploitation of hybrid vigor. We have developed a three-step strategy for establishing a heterotic pattern, which was one of the central unsolved challenges for initiating hybrid breeding programs. The benefits of our approach are demonstrated using data for wheat, but the strategy is relevant for several autogamous crops. Our three-step approach facilitates identification of a heterotic pattern, and thus may contribute to meeting the global challenge of increasing demand for food, feed, and fuel.
Keywords: hybrid breeding, genomic prediction, heterotic pattern
Abstract
Hybrid breeding promises to boost yield and stability. The single most important element in implementing hybrid breeding is the recognition of a high-yielding heterotic pattern. We have developed a three-step strategy for identifying heterotic patterns for hybrid breeding comprising the following elements. First, the full hybrid performance matrix is compiled using genomic prediction. Second, a high-yielding heterotic pattern is searched based on a developed simulated annealing algorithm. Third, the long-term success of the identified heterotic pattern is assessed by estimating the usefulness, selection limit, and representativeness of the heterotic pattern with respect to a defined base population. This three-step approach was successfully implemented and evaluated using a phenotypic and genomic wheat dataset comprising 1,604 hybrids and their 135 parents. Integration of metabolomic-based prediction was not as powerful as genomic prediction. We show that hybrid wheat breeding based on the identified heterotic pattern can boost grain yield through the exploitation of heterosis and enhance recurrent selection gain. Our strategy represents a key step forward in hybrid breeding and is relevant for self-pollinating crops, which are currently shifting from pure-line to high-yielding and resilient hybrid varieties.
Wheat production must be doubled by 2050 to cope with increased demand arising from continuing population growth, increasing meat and dairy consumption, and expanding biofuel use (1). An environmentally sound approach to meeting this goal involves enhancing crop yields per area rather than clearing more land for agriculture (1); however, yield growths in wheat are stagnating in several parts of the world, affecting 37% of the global acreage (2).
Hybrid breeding is a potential disruptive technology in selfing species that could boost yield per area (3) and enhance yield stability. The latter is of particular relevance for climate-smart agriculture and low-yielding environments, where wheat is widely grown (4). Wheat hybrids are currently cultivated on only <1% of the global acreage, mainly because of the failure to implement a cost-efficient hybrid seed production system, which is required for establishment of a competitive hybrid breeding program (5). Recently, considerable progress has been achieved in developing alternative, more economically feasible hybridization systems, such as the functional characterization of potential cytoplasmic male sterility systems (6). Moreover, a proof-of-concept study has demonstrated the use of a split-gene system for hybrid wheat production (5), and a transgenic construct-driven system for production of non-genetically modified hybrid maize has been deregulated (7), which is also of interest for wheat. Consequently, it is projected that the barriers to economically feasible hybrid wheat production can be overcome in the next 10–15 years (8).
The success of hybrid wheat breeding depends crucially on the clustering of suitable germplasm into heterotic groups and on the identification of a high-yielding heterotic pattern (5). A heterotic group is a set of genotypes displaying similar hybrid performance when crossed with individuals from another, genetically distinct germplasm group (9). A specific pair of two heterotic groups expressing pronounced hybrid performance in their cross is termed a heterotic pattern. A heterotic pattern is improved by exploiting genetic variation generated within heterotic groups (10). Breeding hybrids in such a manner promotes genetic divergence among parents (11), optimizes the exploitation of heterosis and hybrid performance, and simplifies the identification of superior single crosses (12).
The heterotic pattern used for maize breeding in the US corn belt, the cradle of hybrid breeding, did not exist initially (11). The germplasm was not structured into heterotic groups, but with the introduction of single-cross hybrids, available inbred lines were clustered into a female pool and a male pool according to production traits, such as seed yield. With ongoing hybrid breeding, the male and female groups coevolved and diverged (13), most likely owing to differential fixation of quantitative trait locus (QTL) alleles caused by dominance or overdominance (14).
The great success of maize hybrids in the US corn belt stimulated the initiation of hybrid breeding programs for several other outcrossing crops, including sunflower, sugar beet, and rye. Heterotic patterns for these second-generation hybrid crops were established empirically by testing hybrid combinations among potential parental lines in field trials. Importantly, however, because the number of all pairwise single crosses is a quadratic function of the number of parents, evaluating all hybrid combinations in field trials is not manageable for most crops. Thus, the picture of the combining ability of potential parents in a hybrid breeding program has always been incomplete.
Various approaches to improving the overall view of combining ability in the absence of exhaustive crossing schemes have been proposed. Molecular marker-based genetic distance has been promoted as a proxy for heterosis and ultimately hybrid performance, although this is not the case for unrelated parental lines (12). Alternatively, genome-wide approaches can be used to predict hybrid performance (3, 10, 14). Genomic prediction is particularly promising for the complex trait of grain yield, because pedigree information, cosegregation, and linkage disequilibrium between markers and QTLs are jointly exploited. To date, genomic prediction has not been used to identify heterotic groups for hybrid breeding, however.
When data on hybrid performance of all pairwise single crosses are available, efficient algorithms are needed to sift through the plethora of potential groupings to identify the most promising heterotic pattern. An extended enumeration algorithm has been suggested for this search (15), but this class of algorithms is too computationally intensive for large populations. Moreover, optimizing hybrid performance focuses on the short-term success of a heterotic pattern. This short-term success arises mainly through high selection intensities (16) and a low number of elite founder individuals in the heterotic groups. In contrast, long-term selection gain benefits from genetic variance, which is associated with a large effective population size of the heterotic pattern. Consequently, criteria for maximizing high short-term selection gains without promptly diminishing genetic variance are required in the search for heterotic patterns, but such approaches are not yet in place.
Here we report a unified quantitative genetic framework for identifying a promising heterotic pattern considering short- and long-term selection gains. To demonstrate the scope of this framework, we assembled the largest phenotypic, metabolomic, and genomic hybrid wheat dataset documented to date, comprising 1,604 single crosses established from a diverse set of 135 Central European elite lines. We first evaluated the potential of genomic- and metabolomic-based hybrid prediction to derive the data required to identify heterotic groups. We then applied our genomic prediction models to predict the performance of all 9,045 possible unique single-cross hybrids. These data served to identify heterotic patterns with variable population sizes, maximizing hybrid performance as determined based on a developed simulated annealing algorithm. We then studied the suitability of several parameters for judging the long-term potential of the selected heterotic pattern. Wheat has been successfully used for the proof of principle presented here, but the developed quantitative genetic framework is generically applicable to other self-fertilizing crops as well.
Results
Hybrid Superiority Demonstrated Through Large-Scale Phenotyping.
We sampled 120 diverse female and 15 male wheat lines adapted to Central Europe (SI Appendix, Table S1), and produced 1,604 single-cross hybrids. We evaluated the genotypes for grain yield in field trials across 11 environments to produce high-quality phenotypic data (Dataset S1). This is reflected by a broad-sense heritability of 73% (SI Appendix, Table S2 and Fig. S1A), which closely corresponds to expectations resulting from other phenotypic variance components reported for Central European wheat populations (17). In total, 97 hybrids significantly (P < 0.05) outperformed the highest-yielding released line variety Tobak, with a maximum surplus of 1 Mg ha−1 (SI Appendix, Fig. S1C). This improvement reflects roughly 15 y of breeding progress (18) in a single year, clearly exemplifying the potential boost to wheat grain yield through hybrid breeding.
Wheat breeders do not emphasize grain yield exclusively, but also consider abiotic and biotic stress resistance, as well as food or feed quality. Thus, we created an index comprising grain yield, six abiotic and biotic stress traits (frost tolerance, resistance to brown and yellow rust, Fusarium head blight, powdery mildew, and Septoria tritici blotch), and seven quality characteristics (1,000-kernel weight, gluten content, kernel hardness, protein content, sedimentation volume, starch content, and test weight) (SI Appendix, Fig. S1B). Importantly, the superiority of particular hybrids over the best released line variety is not restricted to grain yield (9%), but is also present, albeit slightly less pronounced (6%), for an index combining the above-listed agronomic and quality traits.
Genomic Prediction Allowed Compilation of the Required High-Quality Hybrid Performance Data to Identify Heterotic Groups.
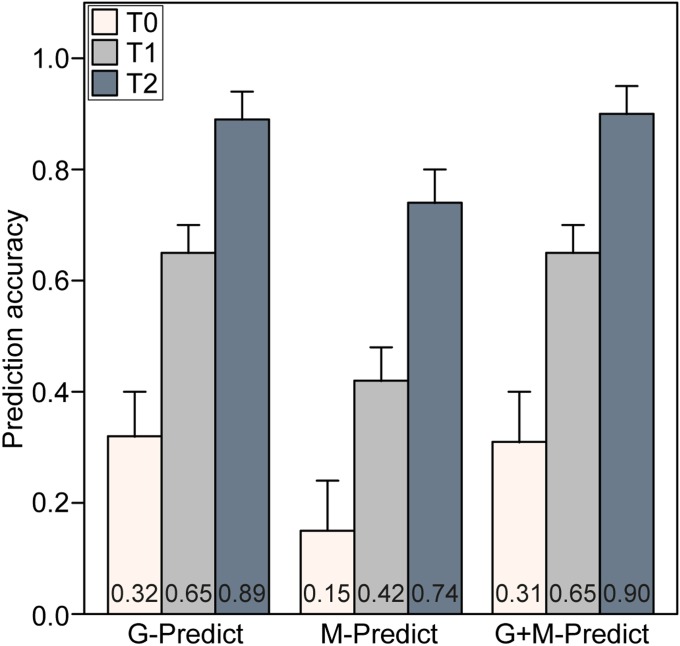
Estimates of the hybrid performance of all 9,045 pairwise single crosses are required to search for heterotic patterns among the 135 parental wheat lines. Because each of the 135 parents was tested in several of the 1,604 hybrids, and male and female lines do not reflect different germplasm pools (SI Appendix, Note a), the prediction accuracy of the remaining 7,441 nonphenotyped hybrids corresponds approximately to that of the T2 scenario obtained in the chessboard-like cross-validation study (SI Appendix, Note b). This is further substantiated by the high reliability values of the hybrid performance estimated for the 7,441 single crosses, comparable to those of the phenotyped hybrids (SI Appendix, Note b). The reliability criterion, which has been proposed in the context of animal breeding (19), is a measure of the prediction accuracy of a particular hybrid determined based solely on its genotypic data (SI Appendix, Note b). The prediction accuracy of the T2 scenario based on additive and dominance effects was high at 0.89 (Fig. 1 and SI Appendix, Note b). This value is higher than previously observed for a population comprising 90 wheat hybrids (20), but corresponds to findings reported for 1,254 factorial crosses in a public maize breeding program (14). Thus, the high prediction accuracy observed in our study can be explained by a large population size of 1,604 single crosses, the phenotyping in 11 environments, as well as efficient exploitation of genetic relatedness for hybrid prediction. The quality of the resulting predicted hybrid performances corresponds to a broad-sense heritability of field trials conducted in seven environments (SI Appendix, Fig. S2); consequently, the hybrid data provide a solid database for identifying heterotic patterns among the 135 lines.
Fig. 1.
Accuracy of genomic (G-Predict), metabolomic (M-Predict), and joint genomic and metabolomics-based (G+M-Predict) prediction of hybrid performance. The results are based on G-BLUP models exploiting additive and dominance effects. T2 test sets included hybrids sharing both parental lines, T1 test sets included hybrids sharing one parental line, and T0 test sets included hybrids with no parental line in common with the hybrids in the related training sets.
If further lines outside the sample space of 135 parents were considered for identifying heterotic groups, then prediction accuracies obtained in the less-related T1 and T0 scenarios were relevant. Prediction accuracies declined with reducing relatedness between the training and test populations, from 0.89 for T2 to 0.65 for T1 and to 0.32 for T0 (Fig. 1). This decreasing trend can be explained by an increasing relevance of exploiting information on linkage disequilibrium between the QTLs and single-nucleotide polymorphisms (SNPs) for genomic prediction for the T0 scenario vs. the T2 scenario. The genetic architecture of grain yield is complex with the absence of a large-effect QTL, as revealed by an association mapping study combined with fivefold cross-validation (SI Appendix, Note c). Thus, exploiting information on linkage disequilibrium between the QTLs and SNPs for hybrid prediction is challenging.
Efficient Designs of Training Populations Increased the Prediction Accuracy Using Significantly Less Resources.
We examined the prediction accuracy after reducing the number of hybrids but keeping the number of parents constant (SI Appendix, Note d). Our findings clearly show that a drastic reduction in the number of hybrids from 1,604 to 360 caused only a marginal 3% loss in prediction accuracy for the T2 scenario. Interestingly, randomly sampling subsets of hybrids yielded higher prediction accuracies compared with targeted designs, such as the nested factorial (1%), balanced incomplete factorial (1%), and top-cross (9%) designs (SI Appendix, Note d). Consequently, in cases of limited resources, random missing designs can facilitate a shift from the T0 scenario to the T2 scenario, thereby tripling the prediction accuracy of the nonphenotyped hybrids from ∼0.3 to ∼0.9 (Fig. 1).
Neither Modeling Epistasis nor Metabolite Profiling Increased Prediction Accuracy.
We examined the prediction accuracy of genomic prediction models, considering both main (i.e., additive and dominance) and epistatic effects. Splitting the total genetic variance into its different components demonstrated the important contribution of additive effects (71%), but also highlighted the relevance of epistatic effects (20% of the genetic variance) (SI Appendix, Fig. S3). Nevertheless, the prediction accuracies for the T0, T1, and T2 scenarios profited only marginally, with a maximum 3% increase through modeling of main and epistatic effects (SI Appendix, Table S3). This can be explained by the fact that the additive and dominance kinship matrices are correlated with the kinship matrices of the epistatic effects (SI Appendix, Table S4), and thus capture much information for hybrid prediction.
Metabolite profiling provides complementary data to genome-based hybrid prediction because of the high level of condensed information that can be collected with large-scale automated analytical platforms (21). Thus, we generated metabolite profiles from flag leaf samples of the parental lines collected in multienvironmental trials at three locations, to show significant (P < 0.05) genetic variances with average heritability estimates of 46% (SI Appendix, Table S5). Despite the high quality of the metabolite profiles and the low correlation between the genetic and metabolic distances (r = 0.02; P < 0.34), the genome-based prediction accuracies could not be improved any further.
Our Simulated Annealing Algorithm Enabled Prediction of a High-Yielding Heterotic Pattern.
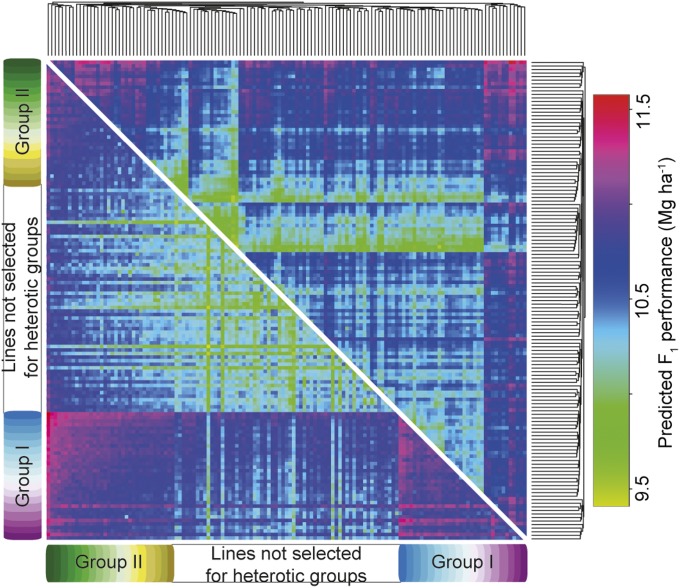
We used the genomic prediction model calibrated based on the grain yield data of the phenotyped individuals and predicted the performance of all 9,045 unique single-cross hybrids among the 135 parental lines (Fig. 2). No obvious groups of lines displaying high hybrid performance could be identified using the complete linkage clustering method. Consequently, we developed a simulated annealing algorithm (SI Appendix, Note e), which enabled the identification of a high-yielding heterotic pattern (Fig. 2).
Fig. 2.
Heat plot of predicted hybrid performance ordered using complete linkage clustering (above the diagonal) and ordered based on the developed simulated annealing algorithm (below the diagonal).
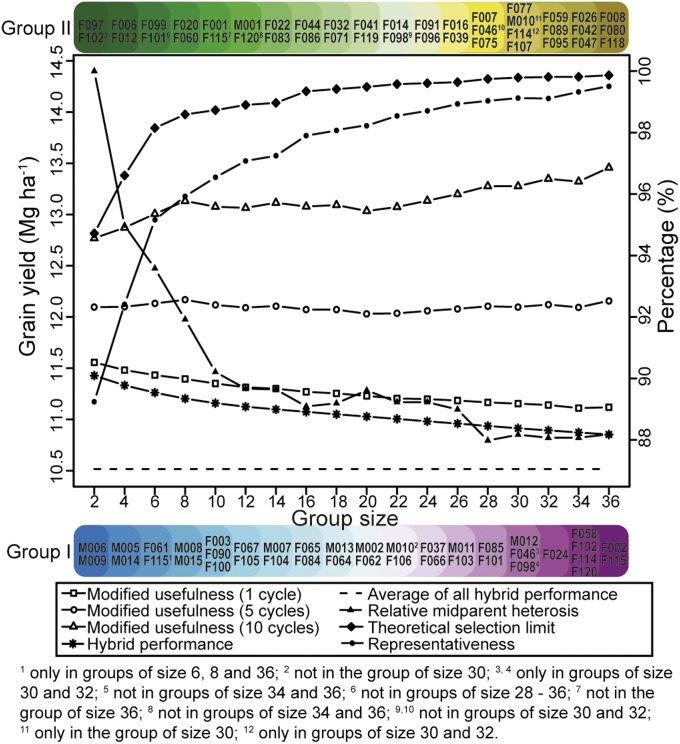
Interestingly, we found that once lines had been clustered into a heterotic group, they mostly remained within that group even when the population size was expanded (Fig. 3). We did note some exceptions, however; for example, line F115 clustered into heterotic group I only for population sizes 6, 8, and 36, but was assigned into heterotic group II for the remaining population sizes. Nonetheless, it is important to note that the groupings were identified based solely on predicted hybrid performance. Therefore, we devised a cross-validation scenario exclusively for the phenotyped hybrids, and confirmed the stability of the groups identified with the simulated annealing algorithm. Approximately 80% of the individuals overlapped between the group identified based on predicted hybrid performance and that identified based on observed hybrid performance (SI Appendix, Note e). The identified heterotic groups outperformed the average performance of all possible 9,045 hybrids with a maximum difference of 0.91 Mg ha−1, greater than that reported for groups identified with alternative clustering approaches (SI Appendix, Note e). The lines clustered into heterotic group I (Fig. 3 and SI Appendix, Note e) often consisted of top performers with respect to general combining ability effects (SI Appendix, Table S1); nevertheless, the population of intragroup hybrids did not outperform the intergroup single crosses for all examined sizes of the heterotic groups (SI Appendix, Note e). Decreasing the number of individuals per heterotic group resulted in enhanced performance of the hybrid populations, owing to higher selection intensities among parental lines. This improved hybrid performance went hand in hand with an increased midparent heterosis (Fig. 3), which was expected because of the highly significant correlation (r = 0.41; P < 0.001) observed between midparent heterosis and hybrid performance for the 9,045 single crosses.
Fig. 3.
Heterotic groups of varying sizes identified maximizing the hybrid performance between them. Shown are hybrid performance (Mg ha−1); standardized midparent heterosis (percentage); representativeness of heterotic groups in relation to the full population of 135 lines (percentage); usefulness (Mg ha−1) after 1, 5, and 10 cycles of selection; and selection limit (Mg ha−1) in relation to the heterotic group size.
Long-Term Success for Grain Yield Can Be Attained with Heterotic Group Sizes of Only 16 Individuals.
The long-term success of heterotic groups depends not only on the initial mean value of the hybrid population, but also on the realized future selection gain. The latter parameter is driven by the design and allocation of resources to the respective hybrid wheat breeding program, in addition to the diversity of the selected lines determining the increase in grain yield in the hybrid population. As a first criterion for judging the long-term success of the selected heterotic pattern, we used the usefulness criterion, which takes both the mean performance of the population and the expected gain of one cycle of selection into consideration (22). We expanded the usefulness by assuming additional 5 and 10 cycles of selection and a constant genetic variance. Previous simulation studies have shown that the assumption of a constant genetic variance is valid within 10 cycles of selection (23). Maximizing the usefulness for one cycle of selection favors small heterotic group sizes, whereas selection favors large heterotic group sizes as the tenth cycle is approached (Fig. 3). Nevertheless, the usefulness criterion showed no linear increase with increasing size of the heterotic groups; for instance, the slope of the usefulness criteria decreased more than twofold in the interval from two to eight parental lines compared with in the interval from 8 to 36 individuals per heterotic group.
As a second criterion, we used the estimated additive and dominance effects of the SNPs and examined the selection limits reached after an infinite number of selection cycles. The selection limit increased with growing size of the heterotic groups, but plateaued at approximately 16 individuals per pool (Fig. 3). Interestingly, this plateau was also observed for the genetic representativeness of the heterotic groups with respect to the total population of 135 lines, which was estimated based solely on the genomic data (Fig. 3). Consequently, these results indicate that starting in Central Europe, a hybrid wheat breeding program with heterotic groups comprising the identified 16 individuals may guarantee long-term success in improving grain yield performance.
Discussion
Breeding Based on the Identified Heterotic Pattern Boosts Midparent Heterosis and Enhances Recurrent Selection Gain.
To highlight the advantages of our approach to the search for an optimal heterotic pattern, we contrasted it with several alternative approaches, including the use of midparent or better-parent heterosis and general combining ability effects or per se performance of parents (SI Appendix, Note e). The trends across alternatives and the resulting conclusions were comparable, and thus we discussed them only for a strategy in which parents were selected based on their per se performance, followed by a random clustering of superior lines into heterotic groups. Interestingly, the selection of parents was similar, with an average of 71% overlap of selected lines in both approaches (Table 1). This similarity can be explained by the moderate to high correlation observed between midparent performance and hybrid performance (r = 0.62; P < 0.001). Despite the overlap of selected lines, the heterotic pattern defined according to the targeted grouping of parents based on the simulated annealing algorithm yielded midparent heterosis values up to 21% higher than values yielded by the scenario of random grouping of lines (Table 1). Surprisingly, the increase in midparent heterosis was due only in part to enhanced genetic diversity between the heterotic groups identified based on the simulated annealing algorithm compared with the per se scenario (Table 1 and SI Appendix, Fig. S4). This can be explained by the use of neutral SNP markers for estimating genetic diversity, which is not significantly correlated with heterosis (r = 0.09; P = 0.18) (SI Appendix, Fig. S5).
Table 1.
Comparison of overlapping genotypes (OG); yield increase, expressed as number of years needed to realize this selection gain (ΔSG, y) (18); increase in midparent heterosis (ΔMPH); increase in average Rogers’ distance (ΔRD); and decrease in the ratio of dominance vs. additive genetic variance (ΔVC) contrasting the heterotic pattern identified based on the simulated annealing algorithm vs. random grouping among the best per se performance parental lines
| Variable | Group size | |||||||||||||||||
| 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 | 22 | 24 | 26 | 28 | 30 | 32 | 34 | 36 | |
| OG, % | 25 | 50 | 50 | 63 | 65 | 67 | 68 | 75 | 78 | 78 | 80 | 77 | 81 | 84 | 83 | 83 | 85 | 83 |
| ΔSG, y | 7.5 | 4.4 | 4.8 | 3.8 | 3.8 | 3.7 | 3.6 | 3.1 | 3.0 | 2.9 | 2.8 | 2.8 | 2.7 | 2.4 | 2.6 | 2.7 | 2.7 | 2.7 |
| ΔMPH, % | 21 | 9 | 8 | 8 | 7 | 7 | 7 | 6 | 6 | 7 | 6 | 6 | 6 | 6 | 7 | 8 | 8 | 8 |
| ΔRD, % | 17 | −2 | 1 | 2 | 1 | 2 | 2 | 3 | 3 | 3 | 2 | 2 | 3 | 3 | 4 | 3 | 3 | 3 |
| ΔVC, % | 95 | 92 | 77 | 74 | 55 | 32 | 45 | 46 | 47 | 42 | 41 | 43 | 39 | 41 | 26 | 25 | 35 | 32 |
Another potential benefit of our targeted strategy for searching for a heterotic pattern relates to the ratio of genetic variance components in the hybrid population, which depends on allele frequencies. Based on quantitative genetic theory, hybrid breeding using divergent heterotic groups would be expected to promote the additive over the dominance genetic variance (12). Accordingly, we observed a higher ratio of the additive versus the dominance genetic variance for the heterotic groups identified based on the simulated annealing algorithm compared with the scenario of random grouping of lines (Table 1). The enhanced relevance of additive genetic variance contributes to an increase in recurrent selection gain (16). Moreover, predictions based on additive effects are more accurate than those based on dominance effects (14). Thus, breeding based on the identified heterotic pattern also increases the prediction accuracies in genomic prediction.
Focusing on Hybrid Performance and Ignoring Hybrid Seed Production Traits Appears Justified.
We focused our approach on maximizing grain yield of the hybrid population while ignoring traits relevant for hybrid seed production. In wheat, the most important limiting factor in a cost-efficient hybrid seed production system is low pollen shedding in the cleistogamic wheat flower (5). Redesigning the wheat flower (7) seems feasible, given the large genetic variation present in genetic resources and even in elite breeding pools (24). This, in combination with advances in molecular breeding tools that enable the efficient modulation of major genes (25), facilitates the exclusive focus on grain yield as the most important trait in the search for promising heterotic patterns.
Large Population Sizes of Heterotic Groups Enable Coevolution Owing to Differential Fixation of QTL Alleles.
Intense selection increases short-term selection gain, but a reduction in the effective population size carries a cost in terms of long-term response (23). This could be of particular relevance for hybrid breeding, where ongoing reciprocal recurrent selection likely leads to divergent chromosomal regions caused by dominance or overdominance (14). Consequently, once a heterotic pattern has been identified and enhanced through interpopulation selection, the introgression of novel germplasm potentially disrupts coevolved gene complexes. This clearly suggests that larger heterotic group sizes are beneficial.
We observed that the selection limit targeting long-term response plateaus at approximately 16 individuals per heterotic group (Fig. 3). At first glance, this population size seems low and suggests that hybrid breeding schemes should be implemented to maximize the response for selection with a predefined rate of inbreeding (26). Interestingly, the effective number of ancestors for the Iowa Stiff Stalk Synthetic and Iodent heterotic groups used intensively in North American maize was around 16 as well (11). Substantial maize yield increases were realized in the United States, with grain yield doubled in the past eight decades through hybrid breeding (13). Considering this, along with the role of mutations generating new variations (27), suggests that a heterotic group size of around 16 individuals guarantees a sustainable long-term selection gain in hybrid wheat breeding for Central Europe. Impending genetic vulnerability to evolving pests and diseases, which cannot be tackled with the latent genetic variation present within heterotic groups, can be counteracted with targeted introgressions of relevant major resistance genes into the heterotic groups.
Expanding the Search for Promising Heterotic Patterns Toward Exotic Germplasm Is Challenging.
For wheat, heterotic groups between adapted and nonadapted lines, such as winter by spring types, have been suggested because of their high genetic divergence, which would be expected to increase heterosis and hybrid performance (28). The proposed random missing mating design for the T2 scenario (SI Appendix, Note d) facilitates a cost-efficient expansion of the diversity of parental lines beyond the sample space of the 135 parents used here. The challenge lies in the production and field evaluation of such a broad array of hybrids. Flowering time is considered a critical adaptive trait for wheat production (29). Sampling of diverse parents adapted to different target environments is often associated with large differences in flowering time; however, hybrid seed production among the parental lines requires synchronization of flowering time, which is optimized if the female line flowers 3–4 d earlier than the male line. Even if hybrids between parents with a large difference in flowering time are produced, precise field evaluation is impeded because adaptation problems, such as improper maturity, mask the genetic potential of hybrids. One approach to expanding the sampling space of parental lines involves relying on genomic predictions for the unrelated T0 or T1 scenarios, but this would lead to a drastic reduction in prediction accuracy (Fig. 1). Consequently, despite its potential, searching for promising heterotic patterns among adapted and exotic germplasm remains challenging.
As one option for enhancing the T0 prediction accuracy, we evaluated the potential of complementing genomic data with metabolite profiles, but found that this failed to increase prediction accuracy (Fig. 1), as was reported previously for maize (10). One reason for this failure might lie in the sampling of flag leafs under field conditions. Further research is needed to investigate the option of improving prediction accuracy through metabolite profiling under strictly controlled conditions.
Besides metabolite profiling, there are other promising genomics data sources with the potential to enhance the prediction accuracy of hybrid performance exploiting, for instance, structural variation (30) or transcriptome profiling (31). It would be of interest to integrate these data sources into the existing genomic selection models to increase the accuracy of prediction of hybrid performance for unrelated genotypes.
The Proposed Strategy for Searching for Heterotic Patterns Is Generally Applicable in Autogamous Crops.
As suggested based on quantitative genetic considerations (9) and empirical evidence in hybrid maize breeding (32), the single most important element of a hybrid breeding program is the recognition and utilization of a heterotic pattern. Here we have developed a three-step approach to identifying a promising heterotic pattern that comprises the following elements: (i) compilation of a full hybrid performance matrix using genomic prediction; (ii) a search for a high-yielding heterotic pattern based on a simulated annealing algorithm; and (iii) assessment of the long-term success of the identified heterotic pattern. We have evaluated this three-step approach using a comprehensive experimental dataset of wheat adapted to Central Europe. Using this germplasm to identify heterotic groups is particularly challenging because of the absence of genetically distinct subpopulations (SI Appendix, Fig. S4), the result of constant exchanges of lines between wheat breeding programs (17). Nevertheless, it reflects the typical scenario for many crops that are in the infancy of hybrid breeding, including rice (33), barley (34), pearl millet (35), and pigeon pea (36). Thus, the framework for the recognition of heterotic groups developed in our study represents a central step forward in the introduction of sustainable hybrid varieties to the market for several important crops, with the final goal of meeting the global challenges of an increasing demand for food, feed, and fuel.
Materials and Methods
Plant Material, Genotyping, Field Data, and Metabolite Profiling.
Our study is based on 135 advanced elite winter wheat lines (SI Appendix, Table S1), which reflect a broad range of diversity present in Central Europe (37). The lines were grouped into a female pool and a male pool according to pollination capability, plant height, and flowering time. The 15 male lines were crossed with the 120 female lines using a factorial mating design. For 1,604 of the potential 1,800 single-cross hybrids, a sufficient amount of seeds was harvested for intensive field trials. Details of hybrid seed production have been published elsewhere (37). We added eight commercial lines and two commercial hybrid varieties (quality class E: Genius; quality class A: As de Coeur (hybrid variety), JB Asano, Julius, Tuerkis; quality class B: Colonia, Hystar (hybrid variety), Kredo, Tobak; quality class C: Tabasco) reflecting the current yield performance in Germany.
We fingerprinted the 120 female and 15 male lines using a 90,000 SNP array based on an Illumina Infinium assay (38). After quality tests, 17,372 high-quality SNP markers were retained (Dryad Digital Repository: doi:10.5061/dryad.461nc). The genomic profiles of the hybrids were deduced using the fingerprints of the parental lines. For a selected set of 20 hybrids, we cross-checked the deduced fingerprints from their parents and observed a very low rate of mismatches, with an average of 1.9%.
We evaluated all genotypes (135 parents, 1,604 hybrids, 10 commercial varieties) for grain yield in 11 environments. Details of the experimental design and data analyses are provided in SI Appendix, Note f. We combined the grain yield data with published data on six abiotic and biotic stress traits (frost tolerance, resistance to brown and yellow rust, Fusarium head blight, powdery mildew, and Septoria tritici blotch) and seven quality characteristics (1,000-kernel weight, gluten content, kernel hardness, protein content, sedimentation volume, starch content, and test weight) and estimated an index (SI Appendix, Fig. S1B). The index was calculated by standardizing the values for each trait through division by the SD and subtraction of the mean value. Grain yield was weighted with 60%, abiotic and biotic stress traits with 20%, and quality traits with 20%.
For each of the 135 parental lines, we sampled 10 flag leafs per replicate at three environments at the time when >60% of the genotypes had reached BBCH-69 (39). The measurement of polar flag leaf extracts followed the protocol outlined by Lippmann et al. (40). Details of the metabolite profiling and metabolite data analyses are provided in SI Appendix, Note g.
Predicting Hybrid Performance and Identifying Heterotic Groups.
We implemented genomic best linear unbiased prediction (G-BLUP) (41) and BayesCπ approaches (42) to predict hybrid performance using genomic as well as metabolomic data. We considered prediction approaches including additive, dominance as well as additive × additive, additive × dominance, and dominance × dominance digenic epistatic effects. Details of model implementation are provided in SI Appendix, Note h. We evaluated the accuracy of predicting grain yield using chessboard-like cross-validation as well as the reliability criterion. Details are provided in SI Appendix, Note b.
We used the predicted hybrid performance matrix of all 9,045 single crosses and searched for a high-yielding heterotic pattern based on our simulated annealing algorithm. Implementation and evaluation of the algorithm are described in detail in SI Appendix, Note e. We assessed the long-term success of the identified heterotic pattern by estimating usefulness, selection limit, and representativeness of the heterotic pattern with respect to a defined base population (SI Appendix, Note e).
Supplementary Material
Acknowledgments
We thank Dr. Renate Schmidt and Dr. Timothy F. Sharbel for fruitful discussions. The wheat dataset for this research was generated within the HYWHEAT project funded by the Federal Ministry of Education and Research of Germany (Grants FKZ0315945D and FKZ0315945B). G.L. was supported by the German Federal Ministry of Food and Agriculture within the ZUCHTWERT Project (Grant FKZ0103010). This paper is dedicated to Prof. Dr. Albrecht E. Melchinger, who pioneered research on predicting hybrid performance including the elaboration of a quantitative genetic framework to study heterosis. Both were pivotal for the first step of our developed three-step strategy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514547112/-/DCSupplemental.
References
- 1.Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA. 2011;108(50):20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray DK, Ramankutty N, Mueller ND, West PC, Foley JA. Recent patterns of crop yield growth and stagnation. Nat Commun. 2012;3:1293. doi: 10.1038/ncomms2296. [DOI] [PubMed] [Google Scholar]
- 3.Xu S, Zhu D, Zhang Q. Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc Natl Acad Sci USA. 2014;111(34):12456–12461. doi: 10.1073/pnas.1413750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327(5967):818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- 5.Kempe K, Rubtsova M, Gils M. Split-gene system for hybrid wheat seed production. Proc Natl Acad Sci USA. 2014;111(25):9097–9102. doi: 10.1073/pnas.1402836111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo D, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013;45(5):573–577. doi: 10.1038/ng.2570. [DOI] [PubMed] [Google Scholar]
- 7.Whitford R, et al. Hybrid breeding in wheat: Technologies to improve hybrid wheat seed production. J Exp Bot. 2013;64(18):5411–5428. doi: 10.1093/jxb/ert333. [DOI] [PubMed] [Google Scholar]
- 8.Fischer T, Byerlee D, Edmeades G. Crop Yields and Global Food Security: Will Yield Increase Continue to Feed the World? Australian Centre for International Agricultural Research; Canberra, Australia: 2014. [Google Scholar]
- 9.Melchinger AE, Gumber RK. In: Overview of Heterosis and Heterotic Groups in Agronomic Crops: Concepts and Breeding of Heterosis in Crop Plants. Larnkey KR, Staub JE, editors. Crop Science Society of America; Madison, WI: 1998. pp. 29–44. [Google Scholar]
- 10.Riedelsheimer C, et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet. 2012;44(2):217–220. doi: 10.1038/ng.1033. [DOI] [PubMed] [Google Scholar]
- 11.van Heerwaarden J, Hufford MB, Ross-Ibarra J. Historical genomics of North American maize. Proc Natl Acad Sci USA. 2012;109(31):12420–12425. doi: 10.1073/pnas.1209275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melchinger AE. Genetic diversity and heterosis. In: Coors JG, Pandey S, editors. The Genetics and Exploitation of Heterosis in Crops. American Society of Agronomy, Crop Science Society of America; Madison, WI: 1999. pp. 99–118. [Google Scholar]
- 13.Duvick D, Smith J, Cooper M. Long-term selection in a commercial hybrid maize breeding program. In: Janick J, editor. Plant Breeding Reviews. Wiley; Hoboken, NJ: 2004. pp. 109–152. [Google Scholar]
- 14.Technow F, et al. Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics. 2014;197(4):1343–1355. doi: 10.1534/genetics.114.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer S, et al. Development of heterotic groups in triticale. Crop Sci. 2010;50(2):584–590. [Google Scholar]
- 16.Hill WG. Applications of population genetics to animal breeding, from Wright, Fisher and Lush to genomic prediction. Genetics. 2014;196(1):1–16. doi: 10.1534/genetics.112.147850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reif JC, et al. Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theor Appl Genet. 2011;123(2):283–292. doi: 10.1007/s00122-011-1583-y. [DOI] [PubMed] [Google Scholar]
- 18.Laidig F, Piepho H-P, Drobek T, Meyer U. Genetic and non-genetic long-term trends of 12 different crops in German official variety performance trials and on-farm yield trends. Theor Appl Genet. 2014;127(12):2599–2617. doi: 10.1007/s00122-014-2402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes BJ, Bowman PJ, Chamberlain AJ, Goddard ME. Invited review: Genomic selection in dairy cattle: progress and challenges. J Dairy Sci. 2009;92(2):433–443. doi: 10.3168/jds.2008-1646. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Zeng J, Fernando R, Reif JC. Genomic prediction of hybrid wheat performance. Crop Sci. 2013;53(3):802–810. [Google Scholar]
- 21.Ward J, Rakszegi M, Bedő Z, Shewry PR, Mackay I. Differentially penalized regression to predict agronomic traits from metabolites and markers in wheat. BMC Genet. 2015;16(1):19. doi: 10.1186/s12863-015-0169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardo R. Breeding for Quantitative Traits in Plants. Stemma Press; Woodbury, MI: 2002. [Google Scholar]
- 23.Walsh B, Lynch M. 2014. Evolution and selection of quantitative traits. Available at nitro.biosci.arizona.edu/zbook/NewVolume_2/newvol2.html. Accessed November 23, 2015.
- 24.Langer SM, Longin CFH, Würschum T. Phenotypic evaluation of floral and flowering traits with relevance for hybrid breeding in wheat (Triticum aestivum L.) Plant Breed. 2014;133(4):433–441. [Google Scholar]
- 25.Araki M, Ishii T. Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 2015;20(3):145–149. doi: 10.1016/j.tplants.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Meuwissen TH. Maximizing the response of selection with a predefined rate of inbreeding. J Anim Sci. 1997;75(4):934–940. doi: 10.2527/1997.754934x. [DOI] [PubMed] [Google Scholar]
- 27.Walsh B. Population- and quantitative-genetic models of selection limits. In: Janick J, editor. Plant Breeding Reviews. Wiley; Hoboken, NJ: 2004. pp. 177–225. [Google Scholar]
- 28.Koekemoer F, van Eeden E, Bonjean A. An overview of hybrid wheat production in South Africa and review of current worldwide wheat hybrid developments. In: Bonjean A, Angus W, editors. The World Wheat Book: A History of Wheat Breeding. Lavoisier; Paris: 2011. pp. 907–950. [Google Scholar]
- 29.Chen A, et al. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci USA. 2014;111(28):10037–10044. doi: 10.1073/pnas.1409795111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton T, et al. Boron toxicity tolerance in barley arising from efflux transporter amplification. Science. 2007;318(5855):1446–1449. doi: 10.1126/science.1146853. [DOI] [PubMed] [Google Scholar]
- 31.Frisch M, et al. Transcriptome-based distance measures for grouping of germplasm and prediction of hybrid performance in maize. Theor Appl Genet. 2010;120(2):441–450. doi: 10.1007/s00122-009-1204-1. [DOI] [PubMed] [Google Scholar]
- 32.Sprague GF. Organization of Breeding Programs. Corn Breeders School; Illinois: 1984. [Google Scholar]
- 33.Wang K, Qiu F, Larazo W, Dela Paz MA, Xie F. Heterotic groups of tropical indica rice germplasm. Theor Appl Genet. 2015;128(3):421–430. doi: 10.1007/s00122-014-2441-5. [DOI] [PubMed] [Google Scholar]
- 34.Mühleisen J, Maurer HP, Stiewe G, Bury P, Reif JC. Hybrid breeding in barley. Crop Sci. 2013;53(3):819–824. [Google Scholar]
- 35.Gemenet DC, et al. Pearl millet inbred and testcross performance under low phosphorus in West Africa. Crop Sci. 2014;54(6):2574–2585. [Google Scholar]
- 36.Saxena KB, Sawargaonkar SL. First information on heterotic groups in pigeonpea. Euphytica. 2014;200(2):187–196. [Google Scholar]
- 37.Longin CF, et al. Hybrid wheat: Quantitative genetic parameters and consequences for the design of breeding programs. Theor Appl Genet. 2013;126(11):2791–2801. doi: 10.1007/s00122-013-2172-z. [DOI] [PubMed] [Google Scholar]
- 38.Wang S, et al. International Wheat Genome Sequencing Consortium Characterization of polyploid wheat genomic diversity using a high-density 90,000-single nucleotide polymorphism array. Plant Biotechnol J. 2014;12(6):787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lancashire PD, et al. A uniform decimal code for growth stages of crops and weeds. Ann Appl Biol. 1991;119(3):561–601. [Google Scholar]
- 40.Lippmann R, et al. Protein and metabolite analysis reveals permanent induction of stress defense and cell regeneration processes in a tobacco cell suspension culture. Int J Mol Sci. 2009;10(7):3012–3032. doi: 10.3390/ijms10073012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91(11):4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 42.Habier D, Fernando RL, Kizilkaya K, Garrick DJ. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics. 2011;12(1):186. doi: 10.1186/1471-2105-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.