Abstract
Cancer stem cells (CSCs), a small and elusive population of undifferentiated cancer cells within tumors that drive tumor growth and recurrence, are believed to resemble normal stem cells. Although surrogate markers have been identified and compelling CSC theoretical models abound, actual proof for the existence of CSCs can only be had retrospectively. Hence, great store has come to be placed in isolating CSCs from cancers for in-depth analysis. On the other hand, although induced pluripotent stem cells (iPSCs) hold great promise for regenerative medicine, concern exists over the inadvertent co-transplantation of partially or undifferentiated stem cells with tumorigenic capacity. Here we demonstrate that the introduction of defined reprogramming factors (OCT4, SOX2, Klf4 and c-Myc) into MCF-10A nontumorigenic mammary epithelial cells, followed by partial differentiation, transforms the bulk of cells into tumorigenic CD44+/CD24low cells with CSC properties, termed here as induced CSC-like-10A or iCSCL-10A cells. These reprogrammed cells display a malignant phenotype in culture and form tumors of multiple lineages when injected into immunocompromised mice. Compared with other transformed cell lines, cultured iCSCL-10A cells exhibit increased resistance to the chemotherapeutic compounds, Taxol and Actinomycin D, but higher susceptibility to the CSC-selective agent Salinomycin and the Pin1 inhibitor Juglone. Restored expression of the cyclin-dependent kinase inhibitor p16INK4a abrogated the CSC properties of iCSCL-10A cells, by inducing cellular senescence. This study provides some insight into the potential oncogenicity that may arise via cellular reprogramming, and could represent a valuable in vitro model for studying the phenotypic traits of CSCs per se.
Keywords: cancer stem cell, reprogramming factors, oncogenesis, MCF-10A, p16INK4a
INTRODUCTION
Recent advances in stem cell and cancer biology have provided strong, although indirect or retrospective, evidence for the existence of cancer stem cells (CSCs) in a variety of both solid and hematopoietic tumors.1–3 In solid tumors, a small subset of CSCs appear to reside within the tumor mass and this population constitutes a reservoir of cancer-initiating cells with the exclusive ability to self-renew and maintain this mass.4 This cell population has come to be implicated as the source of disease recurrence and metastasis during the long period of cancer development.1–3 Most CSCs exhibit resistance to conventional anticancer therapies including chemotherapeutic agents and ionizing radiation.5 The development of novel drugs that target CSC-specific factors should aim to effectively eliminate the CSC population within the tumor, resulting in complete cure of the disease.6 To date, however, evaluation of drugs that specifically target CSCs has been hampered because of the difficulty in isolating CSCs from the bulk of tumor tissues, and the manipulation of pure populations ex vivo.7,8
In common with normal stem cells, CSCs are defined by their double capacity for self-renewal and tissue regeneration through differentiation, and can give rise to phenotypically diverse cells.4 It is quite plausible that CSCs may use the molecular machinery and factors that control normal stem cell function to maintain their oncogenic existence.9–11 It almost goes without saying that deeper comprehension of the characteristics of CSCs and their parallels with normal stem cells will in turn further our understanding of cancer pathogenesis.7,12 To date, the characterization of CSC properties has been limited, and the putative factors directing both normal and transformed stem cell processes have not been elucidated, largely because of the difficulty in obtaining CSCs in vitro.
Induced pluripotent stem cells (iPSCs) are generated from nonpluripotent cells, typically adult somatic cells, through the ectopic expression of the transcription factors OCT4 and SOX2 combined with either Klf4 and c-Myc, or Lin28 and Nanog.13,14 The iPSCs acquire the features of embryonic stem cells (ESCs) including immortal cell growth and pluripotency, thus enabling their self-renewal and differentiation into multiple lineages.13,14 Although it is widely believed that iPSC technology has great therapeutic potential and other medical applications, the molecular events occurring during and after the reprogramming process in these cells have not been clarified. Furthermore, the delineation of molecular signatures to detect malignant transformation among human iPSCs represents an important new challenge in both stem cell and cancer biology. In this regard, several recent studies describe attempts to create CSC-like cells via the ectopic expression of reprogramming factors in already transformed cell lines including gastrointestinal cancer, chronic myeloid leukemia and melanoma.15–17 In these studies, however, it is believed that CSC-like properties pre-existed in the original tumorigenic cell line, and could be selected for during the reprogramming step.
In the current study, we used iPSC technology to reprogram nontumorigenic human MCF-10A mammary epithelial cells. The resultant cells possess the hallmarks of CSCs and generate tumors in an immunosuppressed mouse model comprising cells of multiple lineages. These results help shed light on the possibility for tumorigenicity of epithelial cells by defined reprogramming factors, and provide a potentially valuable system for the study of CSCs in vitro.
RESULTS
Reprogramming of human immortalized mammary epithelial MCF-10A cells by defined reprogramming factors
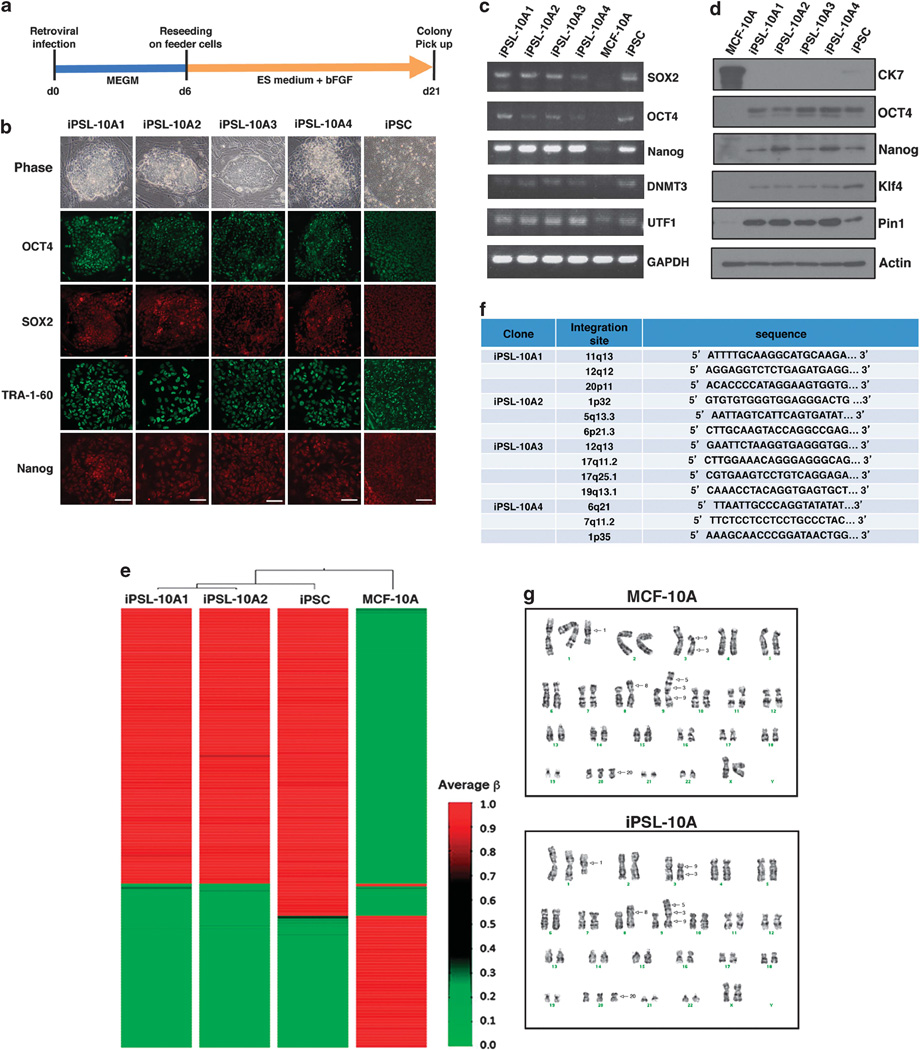
MCF-10A is a mammary epithelial cell line, immortalized by a disruption of the p16 tumor-suppressor gene due to a spontaneous chromosomal translocation.18,19 Like normal human breast epithelial cells, MCF-10A cells have no tumor initiation ability, but have been shown to be more susceptible to oncogenic transformation than primary cells.20 The retroviral-mediated introduction of OCT4, SOX2, Klf-4 and c-Myc into MCF-10A cells, followed by growth in human ESC (hESC) culture medium supplemented with basic fibroblast growth factor on mouse embryonic fibroblast (MEF) feeder cells, gave rise to iPS-like (iPSL) colonies after 14 days (Figures 1a and b). The sole introduction of the c-Myc oncogene did not generate iPS-like colonies (data not shown). The colonies that formed demonstrated well-defined phase-bright borders surrounded by feeder cells and comprising small cells with high nuclear/cytoplasmic ratios and prominent nucleoli, indistinguishable from standard iPSCs (Figure 1b).
Figure 1.
Reprogramming of human MCF-10A mammary epithelial cells. (a) Experimental scheme for the reprogramming of MCF-10A cells. (b) Phase-contrast images and immunofluorescence images of iPS-like colonies from MCF-10A cells (iPSL-10A) and normal human iPSCs stained with antibodies against OCT4, SOX2, TRA-1-60 and Nanog. Scale bar, 500 µm. (c) Semiquantitative reverse transcriptase–PCR (RT–PCR) analysis of iPSC markers in iPSL-10A cell clones 1–4, normal human iPSCs and MCF-10A cells. SOX2 and OCT4 are endogenously derived. (d) Immunoblotting of the stem cell marker proteins in iPSL-10A cell clones 1–4, normal human iPSCs and MCF-10A cells. (e) DNA methylation ‘heat map’ of iPSL-10A cells. DNA methylation analysis was performed using an Illumina Human Methylation 27 Beads Chip (MBL) with genomic DNA extracted from iPSL-10A clones 1 and 2, normal human iPSCs and MCF-10A cells. The β-value was calculated by a quantitative measure of the DNA methylation levels at specific CpG islands. Average β-values were subjected to unsupervised hierarchical clustering based on the Manhattan distance and average linkage. (f) High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM PCR). Genomic DNA was prepared using phenol/chloroform extraction and subjected to LAM PCR. Amplicons were validated by sequencing. (g) Standard G-band chromosome analysis of MCF-10A and iPSL-10A cells. Arrows indicate identifiable aberrations common to both cell types.
These iPSL colonies were continually cultured up to 21 days and 11 alkaline phosphatase-positive representative colonies were picked up. Of these, four clones (designated iPSL-10A1–4) were chosen at random and further analyzed. Immunocytochemical analysis revealed the expression of pluripotent stem cell markers including OCT4, SOX2, Nanog and TRA-1-60 (Figure 1b). Stem cell marker expression was also confirmed in these clones by reverse transcriptase–PCR and immunoblotting (Figures 1c and d). Conversely, expression of the differentiated epithelial cell marker cytokeratin 7 (CK7) was silenced during the reprogramming process (Figure 1d). Additionally, expression of Pin1 was activated in iPSL-10A cells (Figure 1d), as is the case for normal iPS cells21 (Figure 1d). iPSL-10A colonies could be expanded in an undifferentiated state beyond 20 passages (data not shown). Analysis with an Illumina Human Methylation 27 Bead Chip (MBL, Nagoya, Japan) revealed that the DNA methylation patterns of iPSL-10A cells were closer to those of normal iPSCs than to the parental MCF-10A cells (Figure 1e).
We next examined whether the reprogramming in these cells might be associated with the insertion of the transgenes in specific regions of the genome by using linear amplification-mediated PCR and sequencing analysis of proviral integration sites among the iPSL-10A cell clones. The linear amplification-mediated PCR results revealed that the transgenes that were detectable were inserted at different, random chromosomal loci among the four clones (Figure 1f). Moreover, comprehensive karyotyping revealed that iPSL-10A cells were indistinguishable from the parental MCF-10A cells (Figure 1g).
In vitro differentiation of iPSL-10A cells into CSC-like cells
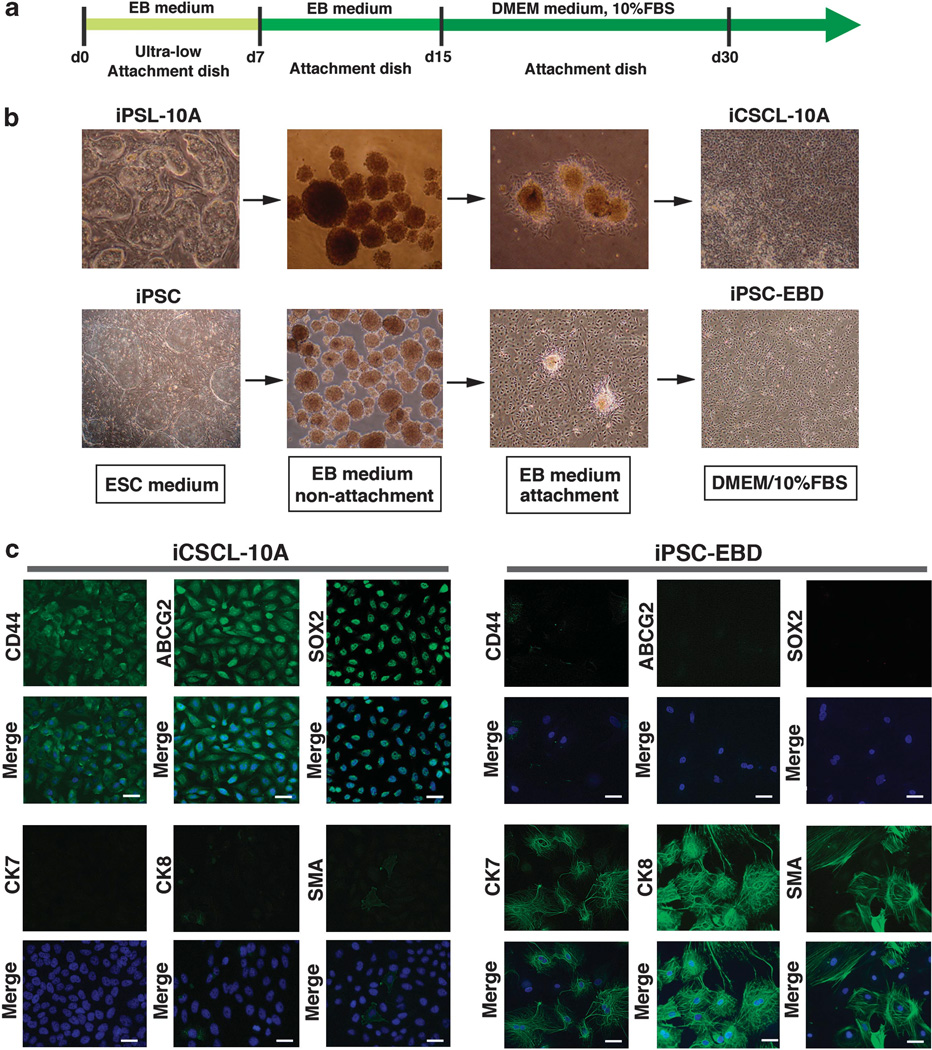
We next attempted to differentiate the iPSL-10A cells in vitro, based on the rationale that a differentiation step would be incorporated into any type of regenerative medicine application involving cell transplantation using iPSCs. A standard and general method for ESC differentiation in vitro is via the formation of cell aggregates in nonadherent spheroids known as embryoid bodies (EBs).22 As for normal iPSCs, the iPSL-10A cells formed EB-like spherical aggregates in suspension culture containing differentiation-promoting medium (Figures 2a and b). iPSL-10A-derived EB-like colonies were subsequently transferred into culture dish-attachment conditions and allowed to continue to differentiate for a further 7 days (Figures 2a and b). The attached cells were then exposed to maintenance cell culture medium for a further 15 days, and the surviving cells designated as iCSCL-10A. As a control, we performed the same procedure for standard iPSCs derived from normal human mammary epithelial cells, the products of which were designated as iPSC-EBD (iPS cells having undergone EB-mediated differentiation). Immunofluorescence analysis revealed that >90% of iCSCL-10A cells express the CSC markers CD44 and ABCG2 as well as the stem cell marker SOX2, but negligible levels of differentiated epithelial markers CK7, CK8 and smooth muscle actin (Figure 2c). In contrast, iPSC-EBD cells did not express any of the three CSC markers, but expressed all three differentiation markers (Figure 2c). Importantly, the majority of parental iPSL-10A cells expressed CD44 and ABCG2 at very low levels. However, a very small population of these cells exhibited levels of expression of both proteins that were comparable to iCSCL-10A cells (Supplementary Figure S1).
Figure 2.
In vitro differentiation of iPSL-10A cells into induced CSCs. (a) Schematic representation of the in vitro differentiation of iPSL-10A and normal iPSCs. (b) Representative phase-contrast images of either iPSL-10A or normal iPSCs during embryoid body (EB)-mediated differentiation. After EBs were transferred onto gelatin-coated attachment plates and allowed to further differentiate for 8 days. These cells were then finally cultured in DMEM/10% FBS up to day 30. (c) Immunofluorescent analysis of lineage marker proteins in cultured iCSCL-10A and iPSC-EBD cells. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 200 µm.
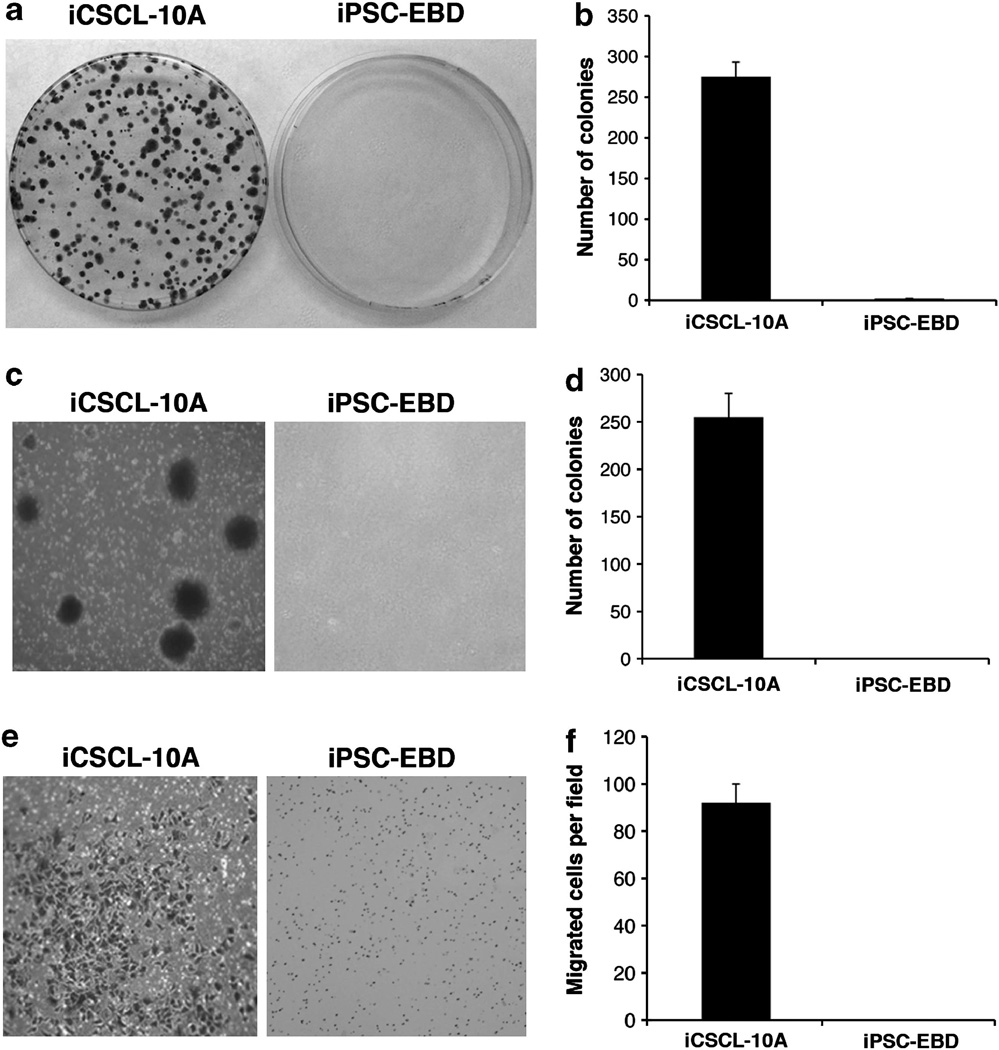
Most interestingly, iCSCL-10A cells were found to have acquired malignant properties in focus formation (Figures 3a and b), colony formation (Figures 3c and d) and cell invasion assays (Figures 3e and f), whereas the iPSC-EBD cells showed no malignant phenotype in any of these assays (Figures 3a–f). These results indicate that the iCSCL-10A cells had undergone malignant transformation following nuclear reprogramming.
Figure 3.
Malignant phenotypes of iCSCL-10A in vitro. (a, b) Focus formation assay of iCSCL-10A and iPSC-EBD cells. Equal numbers of cells (5 × 102) were seeded onto 10 cm plastic dishes. After 10 days, the cells were fixed and stained with crystal violet (a). The numbers of colonies were calculated and scored (mean ± s.d.) from three independent experiments (b). (c, d) iCSCL-10A and iPSC-EBD cells were plated in 0.3% soft agar and cultured for 2 weeks. Representative microscopic fields are presented (c). Colony formation was scored microscopically and the colony numbers (mean ± s.d.) were calculated from three independent experiments (d). (e, f) Cell invasion assays were performed using chemotaxis chambers in transwell tissue culture dishes as described in the Materials and methods. Representative microscopic fields are shown (e). Invasive cells were counted and scored in triplicate. The mean values ± s.d. were calculated from three independent experiments (f).
Characterization of the CSC properties of the iCSCL-10A clones
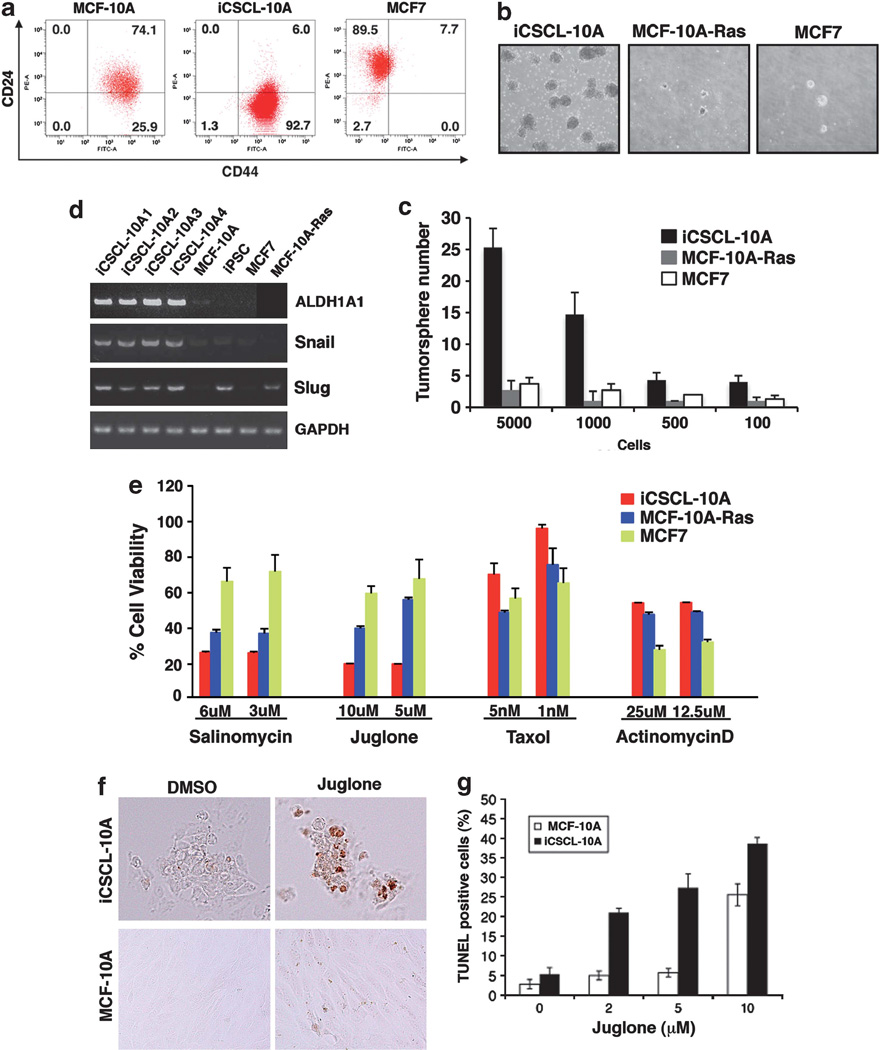
Subsequent to 30 days of differentiation/maintenance of iPSL-10A cells, flow cytometric analysis revealed that each of the resulting iCSCL-10A1–4 cells harbored a population (>90%) with a CD44+/ CD24low marker profile (Figure 4a) that has previously been associated with CSCs.2,3 The iCSCL-10A cells had ability to form tumor spheres as an in vitro assay of the self-renewal capacity of CSCs. Indeed, iCSCL-10A cells showed an ~10-fold higher tumor sphere-forming ability relative to MCF7 or MCF-10A-Ras cells, transformed by the introduction of the HrasV12 oncogene into MCF-10A cells23 (Figures 4b and c).
Figure 4.
Characterization of the CSC properties of iCSCL-10A clones. (a) Flow cytometric analysis of CD44 and CD24 expression in the MCF-10A, iCSCL-10A and MCF7 cell lines. The numbers indicate the percentage of each sub-population according to the CD44/CD24 expression profile. (b, c) Tumor sphere formation assays of MCF-10A-Ras, iCSCL-10A and MCF7 cell lines. Phase-contrast images of tumor spheres are shown (b). Values represent the mean ± s.e.m. (n=3, c). (d) Semiquantitative reverse transcriptase–PCR (RT–PCR) analysis of the expression of CSC- or epithelial-to-mesenchymal transition (EMT)-related genes. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was analyzed as a control. (e) Viability of MCF-10A-Ras, iCSCL-10A and MCF7 cell lines treated with various chemotherapeutic agents for 72 h by MTT assay. Values represent the mean ± s.e.m. (n=3). (f, g) iCSCL-10A and parental MCF-10A cells were treated with Juglone (5 µm) for 24 h and subjected to TUNEL (terminal deoxyribonucleotidyl transferase-mediated dUTP nick end-labeling) assay (f, brown color). TUNEL-positive cells were scored from triplicate independent experiments (g). Values represent the mean ± s.e.m. (n=3).
Reverse transcriptase–PCR analysis revealed that iCSCL-10A cells, but not MCF7 and MCF-10A-Ras cells, expressed aldehyde dehydrogenase 1A1 (ALDH1A1) as a CSC marker24 (Figure 4d). The iCSCL-10A cells also expressed the epithelial-to-mesenchymal transition-related genes Snail and Slug (Figure 4d). Immunofluorescent analysis revealed the nuclear localization (a hallmark of activation) of Gli1 (hedgehog signaling) and Notch1 (Notch signaling), but not β-Catenin (Wnt signaling) in iCSCL-10A cells (Supplementary Figure S2a). Furthermore, the transforming growth factor-β signaling proteins Smad1, Smad3 and Smad5 were preferentially activated in these cells as revealed by immunoblotting analysis with phospho-Smad-specific antibodies (Supplementary Figure S2b).
As Slug has been shown to induce the CSC phenotype in MCF-10A cells,25 we performed a knockdown analysis of this gene in iCSCL-10A cells and found in a tumor sphere assay that slugshRNA-transduced cells exhibited reduced growth and self-renewal ability compared with control cells (Supplementary Figure S3a). Furthermore, both SOX2 and CD44 expression was decreased in these cells (Supplementary Figure S3b).
A major characteristic of CSCs is their resistance to anticancer agents. We next assayed the resistance of iCSCL-10A cells to various chemotherapeutic drugs. The iCSCL-10A cells exhibited increased resistance to the established anticancer chemotherapeutic compounds, Taxol and Actinomycin D, relative to both the control MCF7 breast cancer cells and MCF-10A-Ras cells (Figure 4e). In contrast, iCSCL-10A cells were more sensitive to Salinomycin, a drug that selectively targets CSCs6 and to the selective Pin1 inhibitor Juglone compared with MCF7 or MCF-10ARas cells (Figure 4e). Subsequent TUNEL (terminal deoxyribonucleotidyl transferase-mediated dUTP nick end-labeling) assay revealed that Juglone treatment selectively induces cellular apoptosis in iCSCL-10A cells as compared with parental MCF-10A cells (Figures 4f and g).
It has been shown that Pin1 function is also regulated by the phosphorylation status of both Pin1 and its target proteins.26 Indeed, an immunoblotting analysis revealed that although Pin1 was more highly phosphorylated in iCSCL-10A cells than in iPSCs, potential Pin1-binding sites, recognized by phospho-CDK substrate antibodies, were more prominently phosphorylated in iCSCL-10A cells than in normal iPSCs (Supplementary Figure S4), suggesting a profound role of Pin1 in iCSCL-10A cells.
iCSCL-10A cells form multilineage tumors in vivo
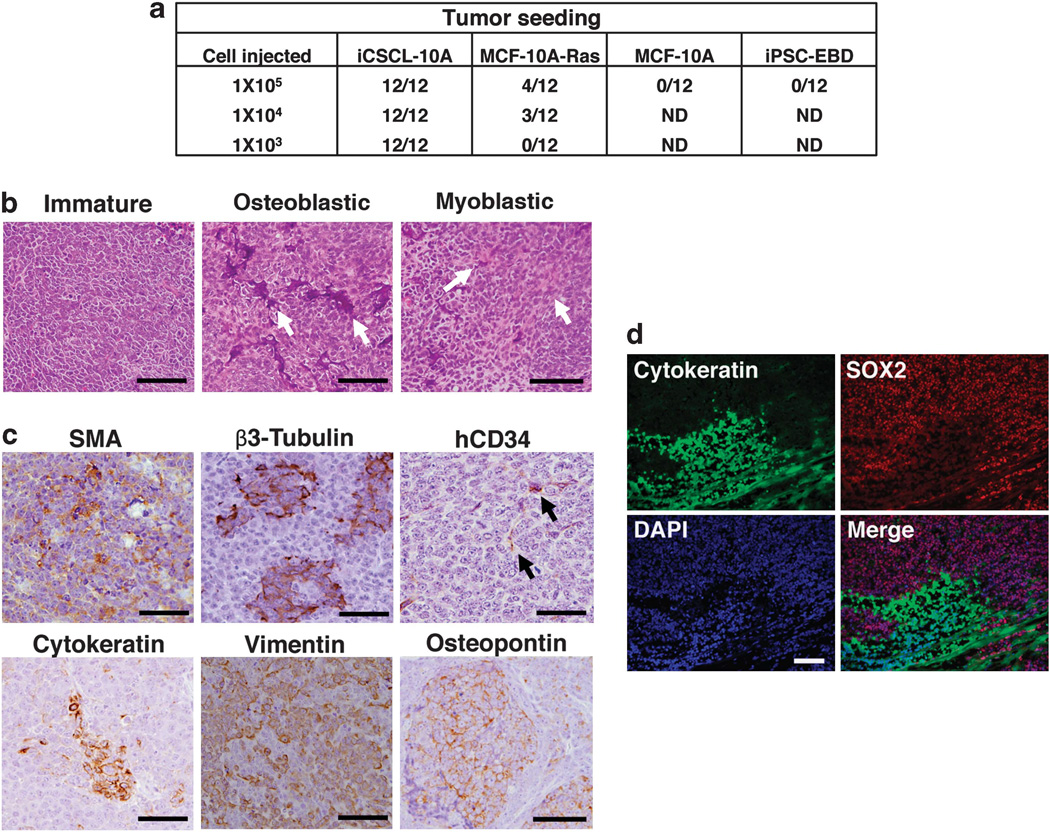
We next assessed the ability of iCSCL-10A cells to form tumors in an immunosuppressed mouse model. We injected iCSCL-10A, MCF-10A-Ras, parental MCF-10A or iPSC-EBD cells subcutaneously into BALB/c nude mice and monitored them for 9 to 12 weeks. Tumors were generated using as few as 1 × 103 iCSCL-10A, which was 10-fold lower than the number of MCF-10A-Ras cells required for tumor seeding (Figure 5a). There was no evidence of tumor formation in the MCF-10A- or iPSC-EBD-injected mice up to 1 × 105 cells (Figure 5a). Histological examinations of tumor tissues revealed that iCSCL-10A cells produced a florid proliferation of small round immature cells with focal mitotic figures (Figure 5b, left). Notably, the tumor contained structures resembling differentiated cells of the bone and muscle lineages (Figure 5b, middle and right). Epithelial (CK), mesenchymal (vimentin), neuronal (β3-tubulin), endothelial (human CD34; does not crossreact with mouse CD34),27 calcifying osteoblastic (osteopontin) and myoblastic (smooth muscle actin)-positive cells were detected by immunohistochemical analysis (Figure 5c). We detected, by immunofluorescence, prominent histological borders of cell populations between putatively less differentiated (SOX2+/AE1/ 3 cytokeratin−) and more differentiated cells (SOX2−/AE1/3 cytokeratin+) within the tumor (Figure 5d).
Figure 5.
iCSCL-10A cells form hierarchically organized tumors in vivo. (a) Tumor-seeding ability of iCSCL-10A, MCF-10A-Ras parental MCF-10A cells and iPSC-EBD. The indicated numbers of each cell type were injected into immunocompromised mice. The tumor-initiation ability per injection was then monitored. (b) Hematoxylin and eosin (H&E) staining of primary tumor tissues. Scale bar, 500 µm. (c) Immunohistochemical analysis of primary tumor tissues derived from iCSCL-10A cells using antibodies targeting hCD34 (endothelial), smooth muscle actin (SMA; myoblastic), β3-tubulin (neural), cytokeratin (CAM5.2, epithelial), vimentin (mesenchymal) and osteopontin (osteoblastic). Scale bar, 500 µm. (d) Immunofluorescent analysis with antibodies targeting SOX2 and cytokeratin (AE1/AE3). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 500 µm.
Cyclin-dependent kinase inhibitor p16INK4a induces cell cycle arrest and senescence in iCSCL-10A cells
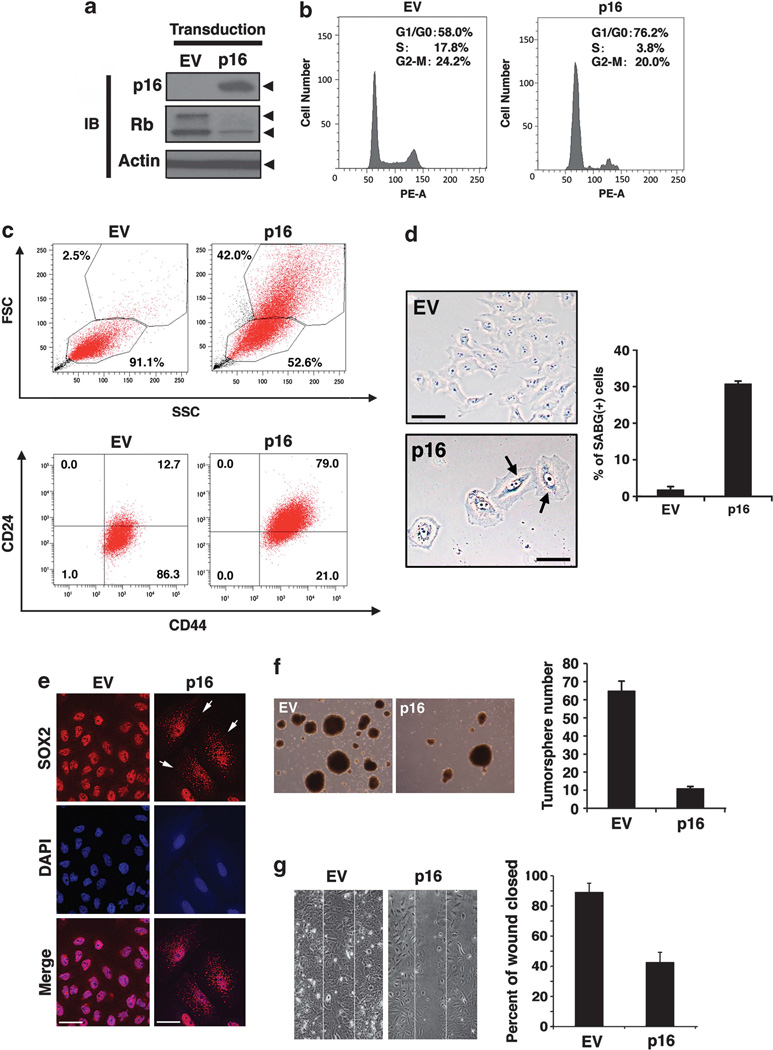
The parental MCF-10A human mammary gland epithelial cells harbor a cytogenetic abnormality involving a chromosome 9 translocation resulting in the deletion of the CDKN2A (p16INK4a) gene. To determine the functional role of p16INK4a in the proliferation and maintenance of CSC-like cells, iCSCL-10A cells were transduced with p16INK4a using a retrovirus vector followed by selection with puromycin. Immunoblotting analysis confirmed the stable expression of the exogenous p16 gene and decreased amounts of phosphorylated Rb (Figure 6a). Cell cycle analysis demonstrated a significantly increased G1 population of p16INK4atransduced cells compared with the control vector-transduced cells (Figure 6b). Cells transduced with p16INK4a exhibited an enlarged, flattened and irregular shape, and flow cytometric analysis revealed this effect in terms of forward scatter and side scatter profiles (Figure 6c, upper panels). These cells also positively stained with senescence-associated β-galactosidase, whereas no such cells were observed among the control vector-transduced population (Figure 6d). Concomitantly, the fraction of the CSC population that was CD44+/CD24low was significantly reduced for the cells transduced with p16INK4a (from 86.3 to 21.0%, Figure 6c, lower panels). In line with these observations, the SOX2 localization profile was found to be significantly shifted from a nuclear to a cytoplasmic distribution, as revealed by immunofluorescence (Figure 6e).
Figure 6.
Cyclin-dependent kinase inhibitor p16 suppresses the CSC properties of iCSCL-10A cells and induces cellular senescence. (a) Immunoblotting analysis of p16 and Rb in iCSCL-10A cells transduced with the p16 vector or with an empty vector (EV) control retrovirus. Actin was used as a loading control. (b) Flow cytometric analysis of the cell cycle status following propidium iodide (PI) staining of iCSCL-10A cells transduced with p16 or EV. (c) Flow cytometric analysis of forward scatter (FSC) versus side scatter (SSC) dot plot (upper panels) and CD44/CD24 expression (lower panels). Note that p16 transduction results in the appearance of large-sized cells. (d) p16-transduced iCSCL-10A cells were subjected to senescence-associated β-galactosidase staining (SABG). Phase-contrast images of the cells are shown. Arrows indicate positive signals shown in blue (photomicrographs). Scale bar, 200 µm. Bars indicate the percentage of SABG-positive cells for each cell species (histogram). Values represent the mean ± s.e.m. (n=3). (e) Re-expression of p16 promotes SOX2 translocation from the nucleus to the cytoplasm. Immunofluorescent analysis of SOX2 in p16-transduced iCSCL-10A cells. Arrows indicate cytoplasmic localization of SOX2. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). (f) p16 transduction abrogates the tumor sphere-forming ability of iCSCL-10A cells. Phase-contrast images of tumor spheres transduced with p16 or EV and quantification of tumor sphere formation. Values represent the means ± s.e.m. (n=3). (g) Effects of p16 on wound healing. Confluent monolayers of the iCSCL-10A cells transduced with either p16 or EV were mechanically wounded using the tip of a pipette. After 6 h, the cells were fixed and images were captured. Wound closures were scored using ImageJ software. Values represent the mean ± s.e.m. (n=3).
The transduction of p16INK4a into iCSCL-10A cells significantly reduced the rate of tumor sphere formation down to ~20% (Figure 6f), indicating that the re-introduction of p16INK4a can suppress the self-renewal properties of iCSCL-10A cells. An important feature of CSCs is their increased mobility. To evaluate whether p16INK4a regulates cell migration in this context, wound healing assays were performed. At 6 h following cell scratching, the extent of wound closure for the empty vector control cells was 90%, whereas that for the p16INK4a-transduced cells was significantly less, at 45% (Figure 6g).
An in vivo tumor-initiating assay revealed no evidence of tumor formation in the iCSCL-10A/ p16INK4a -injected BALB/c nude mouse (1 × 105 cells), whereas control vector-transduced iCSCL-10A cells still formed tumors, indicating that iCSCL-10A cells had lost their tumor-initiating properties following the re-expression of p16INK4a (Supplementary Figure S5). Taken together, these results together indicate that the re-introduction of p16INK4a can indeed attenuate the CSC properties of iCSCL-10A cells.
DISCUSSION
We have demonstrated in the current study that the introduction of defined reprogramming factors and subsequent partial cell differentiation generate cells with a tumorigenic phenotype and CSC-like properties from nontransformed human MCF-10A mammary epithelial cells. These cells show a resistance profile against anticancer agents that is normally associated with CSCs. Moreover, these transformed cells can be maintained in culture in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), permitting easy manipulation and amenability to large-scale drug screening and could therefore provide a valuable basis for the further analysis of CSC characteristics and drug development strategies designed to target CSCs.
Several studies have suggested that the molecular circuitry controlling pluripotency in stem cells is also important for the tumor-initiating ability of cancer cells. In fact, the pluripotent cell markers OCT4, SOX2, Klf4 and Nanog have been shown to be expressed in certain cancer cell types and to play important roles in oncogenesis.28–31 A recent study has reported a crucial role for SOX2 but not OCT4 and Nanog in breast CSCs. SOX2 expression was found in this report to be induced upon the formation of tumor spheres from natural breast tumor cultures and breast carcinoma cell lines.32 SOX2 has been shown to be expressed in a variety of early stage of breast carcinomas and metastatic lymph nodes.33 Hence, high SOX2 expression in iCSCL-10A cells may contribute to the high tumor-initiating ability of these cells.
The current picture of carcinogenesis from iPSCs is ambiguous. Pre-iPS cells have been defined as incompletely reprogrammed multipotent stem cells that arise during the induction of iPSCs.13,34 Like iPSCs, pre-iPS cells have been shown to form tumors in immunosuppressed mouse models, but their histology is not considered to be that of a teratoma13 but rather a phenotype that is reminiscent of the induced CSCs described herein. Furthermore, iPS reprogramming of p53 knockout or p53 mutant mouse embryonic fibroblasts led to the development of cells with a potential to form malignant tumors with sarcoma-like morphology in nude mice.35 Moreover, mouse iPSCs cultured in mouse Lewis lung carcinoma conditioned medium acquire the characteristics of CSCs.36 These data suggest that iPS reprogramming factors might be adequate to transform somatic cells with certain genetic defects or specific culture conditions. However, the iPS-mediated malignant cells analyzed to date come from a rodent source, cells that are believed to be more susceptible to transformation than human cells.37 Meanwhile, the current system using nontumorigenic human mammary epithelial MCF-10A cells may cast new light on the susceptibility of nontumorigenic human mammary epithelial cells to neoplastic reprogramming. How similar the CSC-like cells described in the current paper are to human pre-iPS remains unknown. However, such a comparison may be an important point of departure for future studies concerned with both CSCs and the oncogenic potential of cells produced using iPS technology.
MCF-10A is an immortalized but nontransformed human breast epithelial cell line that is widely used in molecular studies of cancer.19 In addition, although these cells have only a small number of chromosome translocations, one of which results in the depletion of the p16INK4a gene,18 they show a normal mammary epithelial cell morphology and phenotype and can successfully form mammary acini in 3D matrigel cultures.20 These characteristics of MCF-10A cells prompted us to use them for our current model system. As many previous studies have already reported, the INK4A gene locus is completely silenced and p16INK4a is not usually expressed in pluripotent stem cells such as ESCs and iPSCs.38,39 Indeed, several different groups have demonstrated that the INK4A gene locus critically impairs successful reprogramming to pluripotent stem cells and that it represents a principal barrier to iPS cell reprogramming through the induction of reprogramming-induced senescence.40–42 In human somatic cells, the inhibition of p16INK4a enhances the iPSC generation, increasing both the kinetics of reprogramming and the number of emerging iPSC colonies.39 Furthermore, our current study demonstrates that the re-expression of p16INK4a can indeed attenuate the CSC properties and in vivo tumorigenicity of iCSCL-10A cells. Thus, the loss of p16INK4a may affect not only the oncogenic reprogramming but also the maintenance of malignant properties of iCSC-10A cells.
In general, CSCs have increased resistance to conventional anticancer chemotherapeutic agents.5 We observed that iCSCL-10A cells exhibited higher sensitivity to the Pin1-selective inhibitor Juglone compared with MCF7 or MCF-10A-Ras cells, which may reflect the importance of Pin1 in the maintenance of pluripotency of CSC through the binding and stabilization of key ‘stemness’ factors such as Nanog and OCT4 as is the case for human iPSCs and murine ESCs.21,43 This drug-sensitivity profile, together with our finding that Pin1 expression is induced during the nuclear reprogramming process, tempts us to propose that iCSCL-10A cells are distinct from the parental MCF-10A cells as well as from various transformed cancer cell lines with respect to the dependency on Pin1. Considered along with our current results, this leads us to speculate that Pin1 could be an essential factor for the self-renewal and proliferation of CSC, and even perhaps a potential molecular target for CSC-based therapy.
In summary, we here describe the oncogenic transformation of mammary epithelial cells and consequent generation of a potentially valuable model system for the study of CSCs and development of alternative anticancer strategies that target CSCs.
MATERIALS AND METHODS
Cell culture
MCF-10A and MCF7 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). MCF-10A cells cultured in mammary epithelial cell growth medium (MEGM; Sanko Junyaku, Tokyo, Japan). Human primary mammary epithelial cells were purchased from Kurabo Industrial (Osaka, Japan). Human iPSCs and iPSLCs were cultured in hESC culture medium (KNOCKOUT DMEM; Invitrogen, San Diego, CA, USA) supplemented with 20% KNOCKOUT SR (Invitrogen), 1% GlutaMAX (Invitrogen), 100 µm nonessential amino acids (Invitrogen), 1% penicillin/ streptomycin, 50 µm β-mercaptoethanol and 10 ng/ml basic fibroblast growth factor. iCSCL-10A and MEFs were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
Cell reprogramming and transformation
MCF-10A cells were transduced with retroviral vectors encoding nuclear reprogramming factors as described previously.44 Briefly, the retroviral vector plasmids pMXs-hOCT4, pMXs-hSOX2, pMXs-hKlf4, pMXs-hc-Myc (Addgene, Cambridge, MA, USA) and VSV-G were introduced into Plat-E cells using Effectene transfection reagent (QIAGEN, Maryland, MD, USA). After 48 h, virus-containing supernatants were passed through a 0.45 µm filter and supplemented with 10 µg/ml polybrene. MCF-10A cells were seeded at 6 × 105 cells per 100mm dish 24 h before incubation in the virus/polybrene-containing supernatants for 16 h. Cells were then washed and returned to fresh MEGM medium. After 6 days, the cells were plated on irradiated MEFs and the MEGM culture medium was replaced with hESC culture medium 24 h later. Cells were maintained at 37 °C in a 5% CO2 incubator for 21 days.
Alkaline phosphatase staining
Alkaline phosphatase staining was performed using the Leukocyte Alkaline Phosphatase kit (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer’s protocol.
EB formation and spontaneous differentiation
The iPSL-10A cells were dissociated using accutase and seeded on a Matrigel (BD Biosciences, San Diego, CA, USA) coated dish in MEF-conditioned hESC medium for feeder-free culture. The iPSL-10A cells were also dissociated using accutase and plated on ultra low-attachment plates (Cell Seed) at a density of 1 × 104 cells per 6 wells in differentiation medium (KNOCKOUT DMEM supplemented with 20% FBS, 1% GlutaMAX; Invitrogen), 100 µm nonessential amino acids (Invitrogen), 1% penicillin/streptomycin and 50 µm β-mercaptoethanol. After 7 days, the embryoid body-like colonies were transferred to 0.1% gelatin-coated culture dishes and cultured for a further 8 days in the same medium. Finally, medium was replaced by DMEM supplemented with 10% FBS for 15 days before fixation and staining.
In vivo tumor formation assay
Cells were washed twice with antibiotic-free and serum-free cell culture medium and finally resuspended in 0.1 ml of serum-free culture medium. The cell suspension was then mixed with an equal volume of Matrigel (BD Biosciences) and injected subcutaneously into 6-week-old irradiated (4 Gy) BALB/c nude mice (CLEA, Tokyo, Japan). Injections were performed 1 day after irradiation. Tumors were surgically removed 9 to 12 weeks later. Representative tumor tissues were fixed in 3% formalin, embedded in paraffin wax and sectioned at a thickness of 10 µm. Sections were stained with hematoxylin and eosin for pathological examination, or processed for immunohistochemical analysis. All animal protocols were approved by the institutional Administrative Panel on Laboratory Animal Care of Yokohama City University.
RNA isolation and reverse transcriptase–PCR
Total RNA was extracted using TRIzol (Invitrogen). Complementary DNA synthesis was performed with ReverTraAce-α (Toyobo, Osaka, Japan) in accordance with the manufacturer’s instructions. PCR was performed with ExTaq (Takara Bio, Shiga, Japan) using the following primers: human SOX2 (endogenous) fwd 5′-GGGAAATGGGAGGGGTGCAAAAGAGG-3′, rev 5′-TTGCGTGAGTGTGGATGGGATTGGTG-3′; human OCT4 (endogenous) fwd 5′-GACAGGGGGAGGGGAGGAGCTAGG-3′, rev 5′-CTTCCCTCCAACCA GTTGCCCCAAAC-3′; human Nanog fwd 5′-CAGCCCtGATTCTTCCACCAGT CCC-3′, rev 5′-tGGAAGgTTCCCAGTCGGGTTCACC-3′; human DNMT3B fwd 5′-TGCTGCTCACAGGGCCCGATACTTC-3′, rev 5′-TCCTTTCGAGCTCAGTGCA CCACAAAAC-3′; human UTF1 fwd 5′-CCGTCGCTGAACACCGCCCTGCTG-3′, rev 5′-CGCGCTGCCCAGAATGAAGCCCAC-3′; human GAPDH fwd 5′-GTGG ACCTGACCTGCCGTCT-3′, rev 5′-GGAGGAGTGGGTGTCGCTGT-3′; human Snail fwd 5′-TCAAGATGCACATCCGAAGC-3′, rev 5′-AGGACACAGAACCAG AAAATGG-3′; Slug fwd 5′-CAGACCCTGGTTGCTTCAAG-3′, rev 5′-CACAGGAG AAAATGCCTTTGG-3′; human ALDH1A1 fwd 5′-TAAGCATCTCCTTACAGTC AC-3′, rev 5′-TGTTAAGTACTTCAAGAGTCAC-3′.
Immunohistochemistry and immunocytochemistry
For immuncytochemistry, cells were fixed with 4% paraformaldehyde for 15 min at 4 °C, washed with phosphate-buffered saline and then permeabilized using 0.1% Triton X-100 and blocked with 5% goat serum in 0.1% bovine serum albumin. Primary antibodies were diluted in 0.1% bovine serum albumin and incubated with the fixed cells for 1 h at room temperature. For immunohistochemistry, paraffin-embedded tissue sections were subjected to antigen retrieval with microwave irradiation in citrate buffer (pH 6.0). This was followed by incubation with primary antibody at room temperature for 60 min. After incubation with secondary antibody at room temperature for 60 min, the sections were stained with VECTASTAIN Universal Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions. The primary antibodies used in this study were as follows: anti-OCT4 (1:300; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-SOX2 (1:300; Millipore, Billerrica, MA, USA), anti-TRA-1-60 (1:200; eBioscience, San Diego, CA, USA), anti-Nanog (1:300; ReproCELL, Yokohama, Japan), anti-Cytokeratin 7 (1:50, M7018; Dako Cytomation, Düsseldorf, Germany), anti-Cytokeratin 8 (1:50; Becton Dickinson, Mountain view, CA, USA), anti-cytokeratin AE1/AE3 (1:500; Abcam, Cambridge, MA, USA), anti-CD44 (1:100; Cell Signaling, Danvers, MA, USA), anti-ABCG2 (1:100; BioLegend, San Diego, CA, USA), anti-β-Tubulin III (1:100; Sigma), anti-hCD34 (1:100; Abcam), anti-Osteopontin (1:100; Sigma), anti-Vimentin (V9, 1:300; Abcam) and anti-smooth muscle actin (1:100; Sigma).
Cell invasion assay
Cell invasion assay was performed in 24-well format Transwell inserts (8 µm pore, BD Biosciences) coated with 1 mg/ml matrigel (BD Biosciences) as described previously.45 Invasive cells were counted and scored in triplicate (lower chamber).
Soft agar colony formation assay
Soft agar colony formation assays were performed by seeding 5 × 103 cells in 60-mm tissue culture dishes containing 0.3% top low-melting point agarose/0.5% bottom low-melting point agarose. Cells were fed every 4 days and colonies were counted and measured after 2 weeks.
Karyotyping
Karyotyping was performed by Nihon Gene Research Laboratories (Sendai, Japan), as described previously.44 At least 15 metaphase spreads per genotype and from at least two independent cultures per genotype were scored for chromosomal aberrations.
Tumor sphere formation assay
Single cells were seeded in ultra low-attachment dishes (Corning, New York, NY, USA) at a density of 2 × 104 cells/ml and cultured for 7 days in serum-free DMEM/Ham’s F12 nutrient mixture (1:1, v/v) supplemented with 5 mg/ml insulin, 0.5 mg/ml hydrocortisone, 2% B27 and 20 ng/ml epidermal growth factor.
Wound healing assay
Cell layers were gently wounded through the central axis of the plate using a pipette tip. The migration of cells into the wound was observed at 6 h in six random selected microscopic fields. Wound closures were quantified using an image processing and analysis software program (ImageJ 1.40g, Bethesda, MD, USA).
Supplementary Material
ACKNOWLEDGEMENTS
We thank A Kondo, T Taniguchi, Y Kojima, Y Watanabe, M Tanaka and N Sakurai for technical assistance and discussion. This work was in part supported by Grant-in-Aid for Scientific Research on Innovative Areas and the Japan Health Sciences Foundation, and grants from the Uehara Memorial Foundation and Takeda Science Foundation to AR.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 4.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 6.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27:44–46. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- 8.Berry PA, Maitland NJ, Collins AT. Androgen receptor signalling in prostate: effects of stromal factors on normal and cancer stem cells. Mol Cell Endocrinol. 2008;288:30–37. doi: 10.1016/j.mce.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Dey D, Saxena M, Paranjape AN, Krishnan V, Giraddi R, Kumar MV, et al. Phenotypic and functional characterization of human mammary stem/progenitor cells in long term culture. PLoS One. 2009;4:e5329. doi: 10.1371/journal.pone.0005329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 12.Werbowetski-Ogilvie TE, Bosse M, Stewart M, Schnerch A, Ramos-Mejia V, Rouleau A, et al. Characterization of human embryonic stem cells with features of neoplastic progression. Nat Biotechnol. 2009;27:91–97. doi: 10.1038/nbt.1516. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA. 2010;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122(Pt 19):3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowell JK, LaDuca J, Rossi MR, Burkhardt T, Nowak NJ, Matsui S. Molecular characterization of the t(3;9) associated with immortalization in the MCF10A cell line. Cancer Genet Cytogenet. 2005;163:23–29. doi: 10.1016/j.cancergencyto.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 20.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 21.Nishi M, Akutsu H, Masui S, Kondo A, Nagashima Y, Kimura H, et al. A distinct role for Pin1 in the induction and maintenance of pluripotency. J Biol Chem. 2011;286:11593–11603. doi: 10.1074/jbc.M110.187989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desbaillets I, Ziegler U, Groscurth P, Gassmann M. Embryoid bodies: an in vitro model of mouse embryogenesis. Exp Physiol. 2000;85:645–651. [PubMed] [Google Scholar]
- 23.Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci USA. 2009;106:19035–19039. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhat-Nakshatri P, Appaiah H, Ballas C, Pick-Franke P, Goulet R, Jr, Badve S, et al. SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer. 2010;10:411. doi: 10.1186/1471-2407-10-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 27.Scaffidi P, Misteli T. In vitro generation of human cells with cancer stem cell properties. Nat Cell Biol. 2011;13:1051–1061. doi: 10.1038/ncb2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–17978. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- 30.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 31.Clark AT. The stem cell identity of testicular cancer. Stem Cell Rev. 2007;3:49–59. doi: 10.1007/s12015-007-0002-x. [DOI] [PubMed] [Google Scholar]
- 32.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 33.Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Muller F, et al. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. BMC Cancer. 2011;11:42. doi: 10.1186/1471-2407-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarig R, Rivlin N, Brosh R, Bornstein C, Kamer I, Ezra O, et al. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J Exp Med. 2010;207:2127–2140. doi: 10.1084/jem.20100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Kasai T, Li Y, Sugii Y, Jin G, Okada M, et al. A model of cancer stem cells derived from mouse induced pluripotent stem cells. PLoS One. 2012;7:e33544. doi: 10.1371/journal.pone.0033544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 38.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci USA. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 45.Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K, et al. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin Cancer Res. 2005;11:7523–7531. doi: 10.1158/1078-0432.CCR-05-0457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.