Abstract
Genotype Query Tools (GQT) is a new indexing strategy that expedites analyses of genome variation datasets in VCF format based on sample genotypes, phenotypes and relationships. GQT’s compressed genotype index minimizes decompression for analysis, and performance relative to existing methods improves with cohort size. We show substantial (up to 443 fold) performance gains over existing methods and demonstrate GQT’s utility for exploring massive datasets involving thousands to millions of genomes.
For the majority of common human diseases, only a small fraction of the heritability can be attributed to known genetic variation. A widely-held hypothesis posits that the remaining heritability is explained in part by rare, and thus largely unknown, genetic variation in the human population1. Extrapolating from current and forthcoming efforts, it is predicted that more than 1 million human genomes will be sequenced in the near term2. Integrated analyses and community sharing of such population datasets will clearly be critical for future discovery. In aggregate, the resulting datasets will include trillions of genotypes at hundreds of millions of polymorphic loci. Therefore, the development of more effective data exploration and compression strategies is crucial for broad use and future discovery.
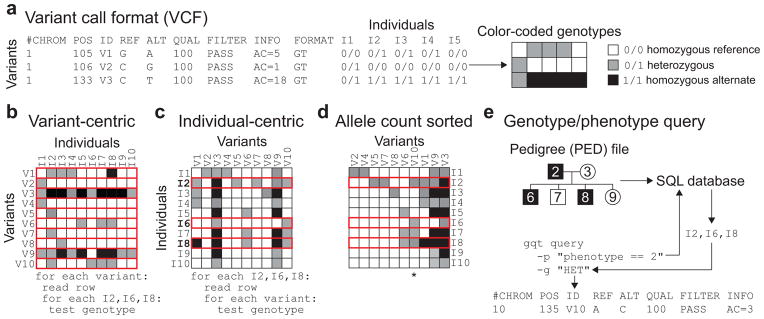
The Variant Call Format (VCF)3 defines a common framework for representing variants, sample genotypes, and variant annotations from DNA sequencing studies (Fig. 1a), and it has become the standard for genome variation research. However, since VCF is intentionally organized from the perspective of chromosomal loci to support “variant-centric” analyses such as “which variants affect BRCA1?”, it is consequently ill-suited to queries focused on specific genotype, phenotype or inheritance combinations such as “which variants are homozygous exclusively in affected women?” (Fig. 1b). Genotype Query Tools (GQT) introduces a new and complementary “individual-centric” strategy for indexing and mining very large genetic variation datasets. GQT creates an index of a VCF file by transposing the genotype data to facilitate queries based on the genotypes, phenotypes, and relationships of one or more of the individuals in the study (Fig. 1c). Once transposed, variant columns are sorted by allele frequency to take advantage of the fact that the majority of genetic variation is extremely rare in the population4,5 (Fig. 1d). Reordering by allele frequency greatly improves data compression since it yields much longer runs of identical genotypes than transposition alone (Supplementary Fig. 1). GQT also utilizes an efficient data compression strategy based upon Word Aligned Hybrid (WAH) compressed6,7,8 bitmap indices of the sample genotype information (see Online Methods and Supplementary Fig. 2–4). The combination of genotype transposition and WAH compression maximizes query performance by allowing direct inspection of genotype data without decompression.
Figure 1.
Creation and data exploration of an individual-centric genotype index. (a) The “variant-centric” VCF standard is essentially a genotype matrix whose rows correspond to variants and columns to individuals. (b) The VCF standard is inefficient for queries across all genotypes and a subset of individuals since each variant row must be inspected (in red) to test all of the genotypes of specific individuals. (c) By transposing the matrix such that rows (data records) now represent the full set of genotypes for each individual, the data better aligns to individual-centric questions and algorithms. (d) Sorting the columns of an individual-centric matrix by alternate allele count (AC) improves compressibility. After the variants have been reorganized based on AC, all genotypes for each sample are converted to Word Aligned Hybrid compressed bitmaps (see Supplementary Note). (e) GQT will create a SQLite database of a PED file describing the familial relationships, gender, ancestry, and custom, user-defined sample descriptions. The resulting database allows GQT to quickly extract the specific compressed bitmap records in the genotype index corresponding to a query. Once the compressed bitmaps for the relevant samples are extracted, they are compared to quickly identify the subset of variant(s) that meet the genotype requirements (in this example all individuals must be heterozygous, yielding variant V10 which is denoted by an asterisk) imposed for the subset of individuals.
In addition to indexing genotypes, GQT can also create a simple database from a PED file describing the names, relationships, multiple phenotypes, and other attributes of the samples in an associated VCF file (Fig. 1e). The sample database complements the GQT index of the original VCF or BCF file and allows GQT to quickly identify the compressed sample genotype bitmaps that are germane to the query. For example, a query could search for variants where all affected individuals (“phenotype==2”) are heterozygous (“HET”) (Fig. 1e). GQT uses the sample database to find compressed genotype bitmaps of the three affected individuals. Once identified, the relevant bit arrays for heterozygous genotypes are AND’ed (Supplementary Fig. 2b) to return the single VCF record in which all three individuals are heterozygous. With this structure, queries can exploit any attribute that is defined in the PED file and one can combine multiple criteria. This functionality enables, as examples, searches for de novo mutations in a multi-generational pedigree (Supplementary Fig. 5), and variants having markedly different minor allele frequencies in different world sub-populations or case versus control samples. Query results are returned in VCF format which supports sophisticated analyses combining GQT queries with other, variant-centric tools such as BEDTOOLS9, VCFTOOLS3 and BCFTOOLS.
GQT’s genotype index is a complement to the existing variant-centric indexing strategies3,10 available for datasets in VCF (or BCF) format. GQT creates two additional indices (BIM and VID; Supplementary Fig. 6) that allow variants satisfying a query to be quickly returned in VCF format. The storage overhead of the GQT indices is minimal with respect to the size of the underlying dataset. Moreover, index size relative to the VCF file continues to diminish as the cohort size grows since most new variation discovered from additional samples will be very rare4,5. Comparisons to PLINK and BCF based on the 1000 Genomes Project (Phase 3) VCF files demonstrated that the GQT compression strategy was on par with the compression scheme used by the BCF format (Supplementary Fig. 7; Supplementary Note). In particular, the GQT genotype index alone (i.e., excluding the ancillary BIM and VID indices) represented merely an additional ~10% (12.08 Gb) storage burden beyond the requirement of the Phase 3 1000 Genomes dataset in BCF format.
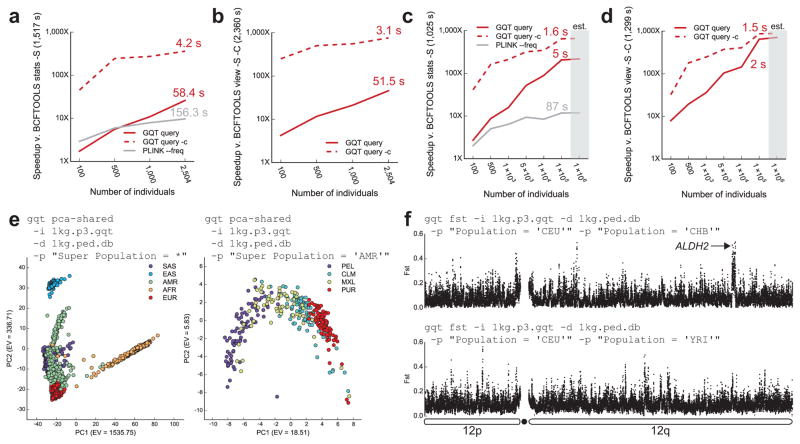
A typical tradeoff for data compression algorithms is the cost of decompressing data prior to analysis11. We designed the GQT indexing strategy precisely to avoid this tradeoff and achieve efficient queries of cohorts involving thousands to millions of individuals. To demonstrate this, we compared the query performance of GQT to both BCFTOOLS and a comprehensive update to PLINK (v1.90). First, we compared the time required to compute the alternate allele frequency among a target set of 10% of individuals from the 1000 Genomes VCF (Fig. 2a). Whereas BCFTOOLS required 1517.5 seconds, both GQT and PLINK were substantially faster, requiring 58.4 seconds (26.0 fold speedup) and 156.3 seconds (9.6 fold speedup), respectively. We emphasize that GQT’s performance advantage improved as the number of individuals increased, whereas PLINK’s performance remained relatively flat. Moreover, matching variants are identified almost instantly and thus, the majority of GQT’s runtime is spent emitting the VCF results. For example, when the GQT “count” option is invoked to simply return the number of matching variants, the runtime dropped to 4.2 seconds. We also compared the time required to identify rare (AAF < 1%) variants among a subset of 10% of the individuals (Fig. 2b). In this case, GQT was up to 45.8 times faster than BCFTOOLS (51.5 seconds v. 2360.5 seconds).
Figure 2.
GQT query performance and applications of the genotype index. (a) Fold speedup for computing the alternate allele frequency (AF) for a targeted 10% of the 2,504 individuals in 1,000 Genomes Phase 3. The baseline was the BCFTOOLS “stats” command. Two versions of GQT output were considered, valid VCF (“GQT query”) and the count of matching variants (“GQT query -c”). (b) Speedup for finding variants having an AF of < 1% in a target 10% of individuals. The baseline was BCFTOOLS “view -C”. PLINK did not directly perform this operation and was excluded. (c) Query performance for simulated genotypes on a 100 Mb genome with between 100 and 100,000 individuals. The speedup for computing the alternate AF count for 10% of individuals is presented. (d) The speedup for finding variants having an AF of < 1% in 10% of individuals. Again, PLINK was excluded. Times reported are for 100,000 simulated genomes. Neither variant nor sample metadata were included. The metrics for 1 million individuals (est.) were estimated using a linear fit. GQT’s runtimes are similar for 2,504 individuals from the 1,000 Genomes and the simulation because the total number of genotypes is nearly identical (2,504 individuals with 84,739,846 variants and 100,000 individuals with 2,052,387 variants, respectively). (e) A principal component analysis of all variants from 1,000 Genomes Phase 3 requiring 207 minutes for 2,504 individuals, and 3 minutes for 347 AMR individuals. (f) Fst analysis of Europeans v. East Asians and Europeans v. Africans on chromosome 12.
When considering the Exome Aggregation Consortium (ExAC, version 0.3) variant dataset (9.36 million exonic variants among 60,706 human exomes), the GQT index was only 0.2% the size of the VCF (28 GB v. 14.1 TB), reflecting a storage requirement of merely 0.38 bits per genotype. Rare variants were found in only 2.1 minutes (9.98 seconds when excluding the time required to report the variants), reflecting a 443-fold improvement over BCFTOOLS (931.4 minutes) (Supplementary Table 1). Based on simulated datasets involving 100 to 100,000 individuals on a 100Mb genome, it is clear that GQT’s relative data compression and query performance continues to improve with larger cohorts. While simulating variants from one million or more individuals was computationally intractable for this study, extrapolation suggested that query performance for a cohort of one million genomes was at least 218-fold faster than BCFTOOLS (Fig. 2c,d).
Since the GQT indexing strategy is fundamentally optimized for questions that involve comparisons of sample genotypes among many variant loci, it is also well suited to many common statistical and population genetics measures. For example, as the basis for principal component analysis among the 2,504 individuals in the 1000 Genomes dataset, GQT produced a similarity matrix using the number of shared, non-reference variants in 207 minutes (Fig. 2e). A similarity matrix for the 347 admixed American (AMR) individuals required merely 3 minutes. GQT also computed the Weir and Cockerham12 Fst statistic for all 84.7 million variants in the Phase 3 1000 Genomes dataset in 74 seconds, reflecting a 146 fold speedup over VCFTOOLS (Fig. 2f). These examples demonstrate how GQT indices could empower other statistical genetics software and serve as a framework for future method development.
While GQT greatly expedites individual-centric analysis, it is currently incapable of searching for variants in specific chromosomal regions, unless the results of GQT queries are filtered to specific regions by variant-centric tools such as Tabix10 and BCFTOOLS. However, we have shown that it is a natural complement to variant-centric indexing strategies that struggle with individual-centric queries. Based on the strengths and tradeoffs of each approach, we envision a general querying interface that integrates both indexing strategies and supports genomic data sharing efforts such as the Global Alliance for Genomics and Health. Based on the efficiency and inherent flexibility of this genotype indexing strategy, we expect GQT to be a potent analysis tool and to enhance existing methods in the exploration of the massive datasets involving millions of genomes that are expected in the near future2.
ONLINE METHODS
Overview of the GQT genotype indexing strategy
Once the genotypes in a VCF file have been transposed from a variant-centric form (Fig. 1b; each row is a variant) to an individual-centric form (Fig. 1c; each row is now an individual), the variant columns are then sorted by allele frequency (Fig. 1d). Each individual’s genotypes are then encoded into a bitmap index comprised of four distinct bit arrays corresponding to each of the four possible (including “unknown”) diploid genotypes (Supplementary Fig. 2). Bitmaps allow efficient comparisons of many genotypes in a single operation using bitwise logical operations, thereby enabling rapid comparisons of sample genotypes among many variants in the original VCF file (Supplementary Fig. 3). Lastly, the bitmap indices are compressed using Word Aligned Hybrid (WAH) encoding6, which achieves near-optimal compression while still allowing bitwise operations directly on the compressed data (Supplementary Fig. 4). This encoding minimizes the disk storage requirements of the index and, by eliminating the need for data inflation, improves query speed since runtime is a function of the compressed input size.
Sorting by allele frequency to improve compression
Long runs of identical genotypes are easily compressed. We have chosen an alternative individual-centric data organization strategy that, while it facilitates queries based on individual genotypes, destroys the inherent compression of the genotype runs in the variant-centric approach. This loss of compression is the direct consequence of the fact that records in the individual-centric approach reflect the genotypes for a given individual at each site of variation in the genome. Runs of identical genotypes are far shorter, on average, than with the variant-centric approach and therefore the individual-centric strategy will yield poor compression. The question then becomes how to leverage the query efficiency of individual-centric data organization while also retaining the opportunity for data compression? GQT solves this by sorting the variant columns of the transposed matrix in order of allele frequency. This results in fewer, longer runs of identical genotypes in each individual’s row of genotypes (Supplementary Fig. 1). For example, we compared the effect of sorting variants on genotype runs using chromosome 20 from Phase 3 of the 1000 Genomes Project. As expected, sorting variants by allele frequency caused both a dramatic increase in the mean length (10.7 versus 23.2) of identical genotype runs and a reduction in the median number of runs per individual (158,993.5 versus 70,718.5). Fewer, longer identical genotype runs allows greater compression of each individual’s (reordered) variant genotypes.
Representing sample genotypes with bitmap indices
The fundamental advantage of individual-centric data organization is the fact that all of an individual’s genotypes can be accessed at once. This enables algorithms to quickly compare all variant genotypes from multiple samples in search of variants that meet specific inheritance patterns, allele frequencies, or enrichment among subsets of individuals. Despite the improved data alignment, comparing sample genotypes can still require substantial computation. For VCF, which encodes diploid genotypes as “0/0” for homozygotes of the reference allele, “0/1” for heterozygotes, “1/1” for homozygotes of the alternate allele, and “./.” for unknown genotypes (Supplementary Fig. 2a), comparing the genotypes of two or more individuals requires iterative tests of each genotype for each individual.
Recognizing this inefficiency, we encode each individual’s vector of genotypes with a bitmap index. A bitmap index (“bitmap”) is an efficient strategy for indexing attributes with discrete values that uses a separate bit array for each possible attribute value. In this case of an individual’s genotypes, a bitmap is comprised of four distinct bit arrays corresponding to each of the four possible (including “unknown”) diploid genotypes. The bits in each bit array are set to true (1) if the individual’s genotype at a given variant matches the genotype the array encodes (Supplementary Fig. 2a). Otherwise, the element is set to false (0). In turn, bitmap encoding facilitates the rapid comparison of individuals’ genotypes with highly optimized bitwise logical operations. As an example, a bitmap search for variants where all individuals are heterozygous involves a series of pairwise AND operations on the entire heterozygote bit array from each individual. The intermediate result of each pairwise AND operation is subsequently compared with the next individual, until the final bit array reflects the variants where all individuals are heterozygous (Supplementary Fig. 2b). Such queries are expedited owing to the ability of modern CPUs to simultaneously test multiple bits (i.e., genotypes) with a single bitwise logical operation.
Efficient comparisons using bitmaps
By using bitwise logical operations we can compare many genotypes in a single operation, rather than comparing each individual value. At a low level, bitwise logical operations are performed on words, which are the fixed-size unit of bits used by the CPU. Modern processors typically use either 32- or 64-bit words. When a bitwise logical operation is performed between two bit arrays (each of which correspond to the genotypes of two individuals), the processor completes this operation on one pair of words at a time. For example, if the word size is 32, then computing the bitwise AND of two bit arrays that are each 320 bits long would require only 10 ANDs. This optimization is equivalent to a 32-way parallel operation with zero overhead. Consider the case where we are searching for the loci where all three individuals are heterozygous among eight variants (Supplementary Fig. 3). When genotypes are encoded in ASCII (as they are in VCF), the algorithm must loop over every individual and every variant to find the common sites. In total, this requires 24 iterations (Supplementary Fig. 3a). In contrast, encoding genotypes with a bitmap allows the same computation to be completed with only three bit-wise AND operations (Supplementary Fig. 3b). In effect, bit-wise logical operations compare all eight genotypes in parallel in a single step. For brevity an 8-bit word is used, and only the heterozygous bit arrays are shown, but the same principles hold for the larger word sizes employed by GQT.
Using Word-Aligned Hybrid to directly query compressed data
While bitwise logical operations can drastically improve query runtime performance, bitmap indices require double the amount of space over the minimum two bits per genotype. To address this issue we can look to compressing that data. While genotype data can be compressed with standard run-length encoding, bitwise logical operations require that the bits associated with variants must be aligned, which is difficult to ensure with run-length encoding (RLE). Instead we used the Word Aligned Hybrid (WAH) encoding strategy, which represents run length in words not in bits. RLE encodes stretches of identical values (“runs”) to a new value where the first bit indicates the run value and the remaining bits give the number of bits in the run (Supplementary Fig. 4a). WAH is similar to RLE, except that it uses two different types of values. The “fill” type encodes runs of identical values, and the “literal” type encodes uncompressed binary (Supplementary Fig. 4a). This hybrid approach address an inefficiency in RLE where short runs map to larger encoded values. The first bit in a WAH value indicates whether it is a fill (1) or literal (0). For a fill, the second bit gives the run value and the remaining bits give the run length in words (not bits, like in RLE). For a literal, the remaining bits directly encode the uncompressed input. Since each WAH-encoded value represents some number of words, and bitwise logical operations are performed between words, these operation can be performed directly on compressed values.
The algorithm that performs bitwise logical operations is straightforward (Supplementary Fig. 4b). To operate on two uncompressed bit arrays we simply move in unison between the two arrays from the first to last word, and find the result of each pair of words. Since each WAH value encodes one or more words, we need to move across each WAH array independently. At each step we track the number of words that have been considered in the current value and only move to the next word once the words have been exhausted. Bitwise logical operations improve the performance of most queries by computing many genotype comparisons in parallel, but some higher-level queries cannot be resolved with these operations alone.
Finding the allele frequency among a set of individuals is one such query. In this case, each bit corresponds to the genotype of a particular individual at a particular genomic position, and the allele frequency for that position is the summation of the corresponding bits across all individuals. Since 32 bits are packed into a single word, this process can be reduced to a series of bitwise sums between words, which, unfortunately, is not a standard operation. While no architecture provides single-instruction support for bitwise sum, the operation does exhibit a high degree of parallelism. The sum of each position is independent of all other positions, which allows (in principle) the sum of all positions to be found concurrently. This classic Single Instruction Multiple Data (SIMD) scenario can be exploited through the use of the vector processor registers and instructions that are supported by the most recent Intel CPUs (Haswell and beyond). These special registers are designed to perform instructions on a list of values in parallel, and by combining several instructions (logical shift, AND, and sum) from the AVX2 instruction set we can perform the bitwise sum of 8 words in parallel. While the 8-way parallelism lags behind what is possible for other operations, it still represents a meaningful speed up for an operation that is expected to be part of many queries. The index we describe above has the ability to identify variants that meet a complex set of conditions among millions of individuals and billions of genotypes in seconds.
32-bit WAH word size
A fundamental choice for WAH-encoding bit arrays is the word size. Modern processors support up to 64 bits, but smaller words of 32, 16, and 8 are also possible, and the choice will affect both the compression ratio and query runtime. Since WAH uses one bit of each word to indicate the type of word (fill or literal), it would seem that larger words would be more efficient. An eight-bit word will have seven useful bits to every overhead bit, while a 64-bit word will have a 63:1 ratio. However, there is a large amount of waste within fill words. Considering that the first two bits of a fill indicate the word type and run value, and the remaining give the length of the run in words, a 64-bit fill word can encode a run that is 1.4×1020 bits long. That is enough bits to encode 46.1 billion human genomes. In fact, we only need 27 bits to cover the full genome, meaning that every 64-bit fill word would have at least 35 wasted bits. This would seem to indicate that smaller words are more efficient, but as the word size decrease the speed up of the bit-wise logical operations also declines. A single operation between two 64-bit words compares 8 times more bits (and their associated genotypes) than an operation between two 8-bit words. Taken together, our test show that 32-bits gives the best balance between size and speed.
Contents of BCF file, PLINK index, and GQT index
Direct compression comparisons must account for the fact that each tool compresses different subsets of the variant and sample genotype sections of a VCF file (Supplementary Fig. 6). By default, the BCF format encodes all of the data and metadata in both sections into binary values, and then compresses those values using blocked LZ77 encoding. PLINK ignores both variant and sample genotype metadata, does not compress the variant data, and simply encodes each genotype with two bits without compression. GQT uses a hybrid strategy for compressing VCF files. It retains all of the variant data and only the genotype values (no metadata) in the genotype section, and stores that data in a “BIM” file. The variant data is compressed with LZ77 encoding. In addition, an index of individual genotypes is created by transposition, sorting and WAH-compression. Since the variants in the BIM file are stored in the same order as the original VCF, and the genotype index columns are ordered by allele frequency, we must maintain a mapping between these two orderings to retain the ability to print results in the same order as the original VCF. This mapping is stored in a “VID” file.
Comparison of GQT index sizes to other file formats
File size comparisons used an uncompressed VCF as a baseline, BCFTOOLS used a compressed binary VCF (BCF) to store both variant and sample data, PLINK used the binary plink format (BED) to store sample data and a BIM file to store variant data, and GQT used a GQT index file to store WAH-encoded sample genotype data as well as a BIM file to store LZ77-compressed variant data.
Performance comparisons
We compared GQT v0.0.1 to PLINK v1.90p and BCFTOOLS (https://github.com/samtools/bcftools) 1.1 in terms of index file size and query runtime against four large-scale cohorts and simulated data sets. The cohorts included: 2,504 human genomes from the 1000 Genomes Project phase 3, 28 mouse genomes from the Mouse Genomes Project, 205 fly genomes from the Drosophila Genetic Reference Panel (DGRP), and 60,706 human exomes from Exome Aggregation Consortium (ExAC). Query comparisons included time to compute the alternate allele frequency count for a target 10% of the population, and time to find rare (details below) variants among a target 10% of the population. Both target sets were comprised of the last 10% of individuals. For all runtime comparisons BCFTOOLS considered a BCF file, PLINK considered a BED and BIM file, and GQT considered a GQT index and BIM file (Supplementary Note). Runtimes for GQT considered two different modes: the default mode that reports all matching variants in full VCF format and the “count” mode (specified by the “-c” option) that only reports the number of matching variants. The “count” mode is a useful operation in practice, and also demonstrates speed without I/O overhead.
Alternate allele count
The baseline runtime for finding the alternate allele count was the BCFTOOLS “stats” command with the “-S” option to select the subset of individuals, the PLINK command was “--freq” with the “--keep” option to select individuals, and the GQT command was “query” (with and without the “-c” option) with the “-g “count(HET HOM_ALT)”” option to specify the allele count function and the “-p “BCF_ID >= N”” option to select the subset (where N was the ID of the range that was considered).
Identifying rare variants
The baseline runtime for selecting the variants was the BCFTOOLS “view” command with the “-S” option to select the subset of individuals and the “-C” option to limit the frequency of the variant, and the GQT command was “query” (with and without the “-c” option) with the “-g “count(HET HOM_ALT)<=F”” option to specify the allele count filter (where F was the maximum occurrence of the variant) and the “-p “BCF_ID >= N”” option to select the subset (where N was the ID of the range that was considered). In both cases the limit was set to either 1% of the subset size or 1, whichever was greater. PLINK was omitted from this comparison because third-party tools are required to complete this operation, and in our opinion it is not fair to assign the runtime of those tools to PLINK.
Principal component analysis (PCA)
Using the “pca-shared” command, GQT computed a score for each pair of individuals in the target population that reflected the number of shared non-reference loci between the pair. This score was calculated in two stages. First, an intermediate OR operation of the HET and HOM_ALT bitmaps within each individual produced two bitmaps (one for each member of the pair) that marked non-reference loci. Then, an AND of these two bitmaps produced a final bitmap that marked the sites where both individuals were non-reference. GQT then counted the number of bits that were set in this bitmap and reported the final score. The “pca-shared” command also takes the target population as a parameter. Here, two cases are considered (Fig. 2e): all 2504 individuals, which included the South Asian (SAS), East Asian (EAS), Admixed American (AMR), African (AFR), and European (EUR) “super populations”, and only the 347 individuals in the Admixed American (AMR) super population, which included: Peruvians from Lima, Peru (PEL), Columbians from Medellin, Columbia (CLM), Mexican Ancestry from Los Angeles, USA (MXL), and Puerto Ricans from Puerto Rico (PUR).
This analysis considered only the autosomes within the 1000 Genomes phase 3 variants. The result of the GQT “pca-shared” command is the upper half of a square symmetrical matrix. The Python script “fill_m.py” fills out a full matrix by reflecting those values, and another Python script “pca_light.py” calculates the Eigen vectors and values using the Numpy scientific computing library and plots the results.
The resulting GQT commands were:
gqt pca-shared \ -i ALL.phase3.autosome.vcf.gz.gqt \ -d integrated_call_samples.20130502.ALL.spop.ped.db \ -p “*” \ -f “Population” \ -l ALL.phase3.autosome.vcf.gz.gqt.pops \ > ALL.phase3.autosome.vcf.gz.gqt.o gqt pca-shared \ -i ALL.phase3.autosome.vcf.gz.gqt \ -d integrated_call_samples.20130502.ALL.spop.ped.db \ -p “Super_Population = ‘AMR’” \ -f “Population” \ -l ALL.phase3.autosome.vcf.gz.gqt.AMR.pops \ > ALL.phase3.autosome.vcf.gz.gqt.AMR.o
Fixation index
Fst is a widely used measurement of the genetic difference between populations, and here we focused on the method proposed by Weir and Cockerham12. While this metric has many parameters, it is fundamentally based on the frequency of an allele and the proportion of individuals that are heterozygous for that allele in each population. GQT can quickly calculate both metrics for various populations across the whole genome. Allele frequency is calculated by considering the bits marked in both the HET and HOM_ALT bitmaps. Since each bit in a bitmap corresponds to a specific variant, calculating the allele frequency of all variants involves incrementing the associated counter for each set bit. WAH encoding makes this process more efficient by collapsing large stretches of reference alleles (which are represented by zeros in the HET and HOM_ALT bitmaps) into a small number of words that can be quickly skipped. We further accelerate the processing of each word by using the AVX2 vector-processing instruction set. Vector processing instructions are a set of special CPU instructions and registers that exploit data-level parallelism by operating on a vector of values with a single operation. These instructions allow us to consider 8 bits in parallel, and therefore compute the resulting sum of each word in 4 (as apposed to 32) operations. The proportion of individuals that are heterozygous for an allele is computed in a similar manner, except only the HET bitmaps are considered.
This analysis considered the 1000 Genomes phase 3 variants that were bi-allelic and had a minimum alternate allele frequency of 1%. The GQT “fst” command can consider two or more populations with the “-p” option. Here we considered two cases (Fig. 2f): the Utah Residents with Northern and Western European Ancestry (CEU) versus the Han Chinese in Beijing, China populations (“-p “Population = ‘CHB’” -p “Population = ‘CEU’”“), and the Utah Residents with Northern and Western European Ancestry (CEU) versus the Yoruba in Ibadan, Nigeria populations (“-p “Population = ‘CHB’” -p “Population = ‘YRI’””). In both cases values were smoothed using the mean Fst value over a 10kb window with a 5kb step. The comparison to VCFTOOLS considered version 0.1.12 and the “--weir-fst-pop” options for the CHB and CEU populations.
The resulting GQT commands were:
gqt fst \
-i \
ALL.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz.gqt \
-d 1kg.phase3.ped.db \
-p “Population = ‘CHB’” \
-p “Population = ‘CEU’” \
> CHB_vs_CEU.gqt.fst.vcf
gqt fst \
-i \
ALL.phase3_shapeit2_mvncall_integrated_v5a.20130502.genotypes.vcf.gz.gqt \
-d 1kg.phase3.ped.db \
-p “Population = ‘YRI’” \
-p “Population = ‘CEU’” \
> YRI_vs_CEU.gqt.fst.vcf
Experimental Data sets
1000 Genomes phase 3: Individual chromosome VCF files were retrieved from [2] (last accessed December 10, 2014) and combined into a single file using the BCFTOOLS “concat” command. To understand how each tool scaled as the number of samples and variants increased, we subsampled the full data set (which included 2,504 individuals) to create new sets with 100, 500, and 1,000 individuals. To create each data set size, we randomly selected the target number of samples, then used the BCFTOOLS “view” command with the “-s” option to return just the genotypes of the target samples. We then recomputed the allele frequency of each variant with the BCFTOOLS “fill-AN-AC” plugin, and filtered all non variable sites with the BCFTOOLS “view” command and the “-c 1” option.
Exome Aggregation Consortium (ExAC). Version 3 of the ExAC dataset was analyzed and run times were measured on the computing infrastructure at the Broad Institute.
Mouse Genomes Project. Data was retrieved in VCF format from (ftp://ftp-mouse.sanger.ac.uk/current_snps/mgp.v4.snps.dbSNP.vcf.gz) and was last accessed November 25, 2014.
Drosophila Genetic Reference Panel. Data was retrieved in VCF format from (http://dgrp2.gnets.ncsu.edu/data/website/dgrp2.vcf) and was last accessed November 25, 2014. CEPH 1473 pedigree. A VCF file of variants in the CEPH 1473 pedigree that was sequenced as part of the Illumina Platinum Genomes Project was downloaded from ftp://ftp-trace.ncbi.nih.gov/giab/ftp/data/NA12878/variant_calls/RTG/cohort-illumina-wgs.vcf.gz.
Simulated data sets
Genotypes were simulated using the MaCS13 simulator version 0.5d with the mutation rate and recombination rate per site per 4N generations set to 0.001, and the region size set to 100 megabases. Since our simulation considered between 100 and 100,000 diploid samples, and MaCS only simulates haplotypes, we simulated 2× haplotypes for each case and combined 2 haplotypes to create a single diploid genome. It was computationally prohibitive to produce a data set for 1 million individuals (the 100,000 simulation ran for over four weeks), so we used a simple linear fit to estimate the file size and runtimes for 1 million individuals.
Supplementary Material
Acknowledgments
We are grateful to C. Chiang for conceptual discussions, I. Levicki for helpful advice on AVX2 operations, both S. McCarthy and P. Danecek for their guidance with htslib, and Z. Kronenberg for advice on population genetics measures. The authors would also like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. This research was supported by an NHGRI award to A.R.Q. (NIH R01HG006693).
Footnotes
AUTHOR CONTRIBUTIONS
R.M.L. designed and wrote GQT and analyzed the data. N.K. wrote a fast output method. K.J.K. analyzed ExAC data. A.R.Q. conceived and designed the study. R.M.L. and A.R.Q. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
COMPUTING ENVIRONMENT
GQT is a tool written in C, which uses the htslib (https://github.com/samtools/htslib) to interact with VCF and BCF files and zlib (http://www.zlib.net/) to compress and inflate variant metadata. All experiments were run on Ubuntu Linux v3.13.0-43, with gcc v4.9.2, 4 Intel Core i7-4790K 4.00GHz CPUs with the Haswell microarchitecture, and a 550 MB/s read/write solid-state hard drive.
CODE AVAILABILITY
All source code for the GQT toolkit is available at https://github.com/ryanlayer/gqt. Furthermore, all commands used for the experiments conducted in this study are available at https://github.com/ryanlayer/gqt_paper.
References
- 1.Zuk O, et al. Proc Natl Acad Sci. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens ZD, et al. PLOS Biol. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danecek P, et al. Bioinforma Oxf Engl. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keinan A, Clark AG. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium et al. Nature. 2012;491:56–65. [Google Scholar]
- 6.Kesheng Wu, Otoo EJ, Shoshani A. 2002:99–108. doi: 10.1109/SSDM.2002.1029710. [DOI] [Google Scholar]
- 7.Siqueira TLL, de A, Ciferri CD, Times VC, de Oliveira AG, Ciferri RR. J Braz Comput Soc. 2009;15:19–34. [Google Scholar]
- 8.Liu YB, et al. J Korean Astron Soc. 2014;47:115–122. [Google Scholar]
- 9.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H. Bioinforma Oxf Engl. 2011;27:718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziv J, Lempel A. IEEE Trans Inf Theory. 1977;23:337–343. [Google Scholar]
- 12.Weir BS, Cockerham CC. Evolution. 1984;38:1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen GK, Marjoram P, Wall JD. Genome Res. 2008;19:136–142. doi: 10.1101/gr.083634.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.