Abstract
The postsynaptic density (PSD) is a protein-rich network important for the localization of postsynaptic glutamate receptors (GluRs) and for signaling downstream of these receptors. Although hundreds of PSD proteins have been identified, many are functionally uncharacterized. We conducted a reverse genetic screen for mutations that affected GluR localization using Drosophila genes that encode homologs of mammalian PSD proteins. 42.8% of the mutants analyzed exhibited a significant change in GluR localization at the third instar larval neuromuscular junction (NMJ), a model synapse that expresses homologs of AMPA receptors. We identified the E3 ubiquitin ligase, Mib1, which promotes Notch signaling, as a regulator of synaptic GluR localization. Mib1 positively regulates the localization of the GluR subunits GluRIIA, GluRIIB, and GluRIIC. Mutations in mib1 and ubiquitous expression of Mib1 that lacks its ubiquitin ligase activity result in the loss of synaptic GluRIIA-containing receptors. In contrast, overexpression of Mib1 in all tissues increases postsynaptic levels of GluRIIA. Cellular levels of Mib1 are also important for the structure of the presynaptic motor neuron. While deficient Mib1 signaling leads to overgrowth of the NMJ, ubiquitous overexpression of Mib1 results in a reduction in the number of presynaptic motor neuron boutons and branches. These synaptic changes may be secondary to attenuated glutamate release from the presynaptic motor neuron in mib1 mutants as mib1 mutants exhibit significant reductions in the vesicle-associated protein cysteine string protein and in the frequency of spontaneous neurotransmission.
Keywords: Postsynaptic Density, Glutamate Receptors, Synapse, Synaptic Transmission, Drosophila Neuromuscular Junction
Introduction
Proper formation and maintenance of glutamatergic synapses is required for diverse neurobiological processes including movement (Girault, 2012), visual processing (Self et al., 2012), and learning and memory (Hu et al., 2007; Matsuo et al., 2008; Sanderson & Bannerman, 2012). Once established, these synapses are plastic and modify themselves as a result of changes in activity. Synaptic plasticity occurs as a result of changes in presynaptic neurotransmitter release probability, the localization and synthesis of synaptic proteins, and remodeling of the synaptic cytoskeleton (reviewed in (Huganir & Nicoll, 2013; Padamsey & Emptage, 2014)). The localization of postsynaptic ionotropic glutamate receptors (GluRs) opposite of presynaptic release sites is particularly important for synaptic transmission as it determines the postsynaptic response (Xie et al., 1997; DiAntonio et al., 1999; Franks et al., 2003; Raghavachari & Lisman, 2004; Lisman et al., 2007).
Excitatory postsynaptic GluRs are components of the postsynaptic density (PSD), a specialized network of proteins that links receptors to the cytoskeleton and downstream signaling pathways. The PSD, localized to mammalian small postsynaptic protrusions or dendritic spines, is estimated to contain hundreds of different proteins (Satoh et al., 2002; Jordan et al., 2004; Peng et al., 2004; Yoshimura et al., 2004; Collins et al., 2006; Dosemeci et al., 2006; Bayes et al., 2011), many of which are represented by multiple copy numbers (Chen et al., 2008; Shinohara, 2011). PSD proteins can be broadly grouped as cell adhesion molecules, cytoskeletal proteins, metabolic proteins, transmembrane proteins, trafficking/motor proteins, scaffold proteins, and enzymes like GTPases and kinases/phosphatases (Okabe, 2007). In mammals, dysfunction of the PSD is linked to neurodegenerative diseases (for review see (Gong & Lippa, 2010)), autism/autism spectral disorders (Feyder et al., 2010; Bangash et al., 2011), schizophrenia (Hashimoto et al., 2007; Cheng et al., 2010), mental impairments (Raymond & Tarpey, 2006; Zanni et al., 2010), and drug abuse (Moron et al., 2007; Okvist et al., 2011).
The composition and size of the PSD are dynamically regulated by synaptic activity. Long-term potentiation (LTP), a process that enhances synaptic efficacy and is thought to be the cellular basis of learning and memory (Neves et al., 2008; Takeuchi et al., 2014), results in the redistribution of the AMPA receptor subunit, GluA1, and NMDA receptor subunit, GluN1, to dendritic areas of the rat dentate gyrus (Kennard et al., 2014). The increased surface localization of GluA1 is mediated by remodeling of the actin cytoskeleton (Gu et al., 2010; Kerr & Blanpied, 2012) and may be linked to altered localization of scaffolding proteins within the PSD (MacGillavry et al., 2013; Bosch et al., 2014; Meyer et al., 2014). LTP also results in expansion of the PSD (Chen et al., 2007; Bosch et al., 2014) and enlargement of dendritic spines (Matsuzaki et al., 2004; Harvey & Svoboda, 2007), both of which require local translation of PSD components (Bramham, 2008; Bosch et al., 2014).
PSD proteins are remarkably conserved with orthologs across archaeal, bacteria, and eukaryote kingdoms (Emes et al., 2008; Alie & Manuel, 2010; Emes & Grant, 2011). We have previously identified Drosophila orthologs for approximately 96% of published mammalian PSD proteins (Liebl & Featherstone, 2008). The functional role of many of these proteins is currently unidentified in any species. Therefore, we performed a reverse genetic screen to determine whether mutations in Drosophila PSD orthologs affect the synaptic localization of GluRs at the neuromuscular junction (NMJ) using immunocytochemistry. The Drosophila larval NMJ contains ionotropic GluRs that are homologous to AMPA receptors (Menon et al., 2013). We uncovered a novel function for the E3 ubiquitin ligase, Mind Bomb1 (Mib1), a component of the Notch signaling pathway, in the regulation of postsynaptic GluR localization. Mib1 regulates the clustering of postsynaptic GluRs, the frequency of spontaneous neurotransmission, and synaptic levels of the presynaptic protein cysteine string protein (CSP).
Results
Reverse genetic screen for gene products that regulate GluR localization
The PSD is a dense protein network opposed to presynaptic release sites that helps provide the structural basis for synaptic regulation and plasticity (Collins et al., 2006; Dosemeci et al., 2006). Hundreds of PSD proteins have been identified and the Drosophila genome encodes orthologs for 95.8% of these proteins (Liebl & Featherstone, 2008). Many of these genes are functionally uncharacterized. Therefore, we conducted a reverse genetic screen of genes that encode homologs of mammalian PSD proteins to identify mutants with altered postsynaptic GluR expression and/or localization at the 6/7 NMJ of third instar Drosophila larvae. This NMJ is innervated by two glutamatergic motor neurons that arborize on muscles by forming a series of distinct swellings or boutons (Jan & Jan, 1976; Johansen et al., 1989; Ruiz-Canada & Budnik, 2006). Drosophila NMJ GluRs are similar to non-NMDA receptors including AMPA receptors and are tetramers that contain either the GluRIIA or GluRIIB subunits along with GluRIIC (Marrus et al., 2004), GluRIID (Featherstone et al., 2005), and GluRIIE (Qin et al., 2005).
We examined 130 different mutations that corresponded to 144 mammalian PSD proteins (Table S2) for altered synaptic localization of the GluRIIA subunit. 18 mutations (12.5%) were lethal prior to the third instar larval stage and, therefore, were not analyzed. Of the remaining mutants analyzed, 48 (42.8%) exhibited phenotypes that significantly affected the localization of postsynaptic GluRs as indicated by a significant change in relative GluRIIA fluorescence intensity. The majority of these mutations (42/48 or 87.5%) resulted in a significant reduction in postsynaptic GluRs containing GluRIIA. Conversely, six mutations (6/48 or 12.5%) produced an increase in synaptic GluRIIA.
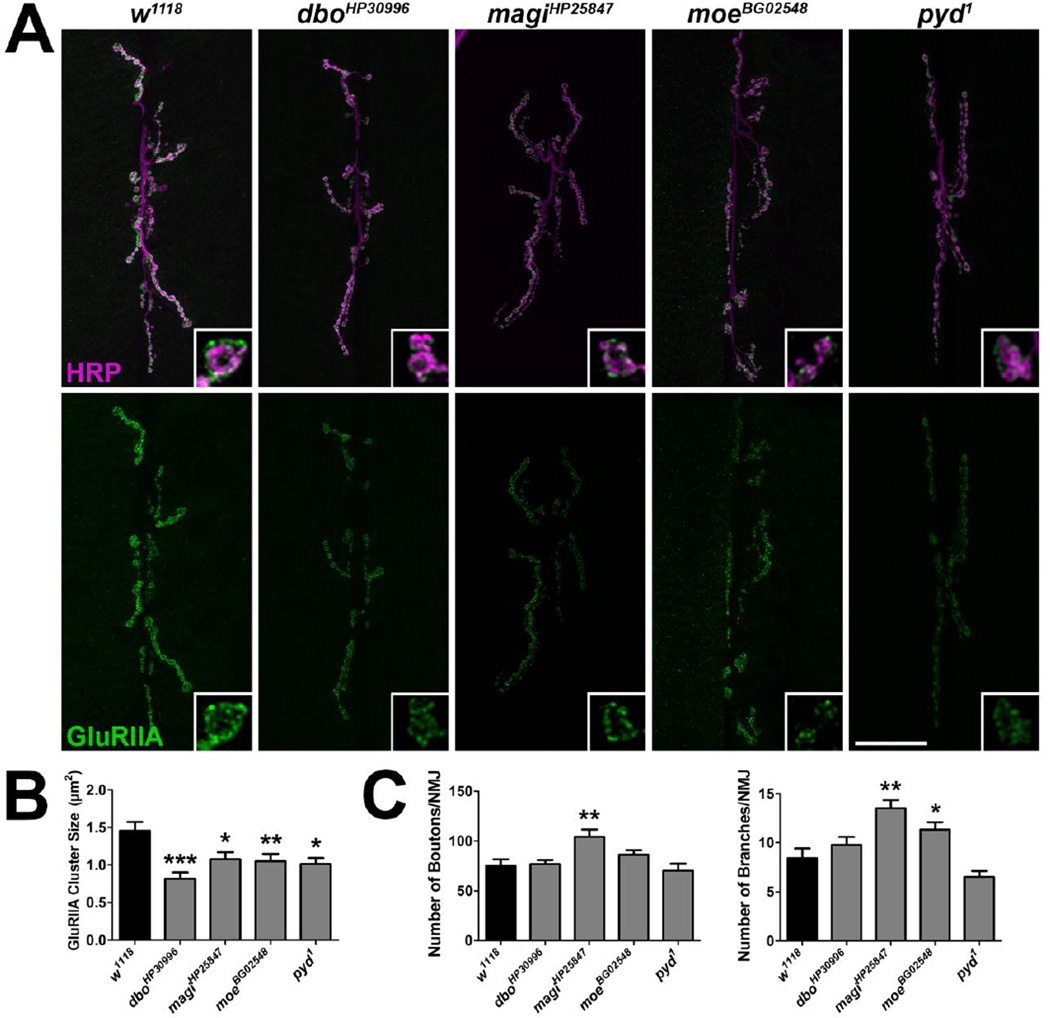
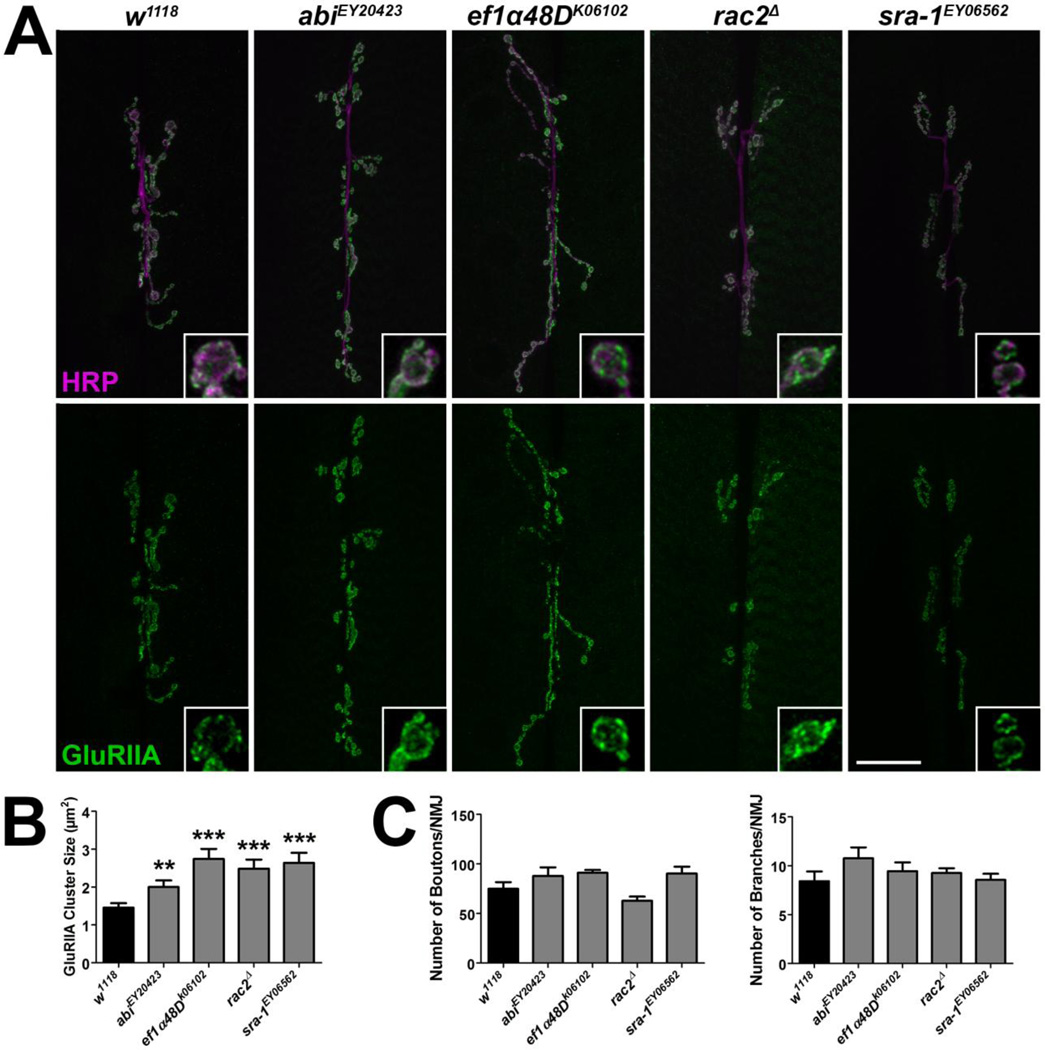
We found that mutations in genes encoding cell adhesion molecules, cytoskeletal proteins, metabolic proteins, transmembrane proteins, trafficking/motor proteins, scaffold proteins, and enzymes led to significant changes in GluRIIA synaptic fluorescence (Tables 1, S2). To further explore these synaptic phenotypes, subsets of mutants were examined for changes in GluRIIA cluster sizes. Postsynaptic GluRIIA-containing receptors localize in clusters or puncta in apposition to presynaptic active zones (Petersen et al., 1997), sites of neurotransmitter release. The size and intensity of these clusters parallels the function of the synapse (Featherstone et al., 2002). Although GluRIIA cluster sizes correlated with relative GluRIIA fluorescence intensity in the mutants identified in the screeen (Figs. 1A–B, 2A–B), there were not consistent changes observed in the morphology of the presynaptic motor neuron (Figs. 1C, 2C).
Table 1.
Classification of mutations identified in the reverse genetic screen that significantly affected synaptic GluRIIA levels
| Function of Gene Product | Percentage |
|---|---|
| Cell adhesion molecules | 8.3% |
| Cytoskeleton and related | 18.8% |
| GTPases and regulators | 25.0% |
| Kinases and phosphatases | 10.4% |
| Metabolic | 4.2% |
| Other | 8.3% |
| Receptors/channels and transmembrane proteins | 6.3% |
| Scaffold protein | 16.7% |
| Trafficking/motor proteins | 2.1% |
Figure 1. Mutations in Drosophila genes encoding homologs of mammalian PSD proteins lead to a reduction in GluRIIA cluster sizes.
(A) Confocal images of third instar larval 6/7 NMJs immunolabeled with α-HRP to label presynaptic motor neurons (magenta) and α-GluRIIA (green) to label postsynaptic glutamate receptor clusters containing the GluRIIA subunit. Inset panels show high resolution terminal boutons. Scale bar = 20 µm. (B) Histogram showing GluRIIA cluster sizes for genotypes represented in A. (C) Quantification of the number of boutons (left) and branches (right) indicative of presynaptic motor neuron morphology.
Figure 2. Mutations in Drosophila genes encoding homologs of mammalian PSD proteins lead to an increase in GluRIIA cluster sizes.
(A) Representative confocal micrographs of 6/7 NMJs immunolabeled with α-HRP to label presynaptic motor neurons (magenta) and α-GluRIIA (green) to label GluRIIA-containing glutamate receptor clusters. Inset panels show high resolution terminal boutons. Scale bar = 20 µm. (B) Quantification of GluRIIA cluster sizes for genotypes shown in A. (C) Quantification of characteristics representative of presynaptic motor neuron morphology including the number of boutons (left) and branches (right).
Mib1 positively regulates GluR clustering
One mutation that led to a reduction in synaptic GluRIIA was in mind bomb1 (mib1), which was also identified in a similar forward genetic screen in our lab. Drosophila Mib1 is 66.6% identical and 76.9% similar to human Mib1 (http://blast.ncbi.nlm.nih.gov/ using NP_678826.2 and NP_065825.1 accession numbers, respectively). Mib1 is an E3 ubiquitin ligase localized to the PSD (Choe et al., 2007) that promotes Notch signaling by regulating endocytosis of the Notch ligands Delta (Koo et al., 2005a) and Jagged/Serrate (Lai et al., 2005; Le Borgne et al., 2005; Koo et al., 2007). Although Mib1 is important for neuronal differentiation in both the central (Haddon et al., 1998; Ossipova et al., 2009; Yamamoto et al., 2010) and peripheral (Kang et al., 2013) nervous systems, we did not observe differences in the sizes of the ventral nerve cord or muscles in mib1 mutants (data not shown). Similarly, there were no significant differences in synaptic or muscle acetylated tubulin levels or the sarcomeric structure of the muscle as indicated by phalloidin labeling in mib1 mutants (data not shown). Therefore, we sought to characterize the role of Mib1 in GluR localization.
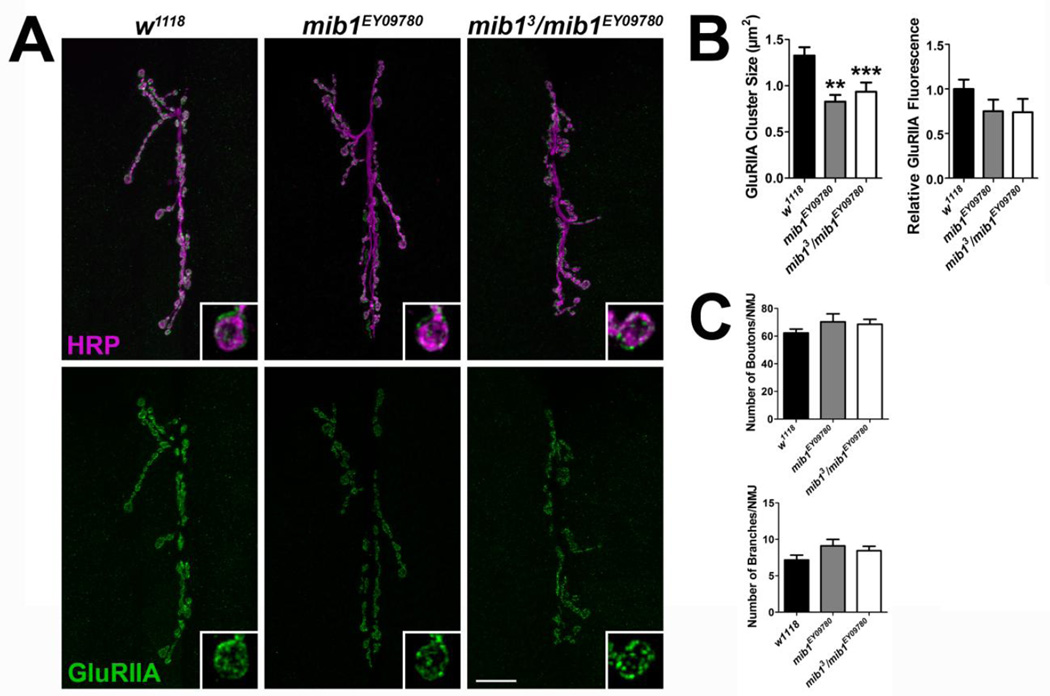
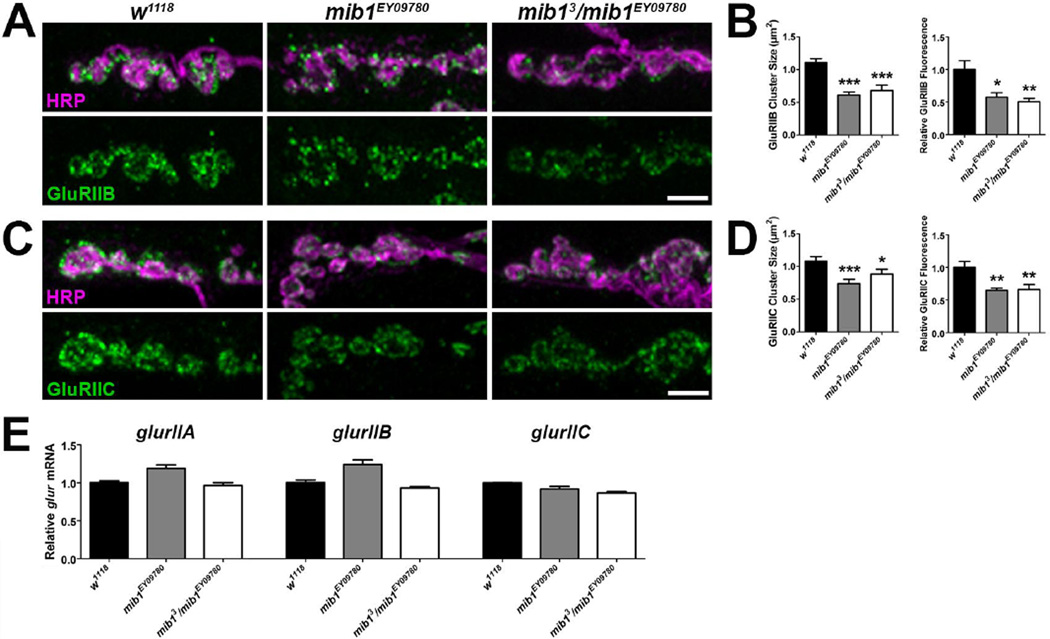
Two mutant alleles were employed to assess the synaptic role of Mib1 including mib1EY09780, which contains a transposable element in the 5’ end of the mib1 coding sequence, and mib13, which is a null mutation that introduces an early stop codon (Le Borgne et al., 2005). The latter causes early larval lethality. Therefore, mib13/ mib1EY09780 transheterozygous mutants were used in our experiments. Both mib1EY09780 and mib13/ mib1EY09780 mutants exhibited a significant reduction in GluRIIA cluster sizes compared with controls (Fig. 3A–B). The reduction in cluster sizes corresponded to a reduction in relative GluRIIA fluorescence intensity in both mutant genotypes but this was not significant. Although there were slight, consistent increases in the number of motor neuron branches and boutons, these increases were not significant (Fig. 3C). Similar to GluRIIA, there were significant reductions in GluRIIB (Fig. 4A–B) and GluRIIC (Fig. 4C–D) cluster sizes in mib1EY09780 and mib13/ mib1EY09780 mutants and this corresponded to a significant reduction in relative fluorescence for each subunit.
Figure 3. Mib1 is important for the clustering of GluRIIA-containing receptors.
(A) Control and mib1 mutant confocal images showing representative 6/7 NMJs from third instar larvae. Preparations were immunolabled with α-HRP to label presynaptic motor neurons (magenta) and α-GluRIIA (green) to label GluRIIA-containing glutamate receptor clusters. Inset panels show high resolution terminal boutons. Scale bar = 20 µm. (B) Histograms showing quantification of GluRIIA cluster sizes (left) and GluRIIA relative fluorescence intensity (right) for genotypes shown in A. (C) Quantification of characteristics representative of presynaptic motor neuron morphology including the number of boutons (top) and branches (bottom).
Figure 4. Mib1 positively regulates synaptic levels of GluRIIB and GluRIIC but does not affect glur transcript levels.
High resolution confocal micrographs showing third instar larvae terminal boutons of the 6/7 NMJ immunolabeled with α-HRP to label presynaptic motor neurons (magenta) and α-GluRIIA (green) to label GluRIIB-containing glutamate receptor clusters (A) or GluRIIC-containing glutamate receptor clusters (C). Scale bar = 5 µm. (B) Quantification of GluRIIB cluster sizes (left) and relative fluorescence intensities (right) for genotypes shown in A. (D) Quantification of GluRIIC cluster sizes (left) and relative GluRIIC fluorescence intensity (right) for genotypes shown in B. (E) Quantification qRT-PCR ΔΔC(t) values normalized to the control, w1118, using gene-specific primers for glurIIA (left), glurIIB (middle), and glurIIC (right).
Notch signaling is initiated by Notch binding to its ligand on adjacent cell surfaces. This leads to the proteolytic cleavage of Notch at two sites (van Tetering & Vooijs, 2011) and endocytosis of both the Notch intracellular domain and the ligand in the adjacent cell (Chitnis, 2006; Brou, 2009). The intracellular domain of Notch translocates into the nucleus and binds to transcription factors of the CBF1/Su(H)/Lag1 (CSL) family thereby activating transcription of hundreds of target genes (Borggrefe & Liefke, 2012). To investigate the possibility that Mib1 may influence GluR transcript levels by regulating the transcriptional activity of the Notch signaling pathway, we assessed relative mRNA levels using qRT-PCR. GlurIIA, glurIIB, and glurIIC transcript levels were not significantly altered in mib1 mutants (Fig. 4E). These data indicate that Mib1 likely regulates the localization or postsynaptic stabilization of GluRs but does not affect transcription of glur subunits.
Overexpression of mib1 increases GluR cluster sizes while deleting the mib1 ring finger domain decreases GluR cluster sizes
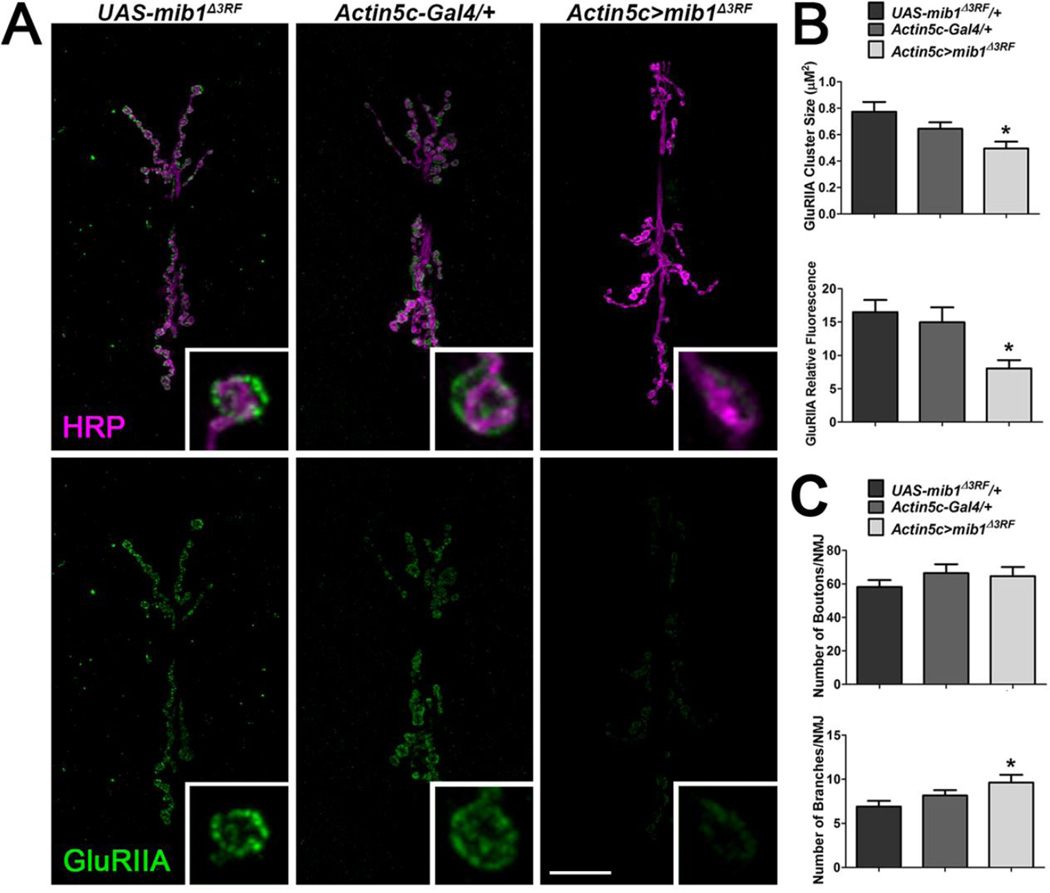
To confirm the role of Mib1 in GluR clustering, we first expressed a mib1 transgene that lacks the region encoding the C-terminal RING finger domains (UAS-mib1Δ3RF). Mib1 contains three RING finger domains (Itoh et al., 2003) that mediate ubiquitination of the Notch ligands Delta (Chen & Casey Corliss, 2004) and Serrate (Lai et al., 2005). Mib1Δ3RF interacts with Delta and Serrate but does not endocytose these ligands thereby inhibiting Notch signaling (Lai et al., 2005). Similar to mib1 mutants, expression of the UAS-mib1Δ3RF transgene in all cells using the Actin5c-Gal4 driver resulted in a significant reduction in GluRIIA cluster sizes and relative fluorescence (Fig. 5A–B) compared with outcrossed controls. Although there was a significant increase in branching of the presynaptic motor neuron in animals expressing mib1Δ3RF in all tissues compared with outcrossed controls, there was no significant change in the number of boutons (Fig. 5C).
Figure 5. Expression of Mib1 lacking the RING finger domains leads to the loss of synaptic GluRIIA.
A mib1 transgene lacking the region encoding the three C-terminal RING finger domains, UAS-mib1Δ3RF, was expressed in all tissues using the Actin5c-Gal4 driver. (A) Representative third instar larvae 6/7 NMJs immunolabeled with α-HRP to label presynaptic motor neurons (magenta) and α-GluRIIA (green) to label postsynaptic glutamate receptor clusters containing the GluRIIA subunit. Inset panels show high resolution terminal boutons. Scale bar = 20 µm. (B) Quantification of GluRIIA cluster sizes (left) and GluRIIA fluorescence intensities (right) for genotypes represented in A. (C) Characterization of presynaptic motor neuron morphology by quantification of the number of boutons (top) and branches (bottom).
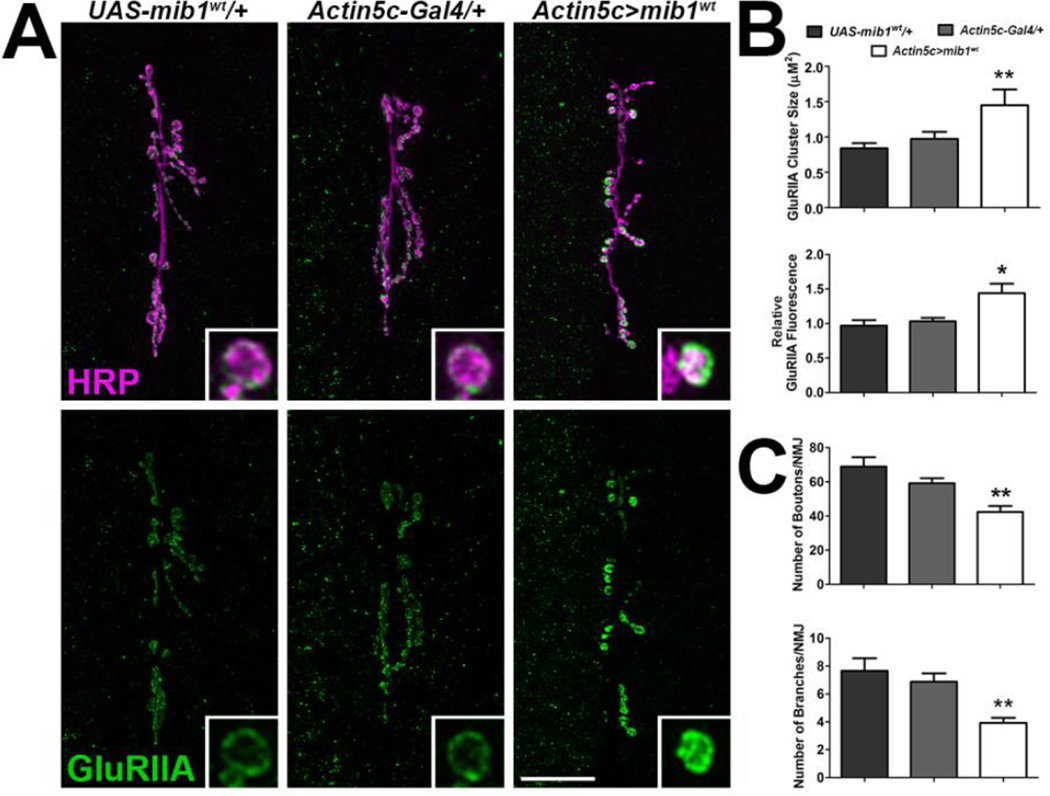
Next, we overexpressed wild type Mib1 using a transgene previously shown to enhance Notch signaling (Lai et al., 2005). There were significant increases in both GluRIIA cluster sizes and relative fluorescence in animals overexpressing mib1 in all tissues compared with outcrossed controls (Fig. 6A–B). Overexpression of mib1 also led to a reduction in the size of the presynaptic motor neuron as indicated by a significant decrease in motor neuron boutons and branches (Fig. 6C). These results collectively suggest that Notch signaling positively correlates with GluR levels at the synapse but negatively correlates with the size of the presynaptic motor neuron.
Figure 6. Overexpression of Mib1 leads to an increase in synaptic GluRIIA and a reduction in the size of the presynaptic motor neuron.
Overexpression of mib1 was achieved by expressing UAS-mib1wt in all tissues using the Actin5c-Gal4 driver. (A) Confocal micrographs showing 6/7 NMJs immunolabeled with α-HRP to label presynaptic motor neurons (magenta) and α-GluRIIA (green) to label postsynaptic GluRIIA-containing glutamate receptor clusters. Inset panels show high resolution terminal boutons. Scale bar = 20 µm. (B) Histograms showing quantification of GluRIIA cluster sizes (left) and GluRIIA fluorescence intensities (right) for genotypes represented in A. (C) Histograms showing quantification of the number of boutons (top) and branches (bottom).
Mib1 Regulates Synaptic Levels of CSP and FasII
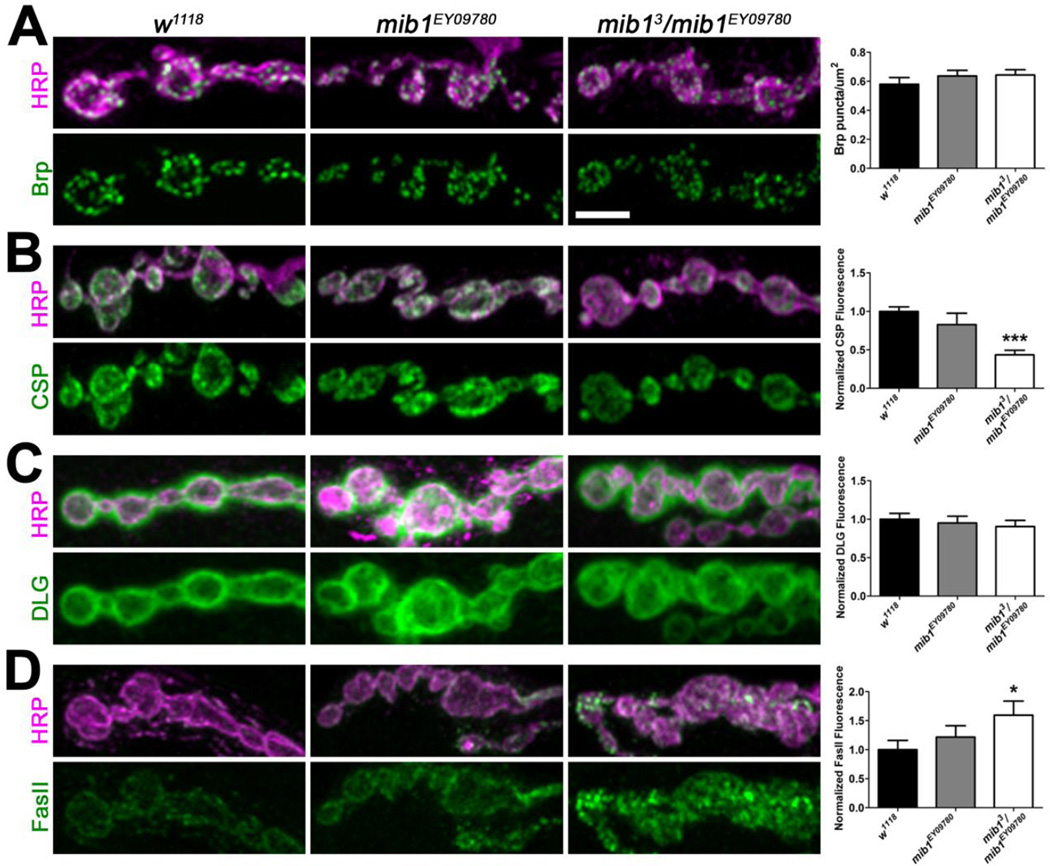
We next examined the levels of several synaptic proteins to determine if Mib1 may influence the localization of other proteins important for synaptic function. Mutations in mib1 did not affect the density of Bruchpilot (Brp; Fig. 7A), which is localized to presynaptic active zones where it helps to organize Ca2+ channels (Kittel et al., 2006; Wagh et al., 2006) and synaptic vesicles (Matkovic et al., 2013). Similarly, the loss of Mib1 did not affect synaptic levels of the scaffold protein discs large (DLG, Fig. 7C), which acts as an adaptor protein required for GluRIIB clustering in embryos (Chen & Featherstone, 2005). Both cysteine string protein (CSP) and Fasciclin II (FasII) levels, however, were significantly affected in mib13/ mib1EY09780 mutants. CSP, a vesicle-associated protein important for evoked neurotransmitter release (Bronk et al., 2005) and presynaptic protein folding (Donnelier & Braun, 2014), was significantly reduced at mib13/ mib1EY09780 mutant NMJs (Fig. 7B). Although there was a slight reduction in CSP levels in mib1EY09780 mutants, it was not significant. Conversely, mib13/ mib1EY09780 but not mib1EY09780 mutants exhibited a significant increase in synaptic levels of the homophilic cell adhesion molecule FasII (Fig. 7D), which regulates synaptic growth (Schuster et al., 1996) and postsynaptic organization during synaptogenesis (Kohsaka et al., 2007). Collectively, these data indicate that, in addition to GluRs, Mib1 is important for the synaptic localization of CSP and FasII.
Figure 7. Mib1 is important for the localization of CSP and FasII at the synapse.
High resolution confocal micrographs showing terminal boutons of 6/7 NMJs from third instar larvae immunolabeled with α-HRP (magenta) and α-Brp (green, A), α-CSP (green, B), α-DLG (green, C), or α-FasII (green, D). Scale bar = 5 µm. Right histograms show quantification of mean normalized fluorescence intensities.
Mib1 regulates the frequency of spontaneous neurotransmission
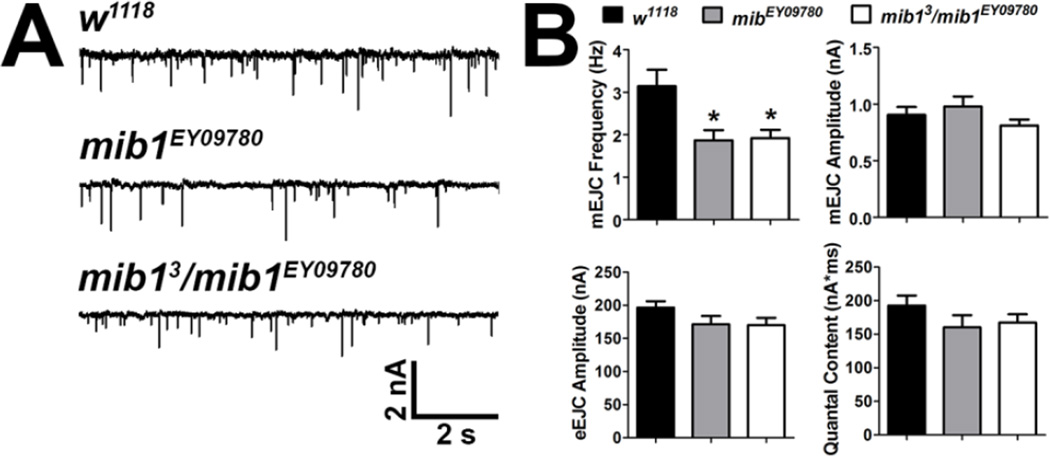
The change in the synaptic localization of GluRs, CSP, and FasII could affect the function of the synapse. Therefore, we assessed the function of mib1 mutant NMJs using two-electrode voltage clamp electrophysiology. Both mib1EY09780 and mib13/ mib1EY09780 mutants exhibited a significant decrease in the frequency of miniature endplate junctional currents (mEJCs) but not in mEJC amplitudes compared with controls (Fig. 8). Although mib1 mutants also exhibited decreased evoked endplate junctional current (eEJC) amplitudes and quantal content, these reductions were not significant. Based on these data, we conclude that the function of mib1 mutant NMJs is affected likely as a result of altered localization of synaptic proteins.
Figure 8. Mib1 negatively regulates spontaneous synaptic transmission.
Spontaneous (mEJCs) and evoked junctional currents (eEJCs) were recorded from third instar larvae after voltage clamp of muscle 6 at −60 mV. (A) Representative mEJCs from control and mib1 mutants. (B) Quantification of mEJC frequency and amplitudes (top) and eEJC amplitudes and quantal content (bottom).
Discussion
The functions of many PSD proteins are poorly characterized. To better understand the relationship between PSD components and the localization of postsynaptic GluRs, we conducted a reverse genetic screen to identify mutations that affected the synaptic localization of GluRs. Drosophila orthologs were previously identified for 95.8% of genes that encode mammalian PSD proteins (Liebl & Featherstone, 2008) and mutations in 130 of these orthologs were examined here. We next focused on one ortholog, mib1, to better understand how it influences the structure and function of synapses.
GluRs are shuttled in and out of the synaptic membrane as a result of changes in synaptic activity ultimately altering the strength of the synapse (Chater & Goda, 2014; Shipton & Paulsen, 2014; Sihra et al., 2014). The synaptic localization of GluRs is directly mediated by components of the PSD including transmembrane proteins associated with GluRs and scaffolding proteins (Jackson & Nicoll, 2011; Verpelli et al., 2012). Other components of the PSD including the actin cytoskeleton and many enzymes that regulate protein interactions influence the localization of GluRs without directly binding to them (Okabe, 2007). 42.8% of the mutations we examined showed significant changes in synaptic GluRIIA localization (Table S2). Given the role of the PSD in GluR localization, we might expect that percentage to be higher. It is important to note, however, that we did not examine GluRIIB localization in these same mutants. The GluRIIA and GluRIIB subunits are mutually exclusive in the GluR tetramer (Marrus et al., 2004; Featherstone et al., 2005; Qin et al., 2005), differentially localized to the synapse (Marrus et al., 2004; Schmid et al., 2008), and are stabilized by unique PSD proteins (Chen & Featherstone, 2005; Chen et al., 2005). Thus, some mutations could affect the localization of GluRIIB without also affecting GluRIIA. In addition, we used P-element mutants in our screen. Because these transposon insertions typically result in hypomorphic mutations (Spradling et al., 1995; LaFave & Sekelsky, 2011), we may have missed phenotypes that would have resulted from the use of null alleles.
Most mutations that affected GluR localization resulted in the loss of synaptic GluRIIA and this is consistent with previous screens (Liebl & Featherstone, 2005). These data suggest that most regulatory proteins promote the trafficking and localization of GluRIIA-containing receptors. Indeed, the PSD proteins KRIP6 and S-SCAM have been shown to be important for the membrane localization of the Kainate receptor subunit GluR6 and the AMPA receptor subunit GluA2 (Laezza et al., 2008; Danielson et al., 2012). Mutations in the Drosophila orthologs diablo (dbo) and magi produce a significant reduction in the synaptic localization of GluRIIA (Fig. 1). The loss of GluRIIA in moesin (moe) mutants may be due to the proposed role of Moe in binding and stabilizing the actin cytoskeleton. Moe and radixin are important for the actin-dependent processes of growth cone extension in rat cultured neurons (Paglini et al., 1998) and Moe is localized specifically to polymerized actin at the Drosophila NMJ (Khuong et al., 2010).
Half of the mutations that produced an increase in synaptic GluRIIA were in genes encoding proteins and GTPases important for remodeling of the actin cytoskeleton. Sra-1 interacts with Abelson interacting protein (Abi-1) (Steffen et al., 2004), which activates WAVE2 to promote actin nucleation (Leng et al., 2005) in non-neuronal cells. Similarly, the Rho family GTPase, Rac2, enhances actin nucleation by activating cofilin (Sun et al., 2007). Remodeling of the actin cytoskeleton is essential for structural changes to the dendritic spine (Fortin et al., 2012) and the stabilization of newly incorporated AMPA receptors (Rudy, 2014). Mutations in abi, rac2, and sra-1 may inhibit actin dynamics such that GluRs are retained in the synapse as we observed (Fig. 2). In support of this, several proteins required for clathrin-dependent endocytosis interact with WASP, WAVE, and Cdc42, proteins that enhance nucleation of the actin cytoskeleton (Saheki & De Camilli, 2012). AMPA receptors are endocytosed during long-term depression (LTD) by clathrin-dependent endocytosis (Anggono & Huganir, 2012; Hanley, 2014).
The Notch signaling protein, Mib1, was one of 42 mutations that resulted in a significant decrease in synaptic levels of GluRIIA. The importance of Notch signaling in cell fate determination is well established in many cell types including neurons (Louvi & Artavanis-Tsakonas, 2006). More recent studies have identified roles for Notch in cell division, axon guidance and synaptogenesis (Giniger, 2012). We sought to better characterize the role of Mib1, which promotes Notch signaling by regulating the endocytosis of the Notch ligands Delta (Koo et al., 2005a) and Jagged/Serrate (Lai et al., 2005; Le Borgne et al., 2005; Koo et al., 2007), in terminally differentiated neurons. We found that, in addition to GluRIIA, Mib1 positively regulates the localization of the GluR subunits GluRIIB and GluIIC. Our data indicate that attenuation of Notch signaling by expressing a ligase-deficient Mib1 (Mib1Δ3RF; (Lai et al., 2005)) in all tissues (Fig. 5) or as a result of mutations in mib1 (Figs. 3–4) produces a loss of synaptic GluRs. Further, mutations in polychaetoid (pyd), which promotes Notch signaling in sensory organ precursors (Chen et al., 1996), significantly reduced synaptic GluRIIA levels (Fig. 1). Conversely, enhanced Notch signaling as a result of overexpressing Mib1 in all tissues (Lai et al., 2005) led to increased synaptic levels of GluRIIA (Fig. 6).
One potentially confounding variable exists in interpreting our overexpression data. Ubiquitous expression of Mib1 could lead to misexpression phenotypes as Mib1 may only be localized to one cell type at the NMJ. Mib1 is expressed in cells containing Notch ligands, which are localized adjacent to cells that express the Notch receptors (Itoh et al., 2003). Notch receptors were previously detected in presynaptic motor neuron cell bodies at the Drosophila NMJ (de Bivort et al., 2009). Expression of Mib1Δ3RF in all tissues, however, would only affect the phenotype of Mib1-expressing cells because the RING finger domains are required Mib1 ubiquitin ligase activity (Lai et al., 2005). The similarity of the phenotypes in mutants deficient in Notch signaling strongly suggests that Notch signaling and Mib1 regulate the localization of GluRs at the NMJ.
This is the first report to show that Notch signaling alters the synaptic levels of glutamate receptors. Although conditional knockout of mib1 impaired memory for hippocampal-dependent tasks and attenuated late LTP and LTD, it did not alter synaptic levels of several GluR subunits including GluA1, GluA2/3, GluN1, GluN2A, or GluN2B (Yoon et al., 2012). Conversely, mib1 mutant zebrafish showed significant reductions in GluR subunit mRNAs for AMPA 2a and AMPA 2b and the glutamate metabolizing gene product, glutamate decarboxylase, as indicated by microarray analyses (Hortopan et al., 2010). Our data suggest that Mib1 likely regulates GluR subunits posttranscriptionally as we did not detect appreciable differences in glur mRNA levels in mib1 mutants (Fig. 4E).
Mib1 may directly regulate the localization of GluRs. AMPA receptor subunits localized to the cell membrane are ubiquitinated after enhanced synaptic activity (Widagdo et al., 2015). The Mib1 paralog, Mib2 (Koo et al., 2005b), directly binds and ubiquitinates the GluN2B but not the GluN1 subunit of the NMDA receptor in an activity-dependent manner. This ultimately decreased NMDA-mediated synaptic currents (Jurd et al., 2008). Our data, however, suggest that Mib1 indirectly regulates the localization of GluR subunits. If Mib1 functioned similar to Mib2 to directly regulate the localization of non-NMDA receptors, we would expect to see an increase in GluR subunits in mib1 mutants and after ubiquitous expression of Mib1Δ3RF. Instead we observe a reduction in synaptic levels of GluRIIA (Fig. 3), GluRIIB, and GluRIIC (Fig. 4) in mib1 mutants. Therefore, we favor the hypothesis that Mib1 attenuates the presynaptic release of glutamate, which, over developmental time, leads to a reduction in synaptic GluR subunit levels.
In support of this hypothesis, we observe a significant reduction in CSP at mib1 mutant NMJs (Fig. 7B). Mouse CSP-α knock out NMJs exhibit deficient presynaptic vesicle endocytosis and a reduction in the size of the readily releasable pool followed by reduced synaptic vesicle exocytosis (Rozas et al., 2012). Similarly, we observe a significant reduction in mEJC frequency in mib1 mutants (Fig. 8C) indicating a reduction in presynaptic glutamate release. Although it may seem counterintuitive that loss of a PSD protein could result in altered levels of CSP, which is primarly localized presynaptically (Kohan et al., 1995), and neurotransmitter release, Mib1 activates Notch signaling in neighboring cells (Koo et al., 2005a; Lai et al., 2005; Le Borgne et al., 2005; Koo et al., 2007). Thus, Mib1 localized to the PSD would activate Notch signaling in the adjacent presynaptic cell. Altered presynaptic Notch signaling resulting from loss of Mib1 activity, could then affect the expression of Notch target genes thereby affecting cellular function.
In summary, we have found that mutations in several genes that encode orthologs of mammalian PSD proteins are important for the proper localization of GluRs at the Drosophila NMJ. The PSD protein, Mib1, positively regulates the synaptic localization of GluRIIA, GluRIIB, and GluRIIC. The localization of GluRs may be secondary to Mib1’s role in localizing presynaptic CSP and regulating the spontaneous release of neurotransmitter.
Experimental Methods
Fly Stocks
Fly stocks were raised at 25°C on Jazz Mix food (Fisher Scientific, St. Louis, MO). Drosophila orthologs and corresponding mammalian PSD proteins were previously identified (Liebl & Featherstone, 2008). Mutant stocks for the reverse genetic screen were identified using FlyBase (http://flybase.org/) and obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/). Identities of the stocks can be found in Table S2. Most of the lines used (65.9%) were homozygous adult viable. However, the remaining lines (34.1%) were homozygous adult lethal and balanced using chromosome-specific GFP-balancers or the TM6 Tb balancer to enable identification of homozygous mutants. Both mib13 and mib1EY09780 alleles and all Gal4 drivers were obtained from the Bloomington Drosophila Stock Center. Stocks containing the UAS-mib1Δ3RF and UAS-mib1wt transgenes were generous gifts from Eric Lai (Lai et al., 2005). Control stocks included w1118 and w1118 outcrossed to Actin5c-Gal4, 24B-Gal4, elav-Gal4, UAS-mib1Δ3RF, and UAS-mib1wt.
Immunocytochemistry and Confocal Microscopy
Third instar larvae were filet dissected on Sylgard-containing petri dishes at room temperature in Roger’s Ringer solution (135 mM NaCl, 5 mM KCl, 4 mM MgCl2, 1.8 mM CaCl2, 5 mM TES, 72 mM sucrose) supplemented with 2 mM glutamate (Augustin et al., 2007). Larval preparations were fixed for 30 min with either Bouin’s fixative (Fisher Scientific, St. Louis, MO) for Brp or GluR antibodies or 4% paraformaldehyde in PBS for all other antibodies. Primary antibodies were diluted in PBTX (PBS + 0.1% Triton and 1% Bovine Serum Albumin) and applied overnight at 4°C after larvae were washed PTX (PBS + 1% Triton). Mouse α-Brp (aka nc82, 1:50), mouse α-CSP (1:200), mouse α-DLG (1:1000), mouse α-FasII (1:5), and mouse α-GluRIIA (1:100) were obtained from the Iowa Developmental Studies Hybridoma Bank (Iowa City, IA). Rabbit α-GluRIIB (1:2000) and rabbit α-GluRIIC (1:5000) were generous gifts from Aaron DiAntonio (Marrus et al., 2004). Mouse α-acetylated tubulin and phalloidin (1:200) were obtained from Sigma Aldrich (Cat #, St. Louis, MO) and Invitrogen (Cat #, Carlsbad, CA), respectively. Additional antibodies including HRP (1:125) and species-specific FITC (1:250) were obtained from Jackson Immunolabs (West Grove, PA), diluted in PBTX, and applied for 2 h at room temperature. Larval preparations were mounted on slides and covered with Vectashield (Vector Labs, Burlingame, CA).
Larval A3 or A4 6/7 NMJ were imaged using a Fluoview 1000 Olympus confocal laser scanning microscope. Imaging parameters were set for controls and subsequently used for all experimental animals. Equal numbers of control and experimental animals were imaged each day. Compressed images of the z-series were used for data analyses.
Electrophysiology
Third instar larvae were filet dissected and secured using VetBond glue (World Precision Instruments, Sarasota, FL) at room temperature in Roger’s Ringer on Sylgard-coated coverslips. Two-electrode voltage clamp recordings were obtained from muscle 6 of A3 or A4 after clamping the muscle membrane potential at −60 mV using an Axoclamp 900A amplifier (Molecular Devices, Sunnyvale, CA). Both the clamp and recording electrodes were filled with 3 M KCl and used if their resistances were 10–20 MΩ. The stimulating electrode was filled with bath saline. For evoked recordings, a 1 Hz, 10V stimulus was delivered by a Grass S88 stimulator with a SIU5 isolation unit (Grass Technologies, Warwick, RI) to recruit both motor neuron axons as previously described (Ehmann et al., 2014). Recordings were digitized with a Digidata 1443 digitizer (Molecular Devices, Sunnyvale, CA). PClamp software (v. 10.4) was used for data analyses. Quantal content was calculated by dividing the eEJC area (nA*ms) by the mEJC area (nA*ms).
qRT-PCR
RNA was extracted with Trizol (Invitrogen, Carlsbad, CA) from 8–12 third instar larvae as previously described (Jowett, 1998). qRT-PCR was performed in single-plex reactions using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad, Hercules, CA), gene-specific primers for gluRIIA, gluRIIB, gluRIIC, and GAPDH, and the Stratagene Mx3000P qPCR System (Agilent Technologies, Santa Clara, CA). 100 ng of total RNA was added to each reaction. Three technical replicates and two biological replicates were performed for each reaction. Relative gluR mRNA levels were obtained by subtracting the GAPDH C(t) value from the GluR C(t) value.
Data Analysis and Statistics
The number of boutons and branches were obtained by manually counting these features using 6/7 NMJs of hemisegments A3 or A4. Branches were defined as an extension of the presynaptic motor neuron with more than one bouton. The density of Brp labeling was quantified by counting the total number of Brp puncta and dividing by the total NMJ area as indicated by HRP labeling using ImageJ (NIH) software. GluR cluster sizes were measured by manually tracing around the GluR puncta overlapping and immediately adjacent to HRP immunloabeling and measuring the area with Image J software as previously described (Featherstone et al., 2002). For all other immunolabeling, immunoreactivity was quantified by measuring the mean fluorescence intensity of the NMJ using Adobe Photoshop software (v. CS2) and subtracting the mean non-NMJ background over an identical area of the neighboring muscle membrane. For DLG and muscle acetylated tubulin, the average background from a non-synaptic, non-muscle area was used. 8–12 animals per genotype were used for analyses for the reverse genetic screen.
Data analyses were conducted with GraphPad Prism (v5.01). Student’s t-tests were used for experiments with a single control. For experiments with more than one control, an ANOVA was performed with a Tukey post hoc test. In figures, p<0.0001 is designated by ***, p<0.001 is designated by **, and p<0.05 is designated by *. Error bars are representative of standard error of the mean values. Summary statistics for all data are reported in Table S1.
Supplementary Material
Highlights.
Identified Drosophila PSD homologs important for glutamate receptor localization.
Further characterized the Notch signaling protein, Mib1.
Mib1 positively regulates glutamate receptor localization at the Drosophila NMJ.
Mib1 may secondarily affect glutamate receptors by influencing glutamate release.
Acknowledgements
We would like to thank Dave Featherstone for his helpful comments on the manuscript. We are grateful for antibodies provided by Aaron DiAntonio and the Iowa Developmental Studies Hybridoma Bank, fly stocks provided by Eric Lai and the Bloomington Drosophila Stock Center, and for the assistance of multiple members of the Liebl Lab. This work was supported by Southern Illinois Edwardsville’s Undergraduate Research and Creative Activities program (to MS) and Research Grants for Graduate Students (to DD) and the National Institutes of Health (NIH R15: NINDS, 1R15NS063315-01 and 3R15NS063315-01S1 to FL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alie A, Manuel M. The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol Biol. 2010;10:34. doi: 10.1186/1471-2148-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Current opinion in neurobiology. 2012;22:461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin H, Grosjean Y, Chen K, Sheng Q, Featherstone DE. Nonvesicular release of glutamate by glial xCT transporters suppresses glutamate receptor clustering in vivo. J Neurosci. 2007;27:111–123. doi: 10.1523/JNEUROSCI.4770-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangash MA, Park JM, Melnikova T, Wang D, Jeon SK, Lee D, Syeda S, Kim J, Kouser M, Schwartz J, Cui Y, Zhao X, Speed HE, Kee SE, Tu JC, Hu JH, Petralia RS, Linden DJ, Powell CM, Savonenko A, Xiao B, Worley PF. Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism. Cell. 2011;145:758–772. doi: 10.1016/j.cell.2011.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bayes A, van de Lagemaat LN, Collins MO, Croning MD, Whittle IR, Choudhary JS, Grant SG. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Liefke R. Fine-tuning of the intracellular canonical Notch signaling pathway. Cell cycle. 2012;11:264–276. doi: 10.4161/cc.11.2.18995. [DOI] [PubMed] [Google Scholar]
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82:444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR. Local protein synthesis, actin dynamics, and LTP consolidation. Current opinion in neurobiology. 2008;18:524–531. doi: 10.1016/j.conb.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Bronk P, Nie Z, Klose MK, Dawson-Scully K, Zhang J, Robertson RM, Atwood HL, Zinsmaier KE. The multiple functions of cysteine-string protein analyzed at Drosophila nerve terminals. J Neurosci. 2005;25:2204–2214. doi: 10.1523/JNEUROSCI.3610-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C. Intracellular trafficking of Notch receptors and ligands. Experimental cell research. 2009;315:1549–1555. doi: 10.1016/j.yexcr.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Chater TE, Goda Y. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Frontiers in cellular neuroscience. 2014;8:401. doi: 10.3389/fncel.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Freedman JA, Bettler DR, Jr, Manning SD, Giep SN, Steiner J, Ellis HM. Polychaetoid is required to restrict segregation of sensory organ precursors from proneural clusters in Drosophila. Mechanisms of development. 1996;57:215–227. doi: 10.1016/0925-4773(96)00548-5. [DOI] [PubMed] [Google Scholar]
- Chen K, Featherstone DE. Discs-large (DLG) is clustered by presynaptic innervation and regulates postsynaptic glutamate receptor subunit composition in Drosophila. BMC biology. 2005;3:1. doi: 10.1186/1741-7007-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Merino C, Sigrist SJ, Featherstone DE. The 4.1 protein coracle mediates subunit-selective anchoring of Drosophila glutamate receptors to the postsynaptic actin cytoskeleton. J Neurosci. 2005;25:6667–6675. doi: 10.1523/JNEUROSCI.1527-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–5372. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Casey Corliss D. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Developmental biology. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Chen X, Winters C, Azzam R, Li X, Galbraith JA, Leapman RD, Reese TS. Organization of the core structure of the postsynaptic density. Proc Natl Acad Sci U S A. 2008;105:4453–4458. doi: 10.1073/pnas.0800897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MC, Lu CL, Luu SU, Tsai HM, Hsu SH, Chen TT, Chen CH. Genetic and functional analysis of the DLG4 gene encoding the post-synaptic density protein 95 in schizophrenia. PLoS One. 2010;5:e15107. doi: 10.1371/journal.pone.0015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A. Why is delta endocytosis required for effective activation of notch? Developmental dynamics : an official publication of the American Association of Anatomists. 2006;235:886–894. doi: 10.1002/dvdy.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, Jin P, Tsai LH, Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- Danielson E, Metallo J, Lee SH. Role of TARP interaction in S-SCAM-mediated regulation of AMPA receptors. Channels. 2012;6:393–397. doi: 10.4161/chan.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bivort BL, Guo HF, Zhong Y. Notch signaling is required for activity-dependent synaptic plasticity at the Drosophila neuromuscular junction. Journal of neurogenetics. 2009;23:395–404. doi: 10.3109/01677060902878481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Petersen SA, Heckmann M, Goodman CS. Glutamate receptor expression regulates quantal size and quantal content at the Drosophila neuromuscular junction. J Neurosci. 1999;19:3023–3032. doi: 10.1523/JNEUROSCI.19-08-03023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelier J, Braun JE. CSPalpha-chaperoning presynaptic proteins. Frontiers in cellular neuroscience. 2014;8:116. doi: 10.3389/fncel.2014.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosemeci A, Tao-Cheng JH, Vinade L, Jaffe H. Preparation of postsynaptic density fraction from hippocampal slices and proteomic analysis. Biochem Biophys Res Commun. 2006;339:687–694. doi: 10.1016/j.bbrc.2005.11.069. [DOI] [PubMed] [Google Scholar]
- Ehmann N, van de Linde S, Alon A, Ljaschenko D, Keung XZ, Holm T, Rings A, DiAntonio A, Hallermann S, Ashery U, Heckmann M, Sauer M, Kittel RJ. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nature communications. 2014;5:4650. doi: 10.1038/ncomms5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, Grant SG. The human postsynaptic density shares conserved elements with proteomes of unicellular eukaryotes and prokaryotes. Front Neurosci. 2011;5:44. doi: 10.3389/fnins.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, Pocklington AJ, Anderson CN, Bayes A, Collins MO, Vickers CA, Croning MD, Malik BR, Choudhary JS, Armstrong JD, Grant SG. Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat Neurosci. 2008;11:799–806. doi: 10.1038/nn.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Rushton E, Broadie K. Developmental regulation of glutamate receptor field size by nonvesicular glutamate release. Nat Neurosci. 2002;5:141–146. doi: 10.1038/nn789. [DOI] [PubMed] [Google Scholar]
- Featherstone DE, Rushton E, Rohrbough J, Liebl F, Karr J, Sheng Q, Rodesch CK, Broadie K. An essential Drosophila glutamate receptor subunit that functions in both central neuropil and neuromuscular junction. J Neurosci. 2005;25:3199–3208. doi: 10.1523/JNEUROSCI.4201-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyder M, Karlsson RM, Mathur P, Lyman M, Bock R, Momenan R, Munasinghe J, Scattoni ML, Ihne J, Camp M, Graybeal C, Strathdee D, Begg A, Alvarez VA, Kirsch P, Rietschel M, Cichon S, Walter H, Meyer-Lindenberg A, Grant SG, Holmes A. Association of mouse Dlg4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorders and Williams' syndrome. Am J Psychiatry. 2010;167:1508–1517. doi: 10.1176/appi.ajp.2010.10040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin DA, Srivastava T, Soderling TR. Structural modulation of dendritic spines during synaptic plasticity. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2012;18:326–341. doi: 10.1177/1073858411407206. [DOI] [PubMed] [Google Scholar]
- Franks KM, Stevens CF, Sejnowski TJ. Independent sources of quantal variability at single glutamatergic synapses. J Neurosci. 2003;23:3186–3195. doi: 10.1523/JNEUROSCI.23-08-03186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E. Notch signaling and neural connectivity. Current opinion in genetics & development. 2012;22:339–346. doi: 10.1016/j.gde.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girault JA. Signaling in striatal neurons: the phosphoproteins of reward, addiction, and dyskinesia. Progress in molecular biology and translational science. 2012;106:33–62. doi: 10.1016/B978-0-12-396456-4.00006-7. [DOI] [PubMed] [Google Scholar]
- Gong Y, Lippa CF. Review: disruption of the postsynaptic density in Alzheimer's disease and other neurodegenerative dementias. Am J Alzheimers Dis Other Demen. 2010;25:547–555. doi: 10.1177/1533317510382893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Lee CW, Fan Y, Komlos D, Tang X, Sun C, Yu K, Hartzell HC, Chen G, Bamburg JR, Zheng JQ. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat Neurosci. 2010;13:1208–1215. doi: 10.1038/nn.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–4644. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- Hanley JG. Actin-dependent mechanisms in AMPA receptor trafficking. Frontiers in cellular neuroscience. 2014;8:381. doi: 10.3389/fncel.2014.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Tankou S, Takeda M, Sawa A. Postsynaptic density: a key convergent site for schizophrenia susceptibility factors and possible target for drug development. Drugs Today (Barc) 2007;43:645–654. doi: 10.1358/dot.2007.43.9.1088821. [DOI] [PubMed] [Google Scholar]
- Hortopan GA, Dinday MT, Baraban SC. Spontaneous seizures and altered gene expression in GABA signaling pathways in a mind bomb mutant zebrafish. J Neurosci. 2010;30:13718–13728. doi: 10.1523/JNEUROSCI.1887-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Real E, Takamiya K, Kang MG, Ledoux J, Huganir RL, Malinow R. Emotion enhances learning via norepinephrine regulation of AMPA-receptor trafficking. Cell. 2007;131:160–173. doi: 10.1016/j.cell.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: the last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Developmental cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. L-glutamate as an excitatory transmitter at the Drosophila larval neuromuscular junction. The Journal of physiology. 1976;262:215–236. doi: 10.1113/jphysiol.1976.sp011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Halpern ME, Johansen KM, Keshishian H. Stereotypic morphology of glutamatergic synapses on identified muscle cells of Drosophila larvae. J Neurosci. 1989;9:710–725. doi: 10.1523/JNEUROSCI.09-02-00710.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- Jowett T. Drosophila: A Practical Approach. Oxford: Oxford University Press; 1998. [Google Scholar]
- Jurd R, Thornton C, Wang J, Luong K, Phamluong K, Kharazia V, Gibb SL, Ron D. Mind bomb-2 is an E3 ligase that ubiquitinates the N-methyl-D-aspartate receptor NR2B subunit in a phosphorylation-dependent manner. J Biol Chem. 2008;283:301–310. doi: 10.1074/jbc.M705580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Lee D, Hong S, Park SG, Song MR. The E3 ligase Mind bomb-1 (Mib1) modulates Delta-Notch signaling to control neurogenesis and gliogenesis in the developing spinal cord. J Biol Chem. 2013;288:2580–2592. doi: 10.1074/jbc.M112.398263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard JT, Guevremont D, Mason-Parker SE, Abraham WC, Williams JM. Redistribution of ionotropic glutamate receptors detected by laser microdissection of the rat dentate gyrus 48 h following LTP induction in vivo. PLoS One. 2014;9:e92972. doi: 10.1371/journal.pone.0092972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JM, Blanpied TA. Subsynaptic AMPA receptor distribution is acutely regulated by actin-driven reorganization of the postsynaptic density. J Neurosci. 2012;32:658–673. doi: 10.1523/JNEUROSCI.2927-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuong TM, Habets RL, Slabbaert JR, Verstreken P. WASP is activated by phosphatidylinositol-4,5-bisphosphate to restrict synapse growth in a pathway parallel to bone morphogenetic protein signaling. Proc Natl Acad Sci U S A. 2010;107:17379–17384. doi: 10.1073/pnas.1001794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science (New York, N.Y. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Kohan SA, Pescatori M, Brecha NC, Mastrogiacomo A, Umbach JA, Gundersen CB. Cysteine string protein immunoreactivity in the nervous system and adrenal gland of rat. J Neurosci. 1995;15:6230–6238. doi: 10.1523/JNEUROSCI.15-09-06230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka H, Takasu E, Nose A. In vivo induction of postsynaptic molecular assembly by the cell adhesion molecule Fasciclin2. J Cell Biol. 2007;179:1289–1300. doi: 10.1083/jcb.200705154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, Lee J, Chitnis AB, Kim CH, Kong YY. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005a;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon KJ, Yoo KW, Lim HS, Song R, So JH, Kim CH, Kong YY. Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem. 2005b;280:22335–22342. doi: 10.1074/jbc.M501631200. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon MJ, Yoon KJ, Im SK, Kim YY, Kim CH, Suh PG, Jan YN, Kong YY. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PLoS One. 2007;2:e1221. doi: 10.1371/journal.pone.0001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laezza F, Wilding TJ, Sequeira S, Craig AM, Huettner JE. The BTB/kelch protein, KRIP6, modulates the interaction of PICK1 with GluR6 kainate receptors. Neuropharmacology. 2008;55:1131–1139. doi: 10.1016/j.neuropharm.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave MC, Sekelsky J. Transcription initiation from within P elements generates hypomorphic mutations in Drosophila melanogaster. Genetics. 2011;188:749–752. doi: 10.1534/genetics.111.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS biology. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci U S A. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl FL, Featherstone DE. Genes involved in Drosophila glutamate receptor expression and localization. BMC neuroscience. 2005;6:44. doi: 10.1186/1471-2202-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl FL, Featherstone DE. Identification and investigation of Drosophila postsynaptic density homologs. Bioinformatics and biology insights. 2008;2:369–381. doi: 10.4137/bbi.s2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman JE, Raghavachari S, Tsien RW. The sequence of events that underlie quantal transmission at central glutamatergic synapses. Nature reviews. Neuroscience. 2007;8:597–609. doi: 10.1038/nrn2191. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nature reviews. Neuroscience. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78:615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J Neurosci. 2004;24:1406–1415. doi: 10.1523/JNEUROSCI.1575-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovic T, Siebert M, Knoche E, Depner H, Mertel S, Owald D, Schmidt M, Thomas U, Sickmann A, Kamin D, Hell SW, Burger J, Hollmann C, Mielke T, Wichmann C, Sigrist SJ. The Bruchpilot cytomatrix determines the size of the readily releasable pool of synaptic vesicles. J Cell Biol. 2013;202:667–683. doi: 10.1083/jcb.201301072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Reijmers L, Mayford M. Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science (New York, N.Y. 2008;319:1104–1107. doi: 10.1126/science.1149967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Carrillo RA, Zinn K. Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip Rev Dev Biol. 2013;2:647–670. doi: 10.1002/wdev.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Bonhoeffer T, Scheuss V. Balance and stability of synaptic structures during synaptic plasticity. Neuron. 2014;82:430–443. doi: 10.1016/j.neuron.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Moron JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R, Devi LA. Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol Cell Proteomics. 2007;6:29–42. doi: 10.1074/mcp.M600184-MCP200. [DOI] [PubMed] [Google Scholar]
- Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nature reviews. Neuroscience. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- Okabe S. Molecular anatomy of the postsynaptic density. Molecular and cellular neurosciences. 2007;34:503–518. doi: 10.1016/j.mcn.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Okvist A, Fagergren P, Whittard J, Garcia-Osta A, Drakenberg K, Horvath MC, Schmidt CJ, Keller E, Bannon MJ, Hurd YL. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol Psychiatry. 2011;69:245–252. doi: 10.1016/j.biopsych.2010.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Ezan J, Sokol SY. PAR-1 phosphorylates Mind bomb to promote vertebrate neurogenesis. Developmental cell. 2009;17:222–233. doi: 10.1016/j.devcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padamsey Z, Emptage N. Two sides to long-term potentiation: a view towards reconciliation. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369:20130154. doi: 10.1098/rstb.2013.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglini G, Kunda P, Quiroga S, Kosik K, Caceres A. Suppression of radixin and moesin alters growth cone morphology, motility, and process formation in primary cultured neurons. J Cell Biol. 1998;143:443–455. doi: 10.1083/jcb.143.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Petersen SA, Fetter RD, Noordermeer JN, Goodman CS, DiAntonio A. Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 1997;19:1237–1248. doi: 10.1016/s0896-6273(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Qin G, Schwarz T, Kittel RJ, Schmid A, Rasse TM, Kappei D, Ponimaskin E, Heckmann M, Sigrist SJ. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J Neurosci. 2005;25:3209–3218. doi: 10.1523/JNEUROSCI.4194-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari S, Lisman JE. Properties of quantal transmission at CA1 synapses. Journal of neurophysiology. 2004;92:2456–2467. doi: 10.1152/jn.00258.2004. [DOI] [PubMed] [Google Scholar]
- Raymond FL, Tarpey P. The genetics of mental retardation. Hum Mol Genet. 2006;15(Spec No 2):R110–R116. doi: 10.1093/hmg/ddl189. [DOI] [PubMed] [Google Scholar]
- Rozas JL, Gomez-Sanchez L, Mircheski J, Linares-Clemente P, Nieto-Gonzalez JL, Vazquez ME, Lujan R, Fernandez-Chacon R. Motorneurons require cysteine string protein-alpha to maintain the readily releasable vesicular pool and synaptic vesicle recycling. Neuron. 2012;74:151–165. doi: 10.1016/j.neuron.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Rudy JW. Variation in the persistence of memory: An interplay between actin dynamics and AMPA receptors. Brain Res. 2014 doi: 10.1016/j.brainres.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Budnik V. Introduction on the use of the Drosophila embryonic/larval neuromuscular junction as a model system to study synapse development and function, and a brief summary of pathfinding and target recognition. International review of neurobiology. 2006;75:1–31. doi: 10.1016/S0074-7742(06)75001-2. [DOI] [PubMed] [Google Scholar]
- Saheki Y, De Camilli P. Synaptic vesicle endocytosis. Cold Spring Harbor perspectives in biology. 2012;4:a005645. doi: 10.1101/cshperspect.a005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson DJ, Bannerman DM. The role of habituation in hippocampus-dependent spatial working memory tasks: evidence from GluA1 AMPA receptor subunit knockout mice. Hippocampus. 2012;22:981–994. doi: 10.1002/hipo.20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Takeuchi M, Oda Y, Deguchi-Tawarada M, Sakamoto Y, Matsubara K, Nagasu T, Takai Y. Identification of activity-regulated proteins in the postsynaptic density fraction. Genes Cells. 2002;7:187–197. doi: 10.1046/j.1356-9597.2001.00505.x. [DOI] [PubMed] [Google Scholar]
- Schmid A, Hallermann S, Kittel RJ, Khorramshahi O, Frolich AM, Quentin C, Rasse TM, Mertel S, Heckmann M, Sigrist SJ. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat Neurosci. 2008;11:659–666. doi: 10.1038/nn.2122. [DOI] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. II. Fasciclin II controls presynaptic structural plasticity. Neuron. 1996;17:655–667. doi: 10.1016/s0896-6273(00)80198-1. [DOI] [PubMed] [Google Scholar]
- Self MW, Kooijmans RN, Super H, Lamme VA, Roelfsema PR. Different glutamate receptors convey feedforward and recurrent processing in macaque V1. Proc Natl Acad Sci U S A. 2012;109:11031–11036. doi: 10.1073/pnas.1119527109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y. Quantification of postsynaptic density proteins: Glutamate receptor subunits and scaffolding proteins. Hippocampus. 2011 doi: 10.1002/hipo.20950. [DOI] [PubMed] [Google Scholar]
- Shipton OA, Paulsen O. GluN2A and GluN2B subunit-containing NMDA receptors in hippocampal plasticity. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369:20130163. doi: 10.1098/rstb.2013.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihra TS, Flores G, Rodriguez-Moreno A. Kainate receptors: multiple roles in neuronal plasticity. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2014;20:29–43. doi: 10.1177/1073858413478196. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci U S A. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TE. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. The EMBO journal. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Magalhaes MA, Glogauer M. Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J Cell Biol. 2007;179:239–245. doi: 10.1083/jcb.200705122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Duszkiewicz AJ, Morris RG. The synaptic plasticity and memory hypothesis: encoding, storage and persistence. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369:20130288. doi: 10.1098/rstb.2013.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G, Vooijs M. Proteolytic cleavage of Notch:"HIT and RUN". Current molecular medicine. 2011;11:255–269. doi: 10.2174/156652411795677972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Schmeisser MJ, Sala C, Boeckers TM. Scaffold proteins at the postsynaptic density. Advances in experimental medicine and biology. 2012;970:29–61. doi: 10.1007/978-3-7091-0932-8_2. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Widagdo J, Chai YJ, Ridder MC, Chau YQ, Johnson RC, Sah P, Huganir RL, Anggono V. Activity-Dependent Ubiquitination of GluA1 and GluA2 Regulates AMPA Receptor Intracellular Sorting and Degradation. Cell reports. 2015 doi: 10.1016/j.celrep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Liaw JS, Baudry M, Berger TW. Novel expression mechanism for synaptic potentiation: alignment of presynaptic release site and postsynaptic receptor. Proc Natl Acad Sci U S A. 1997;94:6983–6988. doi: 10.1073/pnas.94.13.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Morita R, Mizoguchi T, Matsuo H, Isoda M, Ishitani T, Chitnis AB, Matsumoto K, Crump JG, Hozumi K, Yonemura S, Kawakami K, Itoh M. Mib-Jag1-Notch signalling regulates patterning and structural roles of the notochord by controlling cell-fate decisions. Development. 2010;137:2527–2537. doi: 10.1242/dev.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Lee HR, Jo YS, An K, Jung SY, Jeong MW, Kwon SK, Kim NS, Jeong HW, Ahn SH, Kim KT, Lee K, Kim E, Kim JH, Choi JS, Kaang BK, Kong YY. Mind bomb-1 is an essential modulator of long-term memory and synaptic plasticity via the Notch signaling pathway. Molecular brain. 2012;5:40. doi: 10.1186/1756-6606-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88:759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- Zanni G, van Esch H, Bensalem A, Saillour Y, Poirier K, Castelnau L, Ropers HH, de Brouwer AP, Laumonnier F, Fryns JP, Chelly J. A novel mutation in the DLG3 gene encoding the synapse-associated protein 102 (SAP102) causes non-syndromic mental retardation. Neurogenetics. 2010;11:251–255. doi: 10.1007/s10048-009-0224-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.