Abstract
The tumor suppressor p53 is activated upon cellular stresses such as DNA damage, oncogene activation, hypoxia, which transactivates sets of genes that induce DNA repair, cell cycle arrest, apoptosis, or autophagy, playing crucial roles in the prevention of tumor formation. The central regulator of the p53 pathway is Mdm2 which inhibits transcriptional activity, nuclear localization, and protein stability. More than 30 cellular p53-binding proteins have been isolated and characterized including Mdm2, Mdm4, p300, BRCA1/2, ATM, ABL, and 53BP-1/2. Most of them are nuclear proteins; however, not much is known about p53-binding transcription factors. In this review, we focus on transcription factors that directly interact with p53/Mdm2 through direct binding including Dmp1, E2F1, YB-1, and YY1. Dmp1 and YB-1 bind only to p53 while E2F1 and YY1 bind to both p53 and Mdm2. Dmp1 has been shown to bind to p53 and block all the known functions for Mdm2 on p53 inhibition, providing a secondary mechanism for tumor suppression in Arf-null cells. Although E2F1-p53 binding provides a checkpoint mechanism to silence hyperactive E2F1, YB-1 or YY1 interaction with p53 subverts the activity of p53, contributing to cell cycle progression and tumorigenesis. Thus, the modes and consequences for each protein-protein interaction vary from the viewpoint of tumor development and suppression.
Keywords: transcription factor, Dmp1 (Dmtf1), p53, Mdm2, E2F1, YB-1, YY1, Arf, polycomb, apoptosis, cancer
Introduction
The tumor suppressor p53 is activated upon cellular stresses such as DNA damage, oncogene activation, or hypoxia; and initiates a series of transcriptional programs that induces DNA repair, cell cycle arrest, apoptosis, or autophagy (1, 2). A central regulator of the p53 pathway is the Mdm2 protein (Hdm2 in humans) that inhibits transcriptional activity, nuclear localization, and protein stability of p53 (for reviews, 3–5; Fig. 1). The Mdm2 gene is transcriptionally activated by p53 through its direct binding to the p53-responsive elements located within the P2 promoter (4). Mutations in TP53 that disrupt p53 function occur in nearly 50% of human cancers (6, 7), and the alteration of regulators of p53 occurs in most of the remainder, and thus p53 is functionally inactivated in nearly all cancer cells.
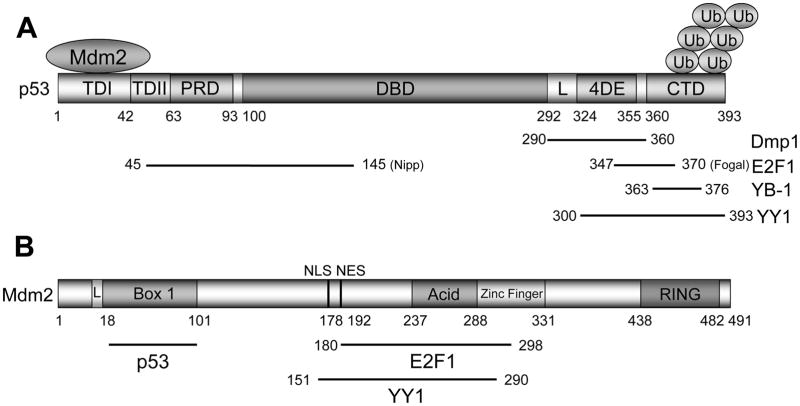
Figure 1. The domain structures of p53 and Mdm2 proteins.
(A) Schematic presentation of wild-type p53. Human p53 consists of 393 amino acids with 5 proposed domains. TD1: transactivation domain 1; TD2: transactivation domain 2; PRD: proline-rich domain; DBD: DNA-binding domain; L: nuclear import signal; 4DE: tetramerization domain; CTD: C-terminal regulatory domain. The 2nd transactivation domain for p53 was mapped between TD1 and PRD (84). Mdm2 quenches p53 transcriptional activity by occluding the p53 TD1 (a.a. 1–42). It also ubiquitinates lysines in the p53 CTD and accelerates nuclear export of p53.
(B) The domain structure for Mdm2. The full-length transcript of the gene encodes a protein of 491 amino acids with a predicted molecular weight of 56kDa. The full-length protein migrates at ~90kDa in SDS-PAGE due to post-translational modification(s) and amino acid composition of the protein (85). This protein contains several structural domains including an N-terminal p53 interaction domain (Box 1). Phosphorylation of S17 in the N-terminal “lid” of Mdm2 is proposed to regulate the binding of p53 (L). The nuclear localization (NLS) and export (NES) signals that are essential for proper nuclear-cytoplasmic shuttling of Mdm2 have been mapped between Box 1 and the acidic domain (a.a. 237–288). The phosphorylation of residues within the acidic domain may stimulate its ability to target p53 for degradation. Another conserved domain within the Mdm2 protein is a zinc finger domain (a.a. 289–331), the function of which is poorly understood. Mdm2 also contains a C-terminal RING domain (a.a. 438–482) that has E3 ubiquitin ligase activity sufficient for auto-ubiquitination. Mdm2 physically interacts with YY1 through a.a. 151–290 that includes the entire acidic residues and N-terminal zinc finger domain.
The activity of Mdm2 is negatively regulated by p19Arf (p14ARF in humans) in response to oncogenic stress (8–12). p19Arf directly binds to Mdm2, and thereby stabilizes and activates p53. The Arf induction by potentially harmful growth-promoting signals forces early-stage cancer cells to undergo p53-dependent and p53-independent cell cycle arrest, apoptosis, or autophagy, thus providing a powerful mode of tumor suppression (8–12). The Arf promoter monitors early stage oncogenic signals in vivo, and hence Arf-null mice are highly prone to spontaneous tumor development (13, 14). The Arf promoter is directly activated by E2Fs and Dmp1 (cyclin D binding myb-like protein 1; Fig. 2A) (15–18) while it is repressed by polycomb repressive complex 1 (PRC1) proteins: BMI1, PCGF1, PCGF2/MEL18, CBX2/7/8 RING1B: PRC2 proteins: EED, SUZ12, EZH2, and long non-coding RNA ANRIL (19).
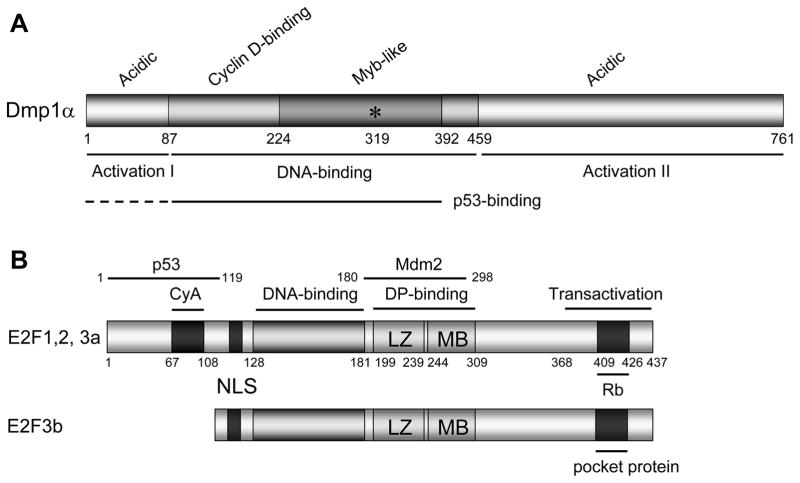
Figure 2. The domain structures of Dmp1α and E2F1-3 proteins.
(A) Schematic presentation of wild-type Dmp1α. Murine Dmp1α consists of 761 amino acids (760 amino acids in humans) with the central DNA-binding domain flanked by two transactivation domains. The DNA-binding domain has three tandem Myb-like repeats. Dmp1 loses its DNA-binding activity by substituting Lys-319 into Glu (shown as an asterisk). D-type cyclins interact with Dmp1 through the amino-terminal DNA-binding domain (a.a. 87–224). Thus, Dmp1-cyclin D complexes do not bind to DNA (16). The p53-binding domains have been mapped to the DNA-binding domain for Dmp1 (a.a. 87–392), thus Dmp1-p53 and Dmp1-DNA interactions are mutually exclusive (31).
(B) The domain structures for E2F1-3. The E2F proteins have a core domain that mediate DNA-binding (a.a. 128–181) or dimerization with DP proteins (leucine-zipper [LZ: a.a. 199–239] and marked box [MB: a.a. 244–309] motifs. The transactivation and pocket protein-binding domains are present only in E2F1-E2F5 (20). Moreover, E2F1, E2F2, and E2F3a+b share a canonical basic nuclear localization signal that is absent in E2F4-E2F5, which have nuclear export signals instead (20). The major p53-binding domain has been mapped around the cyclin A-binding domain of E2F1.
E2Fs are a group of transcription factors (TFs) that regulate cell cycle, DNA repair, replication, and mitochondrial function through making heterodimeric complexes with dimerization partners, DPs (for reviews, 20–22; Fig. 2B). The E2F family proteins are generally grouped by function into two categories: transcription activators and repressors. Activators such as E2F1, E2F2, and E2F3a promote and help cell cycle progression while repressors (E3F3b, E2F4-8) inhibit the cell cycle. Pocket proteins such as pRB and related proteins p107 and p130, can bind to E2F when hypophosphorylated (Fig. 2B). Cyclin D/Cdk4(6) and cyclin E/cdk2 phosphorylate pRB, p107, and p130 allowing them to dissociate from E2F/DP. The inactivating function of cyclin/cyclin-dependent kinases is opposed by the Ser/Thr protein phosphatases PP2A and PP1 (23). In activators, E2F binding with pRB has been shown to mask the transactivation domain responsible for transcription activation (20, 21). E2F activator levels are cyclic, with maximal expression during G1/S. In contrast, E2F repressors stay constant, and they are often expressed in quiescent cells. Activator E2Fs transactivate S phase-promoting genes, such as DNA polymerase α dihydrofolate reductase, and thymidine kinase (20, 21). E2F1, the prototype of activator E2Fs, was found to be a repressor of hTERT transcription by directly binding to its promoter, thereby shortening telomere length by inhibiting hTERT protein expression (24). E2F1 also represses the transcription of tumor suppressor genes such as Dmp1 and ARH1 (25). Among E2F proteins, only E2F1-3a has a nuclear localization signal and interacts with p53.
The Y-box-binding protein YB-1, also known as nuclease-sensitive protein 1 (NSEP1), is a member of the cold shock family of proteins that contain a highly conserved nucleic acid-binding motif (26) named Y-box to which all the family members bind (Fig. 3A), and is very similar to the common CCAAT box. Y-box proteins contain a cold-shock domain, which function as RNA chaperones, RNA splicing, translation, DNA repair, and transcription (26; Fig 3A). YB-1 directly binds to p53, and the interaction of YB-1 with p53 affects the sequence-specific DNA-binding of p53 to its consensus sequences (27).
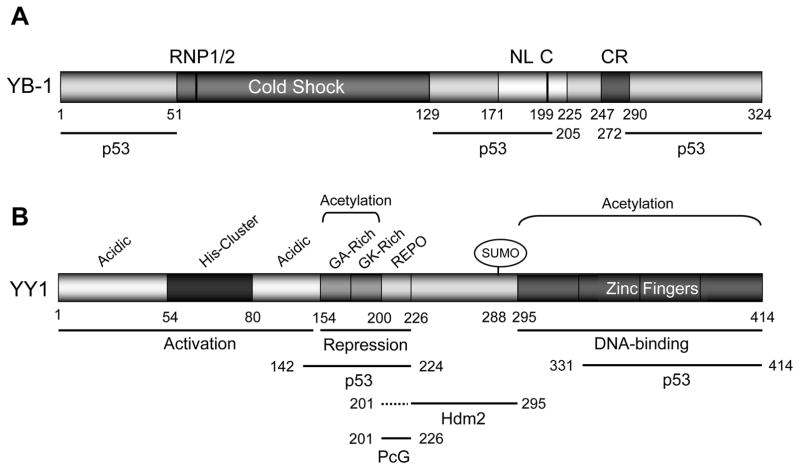
Figure 3. The domain structures of YB-1 and YY1 proteins.
(A) The structure of YB-1. YB-1 has a cold-shock DNA-binding domain with two transactivation domains at both amino- and carboxyl-termini through which it binds to p53. The cold-shock domain has RNP1/2-like motifs consisting of a five-stranded β-barrel structure that creates a surface that may act as a large nucleic acid-binding site. The inactive protein is in the cytoplasm (the cytoplasmic retention [CR] signal is at a.a. 247–290). The nuclear localization signals [NL] have been mapped to a.a. 171–225 by Jürchott et al. (56). This protein needs to be cleaved at a.a. 219–220 [C] before nuclear localization. The p53-binding domains have been mapped to N-terminal, middle, and C-terminal domains (a.a. 1–51, 129–205, and 272–324) (27).
(B) The structure for human YY1. Human YY1 consists of 414 amino acids with amino-terminal transactivation domain containing 11x His cluster with a carboxyl terminal zinc finger domain. The GA, GK-rich, and REPO domains have been shown to be responsible for transcriptional repression. p300 and CBP associated factor (PCAF) mediate the acetylation of residues 171–200 while PCAF also acetylates the C-terminal zinc finger domain of YY1. The REPO domain of YY1 binds and recruits PcG transcriptional repressors. p53 binds to YY1 amino acid residues 142–224, and 331–414; Hdm2 binds to YY1 through amino acid residues 226–295; thus YY1 make a ternary complex with p53 and Mdm2, and inactivating p53-mediated transcription (30).
YY1 (Yin Yang 1) is a nuclear protein that is ubiquitously expressed in all tissues and highly conserved from Xenopus to human (28). The name “Yin Yang” represents its two opposite functions as a TF to act as either a repressor or an activator (28, 29). YY1 is a TF belonging to the GLI-Krüppel class of zinc finger proteins (Fig. 3B). It has fundamental roles in embryogenesis, cell proliferation, and differentiation (28, 29). YY1 exerts its biological effects through its ability to transactivate or repress gene expression, depending upon the genes to which it binds. Recent studies show that YY1 indirectly activates or represses gene expression without DNA-binding, through interaction with histone modifiers and chromatin remodeling proteins (28, 29). In addition to directly binding to p53, YY1 also binds to Arf and Mdm2 and enhances p53 ubiquitination and degradation, thus YY1 is a negative regulator of p53 (30).
This review will summarize the molecular basis and biological consequences of p53 protein-protein interactions with these TFs to demonstrate the differences in how these TFs might function to promote or prevent tumorigenesis. The manipulation of these p53 protein interactions represents a promising therapeutic avenue to promote tumor cell death in cancers that have intact p53 genes, but nevertheless p53 is functionally inactive.
Dmp1
We have identified the Arf regulating transcription factor Dmp1 (cyclin D binding myb-like protein 1; also named Dmtf1; refs. 15–18, 31–42) as a direct interacting molecule for p53 (Fig. 2A; 31). The original form for Dmp1 (Dmp1α) has been isolated in a yeast two-hybrid screen of a murine T cell library with cyclin D2 bait (15). We recently detected endogenous hDMP1-cyclin D1 complex in the lysate of human breast cancer (40). When Dmp1 was cloned, it was expected to be a regulator of the Rb pathway, but later studies have shown that Dmp1 is a critical transcriptional activator for Arf (17, 18). The gene product, p19Arf (or p14ARF) stabilizes nucleoplasmic p53 by binding to Mdm2, sequesters it in the nucleolus, and directly inhibits the ubiquitin ligase activity of Mdm2 (8–12). DMP1 is a tumor suppressor deleted in ~35% of human non-small-cell lung carcinomas and 42% of breast cancer (32–34). Mitogenic signals from oncogenic Ras (35) and HER2/neu (36) have been shown to activate the Dmp1 promoter, while physiological mitogens (25) as well as genotoxic stimuli mediated by NF-κB (37) cause repression. Eμ-Myc, K-rasLA, and HER2/neu-driven tumor development was significantly accelerated in both Dmp1+/− and Dmp1−/− mice with no significant differences in the survival between the two cohorts, suggesting that Dmp1 is haplo-insufficient for tumor suppression (18, 32, 36, 38–40). We recently showed that Dmp1α has the ability to activate both p53 and Rb pathways and prevent tumor development in MMTV-neu mammary tumor model (41). In Eμ-Myc lymphomas, the combined frequencies of p53 mutation and Arf deletion in mice of Dmp1+/− or Dmp1−/− background were much lower than that in Dmp1+/+ littermates, indicating that Dmp1 is a physiological regulator of the Arf-p53 pathway in vivo (39). Of note, the frequency of p53 mutation (~40%) was significantly decreased in both Dmp1+/− and Dmp1−/− backgrounds (<10%) even in K-rasLA lung cancer where the Ink4a/Arf involvement was rare (32). This suggested an Arf-independent mechanism of p53 regulation by Dmp1 in epithelial tissues. Dmp1α regulates genes other than Arf (e.g. Tsp-1, JunB, and Egr1) and contributes to tumor suppression (42).
In our recent study (31), we searched for binding partners for the Dmp1α protein to explain the Arf-independent function of Dmp1 in tumor suppression. We found that Dmp1α physically bound to p53, but not to Mdm2, Arf, c-Myc, or c-Myb (31). Dmp1α binding to p53 neutralized the activity of Hdm2 on p53’s ubiquitination in H1299 cells with ARF deletion. Conversely, DMP1β/γ lack most of the amino acid sequence for the p53 interaction, and thus will not directly affect the activity of the p53 pathway (43). We also showed that Dmp1α neutralized the activity of Hdm2 using recombinant proteins prepared in bacteria (Mdm2, ubiquitin) and Sf9 cells (Dmp1) (31). Dmp1 antagonized nuclear export of p53 by Hdm2 in transfection studies conducted in H1299 cells, which was confirmed in ARF-null MCF7 cells by depleting endogenous hDMP1 using shRNA (31). We mapped the Dmp1-binding domain at the carboxyl-terminus of p53 (Fig. 1A; 31), suggesting that Dmp1-p53 binding does not interfere with p53 binding to target genes or physical interaction between Mdm2 and p53. Conversely, the p53-binding domain was mapped to the DNA-binding domain of Dmp1 (Fig. 2A), and thus binding of Dmp1 to p53 and DNA are mutually exclusive (31).
To study the significance of Dmp1-p53 interaction in vivo, doxorubicin was injected in wild-type and Dmp1-null mice, and binding of p53 to target gene (p21Cip1 and bbc3) promoters was studied by chromatin immunoprecipitation in thymus (31). Significant binding of p53 to the p21Cip1 and bbc3 promoters was found in wild-type, but not in Dmp1-null thymus, suggesting that Dmp1 is indispensable for p53-binding to target genes (31). When injected with doxorubicin, both p21Cip1 and bbc3 increase was more significantly subverted in Dmp1−/− mice than in Arf−/− mice; thus our data indicate that Dmp1 activates p53 target genes independent of Arf in response to the genotoxic drug (31). Together, our data indicate that (i) expression of Dmp1α interferes with the known activities of Hdm2 on the ubiquitination and nuclear localization of p53 in an Arf-independent fashion and (ii) p53-binding to target genes and their inductions are Dmp1-dependent. Interestingly, the most representative molecule that neutralizes all the activities of Mdm2 on p53 is p19Arf, which is a direct target of Dmp1α. Hence Dmp1α is an opponent regulator for Mdm2 by directly transactivating the Arf transcription as well as by direct binding to p53 in Arf-null cells (31).
E2F1
During the cell cycle, the expression of E2F1-3a and cyclin A are both precisely regulated: the levels of these E2Fs are high at the G1/S transition, whereas cyclin A is high at mid-S to G2/M (44). Thus G1/S transition is the time when E2F1-3a are important in cell cycle regulation because cyclin A/CDK2 binds directly to E2F1 and inhibits its DNA-binding by phosphorylation (45). Like pRb, p53 only binds to E2F1, 2, and 3a (Fig. 2B) through the conserved domain in the N-terminus, which is not present in E2F3b, 4–8 (46). The p53-binding domain was mapped in or around the cyclin A-binding domain of E2F1 since 1–119Δ24 mutant did not show any p53-binding (Fig. 2B; 47). Since the p53-binding domain in E2F1-3a also binds to cyclin A, cyclin A and p53 potentially compete for binding to E2Fs.
Nipp et al. found that the transactivation domain for E2F1 was dispensable for p53 induction by E2F (47); therefore it was expected that E2F1-p53 complex does not bind to E2F-consensus sequences, just like Dmp1-p53. Fogal et al. reported that the amino terminal domain in E2F1 (a.a. 1–108) binds to amino acid residues 347–370 of p53 that overlaps its C-terminal nuclear export sequence (NES), enhancing nuclear retention of Ser315-phosphorylated p53, and thus inducing p21Cip1, PIG3, and bax (46). Phosphorylation of p53 at Ser315 destabilizes the p53 tetramer by exposing its NES and increases the likelihood for p53 to be exported into the cytoplasm (46). It should be noted that Ser315 is the only site in p53 that is phosphorylated by cyclin-dependent and aurora kinases that regulate cell cycle progression (46). E2Fs’ preferential binding to Ser315-phosphorylated p53 through protein-protein interaction is cell cycle-dependent and interferes with the degradation of p53 in the cytoplasm, leading to activation of p53 (Fig. 4B). E2F1-p53 interaction thus provides a checkpoint function to provide a feedback loop to silence the activities for E2F1 (46). Zhou et al. recently showed that p53 interacts with E2F1 to form p53-E2F1-DNA complex suppressing E2F1-dependent PLK1 expression promoting apoptosis in response to DNA-damage (48). In this case, the protein complex binds to the E2F target, not the p53 target, thus the mode and significance of p53-E2F1 binding is different dependent on the biological context. Although E2F1 specifically binds to p53 and stimulates its DNA-binding, transactivation, and apoptotic functions of the protein, it does not bind to p63 or p73 (for p63 and p73 review, see ref. 49) showing the specificity of E2F1-p53 interaction.
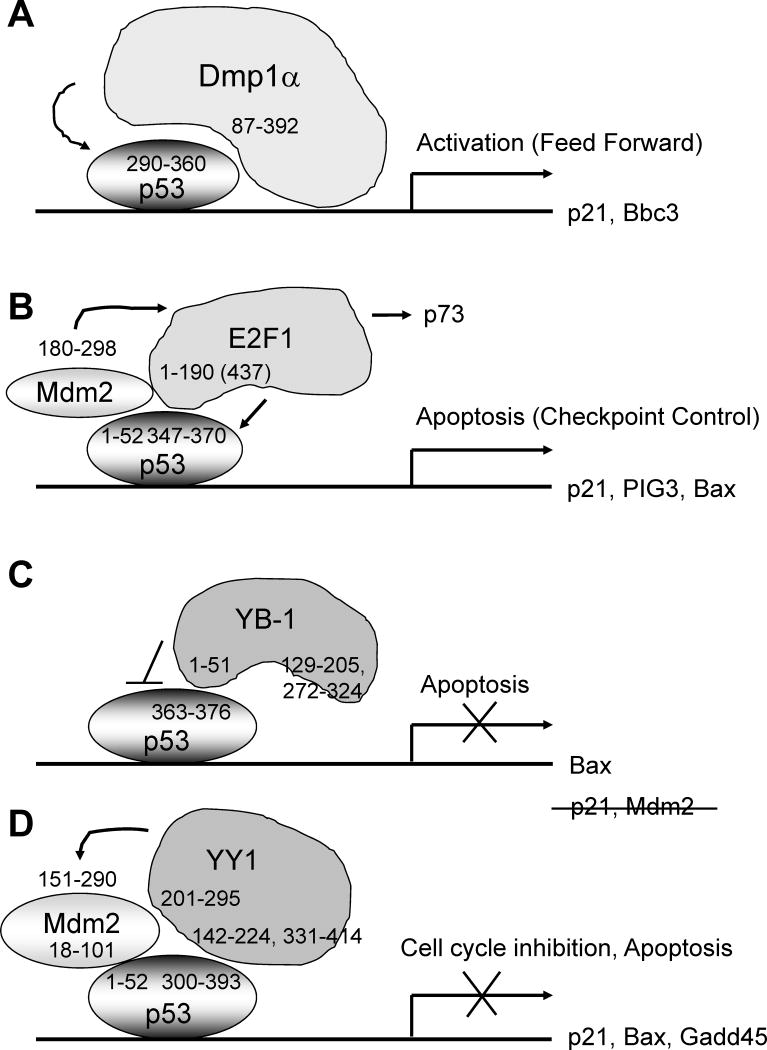
Figure 4. Schematic presentation of Dmp1α-p53, E2F1-p53/Mdm2, YB-1-p53, and YY1-p53/Hdm2 interactions and their biological consequences.
(A) Dmp1-p53 interaction. Dmp1α directly binds to the p53 C-terminus and neutralizes all the known functions for Mdm2 on p53 (31; arrow). This effect is mutually exclusive of DNA-binding of Dmp1α, and thus independent of Arf.
(B) E2F1-p53/Mdm2 interaction. E2F1 bind to p53 C-terminus through the N-terminal domain (major: a.a. 1–190, minor: a.a. 190–437; 47) or a.a. 1–108 (46). This interaction will provide a checkpoint mechanism to silence aberrant E2F1 signaling to apoptosis to prevent carcinogenesis (46). Mdm2 prolongs the half-life of E2F1 by inhibiting ubiquitination (50, arrow). Physical interaction of E2F1 and Mdm2 contributes to TAp73 transcriptional activity (horizontal arrow).
(C) YB-1-p53 interaction. YB-1 outside the cold-shock domain binds to p53 C-terminus to prevent the latter from inducing apoptosis genes (27). Thus YB-1 overexpression enhances tumorigenesis by accumulating cells that are not dying.
(D) YY1-p53/Hdm2 interaction. YY1 binds to both Hdm2 and p53, accelerates the polyubiquitination of p53 (arrow) to subvert the activities of p53. Since YY1 interacts with Hdm2 and p53 through different domains, they can form a ternary complex (30). This YY1-p53 interaction does not induce a checkpoint function to cancel the oncogenic activity of YY1.
E2F1 physically interacts with Mdm2 through a.a. 180–298 (50; Fig. 2B). Since E2F1 uses the DP-binding domain for the physical interaction with Mdm2, it is speculated that the E2F1-Mdm2 complex does not bind to DPs. Mdm2 prolongs the half-life of the E2F1 protein by inhibiting its ubiquitination. Mdm2 displaces the E2F1 E3 ligase SCF (SKP2) through direct binding independent of pRB or ARF (50). Thus stabilization of the E2F1 protein is one p53-independent mechanism of Mdm2-mediated tumorigenesis (50). Physical interaction of E2F1 and Mdm2 in the nucleus contributes to TAp73 transcriptional activity, suggesting a novel regulation pathway of TAp73 (51).
YB-1
YB-1 acts as a regular TF through its property of binding to double-stranded DNA called the Y-box, reverse CCAAT sequences. YB-1 must translocate from the cytoplasm to nucleus for any activities in the nucleus since the majority of the YB-1 protein is in the cytoplasm, much like the mechanism for NF-κB activation (52). Nuclear translocation of YB-1 and transactivation of target genes occur in response to a variety of stresses that include UV exposure, DNA-damaging agents, hyperthermia, and cytokines (53–55). YB-1 must first be cleaved (Fig. 3a) and then only the N-terminal portion translocates to the nucleus, which occurs exclusively at the G1/S phase boundary (55, 56). Nuclear accumulation of YB-1 is associated with transactivation of the cyclin A/B1 genes and a subsequent increase in the cyclin A/B1 proteins (56). This process is dependent on binding to the splicing factor SRp30c, and requires Akt-dependent phosphorylation of YB-1 (57).
It has been hypothesized that YB-1 has a role in promoting cell proliferation and tumorigenesis since Y-box elements are present in the promoters of several genes whose activity is associated with cell division, e.g. epidermal growth factor receptor (EGFR), c-ErbB2, proliferating cell nuclear antigen (PCNA), DNA polymerase α, thymidine kinase, and topoisomerase IIa; among these, PCNA, DNA polymerase α, thymidine kinase are classical E2F targets as well (21). Indeed, Berquin et al. screened a cDNA expression library for genes that mediate EGF-independent proliferation of human mammary epithelial cells (HMECs) (58), and isolated the YB-1 cDNA, which conferred growth factor independence to HMECs.
In addition to transactivation for genes essential for cell proliferation, YB-1 represses the transcription of cell death-promoting genes, such as FAS and p53 by binding possible Y-box sequences (59). YB-1 expression in tumor specimens thus correlates with a progressed clinical stage and poor prognosis of patients in a variety of human cancers, including malignancies of the lung, breast, colon, and prostate (60). Overexpression of YB-1 is frequently observed in drug-resistant tumors since its expression is closely associated with that of P-glycoprotein encoded by the multidrug resistance 1 gene and mitogen-activated protein kinase-interacting seine/threonine kinase 1 gene responsible for impairing trastuzumab resistance (60, 61). The oncogenic activity of YB-1 has been demonstrated in vivo using a mouse model for breast cancer (62).
YB-1 directly binds to p53 both in vitro and in vivo and the interaction of YB-1 with p53 modifies the sequence-specific DNA-binding of p53 to its consensus sequences (27). It has been shown that three independent domains of YB-1 interact with p53 (27; Fig. 3A; a.a. 1–51, 129–205, and 272–324). Conversely, a 14 amino acid sequence at the C-terminal of p53 (a.a. 363–376) is required for its interaction with YB-1 (27; Fig. 1A). YB-1 and p53 also interact with a common transcription factor AP2 to co-regulate the gelatinase A gene that facilitate neo-angiogenesis and distal metastases (63). Importantly, YB-1 prevents p53 from inducing apoptosis by inhibiting p53 from transactivating pro-apoptotic target genes such as APAF-1, Noxa, and Bax (59, 64). The apoptosis stimulating p53-binding proteins, ASPP1 and ASPP2, are the first two common activators of the p53 protein family that selectively enable the latter to regulate specific apoptotic target genes, such as Bax and Pig3. YB-1 thus acts in an opposite manner to ASPP1/2 that binds to p53. Transcriptionally active p53 is required for nuclear localization of YB-1 suggesting that p53 transactivates effector(s) for YB-1 nuclear translocation (64).
Although YB-1 inhibits the ability of p53 to cause cell death, YB-1 does not interfere with the ability of p53 to transactivate p21Cip1 nor Mdm2 (Fig. 4C). In contrast to E2F1-p53 binding, YB-1-p53 interaction does not provide a checkpoint mechanism to quench hyperactive YB-1 signaling in cell cycle progression/tumor development (Fig. 4C). Selective alteration of the p53 activity by nuclear YB-1 provides an explanation for the correlation of YB-1 with drug resistance and poor tumor prognosis. Since inhibition of YB-1 upregulates p21Cip1 expression with p53-dependent apoptosis in human cancer cell lines (59), the YB-1-p53 interaction is an ideal molecular target for drug screenings for future cancer therapy.
YY1
The domain structure for YY1 has been extensively studied (30; Fig. 3B). The viral oncogene product Adenovirus E1A regulates the transcriptional activity of YY1 by converting it from a repressor to an activator, suggesting an important role of YY1 in cell proliferation (28, 29). YY1 plays essential roles in cell proliferation and differentiation by interacting with numerous proto-onco- (e.g. c-Myc, c-Fos, EGFR, c-ErbB2, and Mdm2) and tumor-suppressor genes/proteins (e.g. ARF, p53, p73, and Rb; refs. 28, 29). YY1 contributes to breast cancer development by accelerating p27Kip1 ubiquitination through direct binding (65). The YY1 gene expression itself has been shown to be regulated by prohibitin through an E2F1-binding element (66). YY1 expression is associated with chemo- and immune-resistance in cancer therapy (28, 29), and thus is a novel therapeutic target.
Constitutional ablation of yy1 in mice results in peri-implantation lethality and hence phenotype analysis of the whole animal is not possible (67). The same group later created conditional knockout mice bearing an yy1flox hypomorphic allele (68). Genetic crosses of yy1flox mice with yy1+/− mice allowed them to generate mutant mice expressing 75%, 50%, and 25% of the normal yy1 level. Consistent with the data from tissue culture studies, phenotypic analysis of these mice and corresponding embryonic fibroblasts revealed a critical, dosage-dependent requirement for yy1 in late embryonic development and cell proliferation (68). Specifically, p21Cip1, p27Kip1, p57Kip2, Btg2, Gadd45γ, Dnmt3a, and TGFβ2 genes were upregulated in yy1flox/− and/or yy1−/− murine embryonic fibroblasts while Suv39h2, Polα1, cyclin B1/B2, Plk1, and Cdc6 were downregulated, confirming that yy1 regulates various genes involved in cell cycle progression, cytokinesis, and differentiation as well as apoptosis, oncogenic transformation, and DNA repair (68). Although YY1 has been suggested to play significant roles in oncogenesis, no transgenic studies have been conducted for in vivo tumor development and would therefore be an interesting topic for future studies.
YY1 physically interacts with p53, Mdm2, and ARF. Sui et al. reported that YY1 depletion resulted in p53 accumulation due to a reduction of p53 ubiquitination in cells (30). YY1 directly binds to both Hdm2 and p53 and enhances p53 ubiquitination and degradation, thus YY1 is a negative regulator of p53 (30). GST pull-down assay showed that YY1 binds to C-terminal regulatory region of p53 through two different regions on YY1 (a.a. 142–224, 331–414 [zinc finger domain]; Fig. 3B). Because the p53-binding domain is different from the Hdm2 interaction domain (a.a. 201–295, mainly 226–295, Fig. 3B), and because YY1 and Hdm2 interact with p53 through different regions (a.a. 300–393 for YY1 and a.a. 1–42 for Hdm2; Fig. 1A), it was speculated that YY1, Hdm2, and p53 make a ternary complex, which was proved by glycerol gradient centrifugation (30; Fig. 4D). YY1 promotes p53 polyubiquitination through Hdm2 since the YY1 mutant defective for Hdm2-binding failed to augment the physical interaction between Hdm2 and p53. It was also speculated that the YY1-p53 complex recognizes p53 targets rather than YY1 targets since p53 occupies the DBD of YY1 when bound (Figs. 3B, 4D).
YY1 inactivates p53 through other mechanisms as well. YY1 inhibits p300-mediated p53 acetylation and reduces p53-activated transcription (69). Thus, blocking p53 acetylation decreases its transcriptional activity and facilitates Mdm2-mediated p53 ubiquitination and degradation. YY1 directly binds to p14ARF by relocating it into the nucleolus or by forming tripartite complex with Hdm2 and p53 (30). In summary, these activities of YY1 result in the same consequence: inactivation of p53 (Fig. 4D). The activity of YY1 is regulated by various post-translational modifications such as phosphorylation (28, 29), sumoylation (70), and glycosylation (71). Smurf2 physically interacts with YY1, induces the polyubiquitination of YY1 and shortens the half-life of the protein (72, 73), thus relieving the suppression of p53 activity.
As a member of the polycomb group (PcG) protein family, YY1 has been shown to regulate gene silencing without binding to DNA (74). The functional role of YY1 has been characterized in the developmental studies of Drosophila using two orthologs of YY1, pleiohomeotic (pho) and pho like (phol) (75). Pho and phol recruit the polycomb group silencing complexes to chromatin and establish gene repression (76). The region responsible for recruiting the PcG complex was mapped to the residues 201–226 of human YY1 protein called REPO (Fig. 3B; 29). Enhancer of zeste 2 (Ezh2) was identified as a bona fide oncogene, which has been used as a marker of cancers with aggressive potential (77). Ezh2 has been shown to be essential for cancer progression and invasion (78, 79), and thus increases the possibility of therapeutic failure (80). YY1 recruits Ezh2 to target promoters through the REPO domain; its overexpression in cancers promotes the methyltransferase activity of Ezh2 and causes aberrant epigenetics, which augments cancer progression (29, 81, 82).
YY1 influences the E2F activity through RYBP (Ring1- and YY1 binding protein; a member of the PcG group). RYBP has been identified as a molecule that interacts specifically with the E2F2/E2F3, dependent on the marked-box domain (83, Fig. 2B). The authentic YY1 transcriptional target, the Cdc6 promoter contains adjacent E2F- and YY1-binding sites, and both are required for promoter activity. E2F2, E2F3, YY1, and RYBP bind to the Cdc6 promoter and stimulate the transcription at G1/S phase of the cell cycle (83). Thus the ability of RYBP to mediate an interaction between E2F2/E2F3 and YY1 is an important component of Cdc6 activation.
Role of post-translational modifications in p53/Mdm2-TF interactions
The binding between purified Dmp1α and p53 requires post-translational modification(s) since they bind to each other when they are prepared in Sf9 cells (31). Conversely, E2F1, YB-1, or YY1 binding to p53/Mdm2 does not require any post-translational modification(s) since purified bacterial recombinant proteins bind to p53/Mdm2 (27, 30, 46). Still the Dmp1-p53 binding is physiological since we saw significant binding of endogenous Dmp1 and p53 in thymus from mice injected with doxorubicin (31).
Conclusive remarks, unanswered questions, and future directions
Although these four TFs directly bind to p53, the modes for interaction and the biological consequences are significantly different as summarized in Fig. 4. Dmp1 and YB-1 bind only to p53 while E2F1 and YY1 bind to both p53 and Mdm2. Dmp1-p53 binding blocks all the known functions for Mdm2 on p53 inhibition, providing a secondary mechanism for tumor suppression by Dmp1α. While the E2F1-p53 binding provides a checkpoint mechanism to silence hyperactive E2F1, the interaction of YB-1 or YY1 with p53 subverts the activity of p53, contributing to cell cycle progression and tumorigenesis. In vivo tumor development assays using drug-inducible transgenic or conditional knockout mice for each TF with/without p53-knockout animals will provide physiological insights for such molecular interactions.
Whether or not these TFs bind to gain-of-function mutant p53 at higher or lower affinity than wild-type should be investigated to translate the research findings to clinical levels. Since more than half of human cancers highly express the YB-1 and/or YY1 proteins, inhibit p53, and confer drug resistance, gene silencing or acceleration of protein degradation of these molecules with small chemicals may be a novel strategy in cancer therapeutics.
Acknowledgments
Grant Support: K. Inoue was supported by NIH/NCI 2R01CA106314, ACS RSG-07-207-01-MGO, and KG080179.
We thank Robert D. Kendig, Fumitake Kai, Guangchao Sui, and other members of Dr. Kazushi Inoue’s laboratory for sharing unpublished research findings.
Footnotes
Conflict of interests
The authors have no conflict of interests related to this work.
References
- 1.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–37. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 3.Kruse J-P, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580–9. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eischen CM, Lozano G. The Mdm network and its regulation of p53 activities: a rheostat of cancer risk. Hum Mutat. 2014;35:728–37. doi: 10.1002/humu.22524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan C, Pearsall I, Yeudall A, Deb SP, Deb S. p53: its mutations and their impact on transcription. Subcell Biochem. 2014;85:71–90. doi: 10.1007/978-94-017-9211-0_4. [DOI] [PubMed] [Google Scholar]
- 8.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 9.Sherr CJ, Bertwistle D, DEN Besten W, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. p53-dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–37. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 10.Saporita AJ, Maggi LB, Jr, Apicelli AJ, Weber JD. Therapeutic targets in the ARF tumor suppressor pathway. Curr Med Chem. 2007;14:1815–27. doi: 10.2174/092986707781058869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer. 2010;127:2239–47. doi: 10.1002/ijc.25511. [DOI] [PubMed] [Google Scholar]
- 12.Maggi LB, Jr, Winkeler CL, Miceli AP, Apicelli AJ, Brady SN, Kuchenreuther MJ, Weber JD. ARF tumor suppression in the nucleolus. Biochim Biophys Acta. 2014;1842:831–9. doi: 10.1016/j.bbadis.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–59. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 14.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–22. [PubMed] [Google Scholar]
- 15.Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor DMP1. Mol Cell Biol. 1996;16:6457–67. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci USA. 1999;96:3993–8. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene. 2007;26:4329–35. doi: 10.1038/sj.onc.1210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–9. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–57. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benevolenskaya EV, Frolov MV. Emerging Links between E2F Control and Mitochondrial Function. Cancer Res. 2015;75:619–23. doi: 10.1158/0008-5472.CAN-14-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurimchak A, Graña X. PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues. Cell Cycle. 2015;14:18–30. doi: 10.4161/15384101.2014.985069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang A, Shen C, Zhang B, Rao Z, Wang R, Yang S, Ning S, Mao G, Fang D. E2F1 acts as a negative feedback regulator of c-Myc-induced hTERT transcription during tumorigenesis. Oncol Rep. 2014;32:1273–80. doi: 10.3892/or.2014.3287. [DOI] [PubMed] [Google Scholar]
- 25.Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its repression by E2Fs. Oncogene. 2006;25:7703–13. doi: 10.1038/sj.onc.1209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays. 2003;25:691–8. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto T, Izumi H, Imamura T, Takano H, Ise T, Uchiumi T, Kuwano M, Kohno K. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene. 2000;19:6194–202. doi: 10.1038/sj.onc.1204029. [DOI] [PubMed] [Google Scholar]
- 28.Gordon S1, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–42. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Stovall DB, Inoue K, Sui G. The Oncogenic Role of Yin Yang 1. Critical Reviews in Oncogenesis. 2011;16:163–97. doi: 10.1615/critrevoncog.v16.i3-4.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–72. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Frazier DP, Kendig RD, Kai F, Maglic D, Sugiyama T, Taneja P, Morgan RL, Fry EA, Lagedrost SJ, Sui G, Inoue K. Dmp1 physically interacts with p53 and positively regulates p53’s stabilization, nuclear localization, and function. Cancer Res. 2012;72:1740–50. doi: 10.1158/0008-5472.CAN-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallakin A, Sugiyama T, Taneja P, Matise LA, Frazier DP, Choudhary M, Hawkins GA, D’Agostino RB, Jr, Willingham MC, Inoue K. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007;12:381–94. doi: 10.1016/j.ccr.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue K, Sugiyama T, Taneja P, Morgan RL, Frazier DP. Emerging roles of DMP1 in lung cancer. Cancer Res. 2008;68:4487–90. doi: 10.1158/0008-5472.CAN-07-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maglic D, Zhu S, Fry EA, Taneja P, Kai F, Kendig RD, Sugiyama T, Miller LD, Willingham MC, Inoue K. Prognostic value of the hDMP1-ARF-Hdm2-p53 pathway in breast cancer. Oncogene. 2013;32:4120–9. doi: 10.1038/onc.2012.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–32. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taneja P, Maglic D, Kai F, Sugiyama T, Kendig RD, Frazier DP, Willingham MC, Inoue K. Critical role of Dmp1 in HER2/neu-p53 signaling and breast carcinogenesis. Cancer Res. 2010;70:9084–94. doi: 10.1158/0008-5472.CAN-10-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taneja P, Mallakin A, Matise LA, Frazier DP, Choudhary M, Inoue K. Repression of Dmp1 and Arf promoter by anthracyclins: critical roles of the NF-κB subunit p65. Oncogene. 2007;26:7457–66. doi: 10.1038/sj.onc.1210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inoue K, Wen R, Rehg JE, Adachi M, Cleveland JL, Roussel MF, Sherr CJ. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, ras transformation, and tumorigenesis. Genes Dev. 2000;14:1797–809. [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001;15:2934–39. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu S, Mott RT, Fry EA, Taneja P, Kulik G, Sui G, Inoue K. Cooperation between cyclin D1 expression and Dmp1-loss in breast cancer. Am J Pathol. 2013;183:1339–50. doi: 10.1016/j.ajpath.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fry EA, Taneja P, Maglic D, Zhu S, Sui G, Inoue K. Dmp1α inhibits HER2/neu-induced mammary tumorigenesis. PLoS One. 2013;8(10):e77870. doi: 10.1371/journal.pone.0077870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallakin A, Sugiyama T, Kai F, Taneja P, Kendig RD, Frazier DP, Maglic D, Matise LA, Willingham MC, Inoue K. The Arf-inducing transcription factor Dmp1 encodes transcriptional activator of amphiregulin, thrombospondin-1, JunB and Egr1. Int J Cancer. 2010;126:1403–16. doi: 10.1002/ijc.24938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maglic D, Stovall DB, Cline JM, Fry EA, Mallakin A, Taneja P, Caudell DL, Willingham MC, Sui G, Inoue K. DMP1β, a splice isoform of the tumor suppressor DMP1 locus, induces proliferation and progression of breast cancer. J Pathol. 2015;236:90–102. doi: 10.1002/path.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–62. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 45.Xu M, Sheppard KA, Peng CY, Yee AS, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994;14:8420–31. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fogal V, Hsieh JK, Royer C, Zhong S, Lu X. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J. 2005;24:2768–82. doi: 10.1038/sj.emboj.7600735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nip J, Strom DK, Eischen CM, Cleveland JL, Zambetti GP, Hiebert SW. E2F-1 induces the stabilization of p53 but blocks p53-mediated transactivation. Oncogene. 2001;20:910–20. doi: 10.1038/sj.onc.1204171. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Z, Cao JX, Li SY, An GS, Ni JH, Jia HT. p53 Suppresses E2F1-dependent PLK1 expression upon DNA damage by forming p53-E2F1-DNA complex. Exp Cell Res. 2013;319:3104–15. doi: 10.1016/j.yexcr.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Inoue K, Fry EA. Alterations of p63 and p73 in human cancers. Subcell Biochem. 2014;85:17–40. doi: 10.1007/978-94-017-9211-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene. 2005;24:7238–47. doi: 10.1038/sj.onc.1208814. [DOI] [PubMed] [Google Scholar]
- 51.Kasim V, Huang C, Zhang J, Jia H, Wang Y, Yang L, Miyagishi M, Wu S. Synergistic cooperation of MDM2 and E2F1 contributes to TAp73 transcriptional activity. Biochem Biophys Res Commun. 2014;449:319–26. doi: 10.1016/j.bbrc.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 52.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;32:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 53.Koike K, Uchiumi T, Ohga T, Toh S, Wada M, Kohno K, Kumano M. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997;417:390–4. doi: 10.1016/s0014-5793(97)01296-9. [DOI] [PubMed] [Google Scholar]
- 54.Ohga T, Uchiumi T, Makino Y, Koike K, Wada M, Kuwano M, Kohno K. Direct involvement of the YB-1 binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J Biol Chem. 1998;273:5997–6000. doi: 10.1074/jbc.273.11.5997. [DOI] [PubMed] [Google Scholar]
- 55.van Roeyen CR, Scurt FG, Brandt S, Kuhl VA, Martinkus S, Djudjaj S, Raffetseder U, Royer HD, Stefanidis I, Dunn SE, Dooley S, Weng H, et al. Cold shock Y-box protein-1 proteolysis autoregulates its transcriptional activities. Cell Commun Signal. 2013;11:63. doi: 10.1186/1478-811X-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jürchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. 2003;278:27988–96. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- 57.Basaki Y, Hosoi F, Oda Y, Fotovati A, Maruyama Y, Oie S, Ono M, Izumi H, Kohno K, Sakai K, Shimoyama T, Nishio K, et al. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene. 2007;26:2736–46. doi: 10.1038/sj.onc.1210084. [DOI] [PubMed] [Google Scholar]
- 58.Berquin IM, Pang B, Dziubinski ML, Scott LM, Chen YQ, Nolan GP, Ethier SP. Y-box-binding protein 1 confers EGF independence to human mammary epithelial cells. Oncogene. 2005;24:3177–86. doi: 10.1038/sj.onc.1208504. [DOI] [PubMed] [Google Scholar]
- 59.Lasham A, Moloney S, Hale T, Homer C, Zhang YF, Murison JG, Braithwaite AW, Watson J. The Y-box binding protein YB1 is a potential regulator of the p53 tumor suppressor. J Biol Chem. 2003;278:35516–23. doi: 10.1074/jbc.M303920200. [DOI] [PubMed] [Google Scholar]
- 60.Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dörken B, Royer HD. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–50. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 61.Astanehe A, Finkbeiner MR, Krzywinski M, Fotovati A, Dhillon J, Berquin IM, Mills GB, Marra MA, Dunn SE. MKNK1 is a YB-1 target gene responsible for imparting trastuzumab resistance and can be blocked by RSK inhibition. Oncogene. 2012;31:4434–46. doi: 10.1038/onc.2011.617. [DOI] [PubMed] [Google Scholar]
- 62.Bergmann S, Royer-Pokora B, Fietze E, Jürchott K, Hildebrandt B, Trost D, Leenders F, Claude JC, Theuring F, Bargou R, Dietel M, Royer HD. YB-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer Res. 2005;65:4078–87. doi: 10.1158/0008-5472.CAN-04-4056. [DOI] [PubMed] [Google Scholar]
- 63.Mertens PR, Steinmann K, Alfonso-Jaume MA, En-Nia A, Sun Y, Lovett DH. Combinatorial interactions of p53, activating protein-2, and YB-1 with a single enhancer element regulate gelatinase A expression in neoplastic cells. J Biol Chem. 2002;277:24875–82. doi: 10.1074/jbc.M200445200. [DOI] [PubMed] [Google Scholar]
- 64.Homer C, Knight DA, Hananeia L, Sheard P, Risk J, Lasham A, Royds JA, Braithwaite AW. Y-box factor YB1 controls p53 apoptotic function. Oncogene. 2005;24:8314–25. doi: 10.1038/sj.onc.1208998. [DOI] [PubMed] [Google Scholar]
- 65.Wan M, Huang W, Kute TE, Miller LD, Zhang Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, Deng Z, Wang W, et al. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am J Pathol. 2012;180:2120–33. doi: 10.1016/j.ajpath.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joshi B, Rastogi S, Morris M, Carastro LM, DeCook C, Seto E, Chellappan SP. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401:155–66. doi: 10.1042/BJ20060364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–44. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–81. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci USA. 2004;101:12165–70. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng Z, Wan M, Sui G. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol Cell Biol. 2007;27:3780–92. doi: 10.1128/MCB.01761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799:353–64. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramkumar C, Cui H, Kong Y, Jones SN, Gerstein RM, Zhang H. Smurf2 suppresses B-cell proliferation and lymphomagenesis by mediating ubiquitination and degradation of YY1. Nat Commun. 2013;4:2598. doi: 10.1038/ncomms3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeong HM, Lee SH, Yum J, Yeo CY, Lee KY. Smurf2 regulates the degradation of YY1. Biochim Biophys Acta. 2014;1843:2005–11. doi: 10.1016/j.bbamcr.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 74.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–58. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 76.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci USA. 2006;103:19296–301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 79.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 80.Berezovska OP, Glinskii AB, Yang Z, Li XM, Hoffman RM, Glinsky GV. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle. 2006;5:1886–901. doi: 10.4161/cc.5.16.3222. [DOI] [PubMed] [Google Scholar]
- 81.Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249–57. doi: 10.1093/jb/mvu054. [DOI] [PubMed] [Google Scholar]
- 82.Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y, Bonalli M, McCabe MT, Creasy CL, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015 Jan 22;6:6051. doi: 10.1038/ncomms7051. [DOI] [PubMed] [Google Scholar]
- 83.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–86. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–65. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- 85.Cheng TH, Cohen SN. Human MDM2 isoforms translated differentially on constitutive versus p53-regulated transcripts have distinct functions in the p53/MDM2 and TSG101/MDM2 feedback control loops. Mol Cell Biol. 2007;27:111–9. doi: 10.1128/MCB.00235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]