Abstract
Background
HIV protein Nef plays a key role in impairing cholesterol metabolism in both HIV infected and bystander cells. The existence of a small cohort of patients infected with Nef-deficient strain of HIV presented a unique opportunity to test the effect of Nef on lipid metabolism in a clinical setting.
Methods
Here we report the results of a study comparing six patients infected with Nef-deficient strain of HIV (ΔNefHIV) with six treatment-naïve patients infected with wild-type HIV (WT HIV). Lipoprotein profile, size and functionality of high density lipoprotein (HDL) particles as well as lipidomic and microRNA profiles of patient plasma were analyzed.
Results
We found that patients infected with ΔNefHIV had lower proportion of subjects with plasma HDL-C levels <1 mmol/l compared to patients infected with WT HIV. Furthermore, compared to a reference group of HIV-negative subjects, there was higher abundance of smaller under-lipidated HDL particles in plasma of patients infected with WT HIV, but not in those infected with ΔNefHIV. Lipidomic analysis of plasma revealed differences in abundance of phosphatidylserine and sphingolipids between patients infected with ΔNefHIV and WT HIV. MicroRNA profiling revealed that plasma abundance of 24 miRNAs, many of those involved in regulation of lipid metabolism, was differentially regulated by WT HIV and ΔNefHIV.
Conclusion
Our findings are consistent with HIV protein Nef playing a significant role in pathogenesis of lipid-related metabolic complications of HIV disease.
Keywords: HIV, Lipoproteins, Lipids, HDL Metabolism, Dyslipidemias, MicroRNA, Nef
1. Introduction
HIV disease is characterized by severe metabolic complications including dyslipidemia and atherosclerosis [1, 2]. Adverse side-effects of antiretroviral regimens were originally blamed for these complications, however, as development of better treatment regimens with reduced effect on lipid metabolism did not eliminate dyslipidemia [3] and high risk of atherosclerosis [4], it is becoming increasingly clear that HIV disease itself makes a substantial contribution to the pathogenesis of these complications. We have demonstrated that HIV protein Nef inhibits cholesterol efflux causing cholesterol accumulation in HIV-infected macrophages [5]. The same effect was observed in uninfected cells treated with recombinant Nef or with plasma containing soluble Nef released from infected cells [6]. Furthermore, recombinant Nef injected in mice in vivo caused atherosclerosis and dyslipidemia supporting a key role of Nef in pathogenesis of HIV-associated metabolic abnormalities [7]. However, no clinical evidence supporting the role of Nef in lipid dysregulation is available.
In this study we analyzed plasma samples of six patients infected with Nef-deficient strain of HIV-1 (ΔNefHIV), all members of the Sydney Blood Bank Cohort (SBBC). Pathogenicity and immunogenicity of this strain have been described in previous publications [8–11] and summarized in a recent review [12]. In brief, all patients were infected with the same strain of HIV-1 through blood transfusion; they were slow-progressors or non-progressors and remained asymptomatic for an extended period of time [12]. Lipid metabolism in these patients was never investigated and, considering a potentially key role of Nef in the pathogenesis of HIV related impairment of lipid metabolism, these patients provided a unique opportunity to elucidate the role of Nef in HIV-associated metabolic disorders in a clinical setting.
2. Methods
2.1. Patients
Patients infected with Nef-deficient strain of HIV-1 (ΔNefHIV, n=6) were all members of the SBBC cohort. Clinical and immunological parameters of these patients were originally described in several publications [8–11]; age, sex, CD4+ cell count and viral load values for these patients are shown in Table 1 in comparison to the same parameters in WT HIV subjects. All patients, except patient D36, were not receiving antiretroviral therapy. Two subjects (C49, C64) were postmenopausal females, all other patients were males.
Table 1.
Parameters of HIV Disease and Plasma Lipoprotein Profile
| Subject | CD4+ (cells/μl)1 | Viral load (104/ml)1 | Age | Sex | TC (mmol/l) | LDL-C (mmol/l) | HDL-C (mmol/l) | TG (mmol/l) | apoA-I (g/l) | HDL-C/apoA-I | apoB (g/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV Negative | |||||||||||
| C1 | ND | - | 40 | M | 4.7 | 2.9 | 1.4 | 0.8 | 2.19 | 0.64 | 0.7 |
| C2 | ND | - | 37 | M | 4.9 | 2.7 | 1.3 | 2.1 | 1.75 | 0.74 | 0.9 |
| C3 | ND | - | 39 | M | 4.0 | 2.3 | 1.4 | 0.7 | 1.86 | 0.75 | 0.8 |
| C4 | ND | - | 44 | M | 4.2 | 1.1 | 1.5 | 3.6 | 2.24 | 0.67 | 0.4 |
| C5 | ND | - | 42 | M | 3.9 | 2.3 | 1 | 1.2 | 1.76 | 0.57 | 0.8 |
| C6 | ND | - | 42 | M | 4.3 | 2.8 | 1.2 | 0.8 | 1.86 | 0.65 | 0.9 |
|
| |||||||||||
| Mean±SD | ND | - | 41±3 | 4.3±0.4 | 2.4±0.7 | 1.3±0.2 | 1.5±1.1 | 1.9±0.2 | 0.67±0.07 | 0.8±0.2 | |
|
| |||||||||||
| WT HIV | |||||||||||
| H1 | 588 | 0.07 | 50 | M | 4.2 | 2.5 | 0.8 | 1.8 | 1.2 | 0.67 | 0.7 |
| H2 | 774 | 0.005 | 58 | M | 6.0 | 3.6 | 1.8 | 1.3 | 2.0 | 0.90 | 0.8 |
| H3 | 400 | 8.4 | 51 | M | 4.5 | 2.6 | 1.5 | 0.7 | 1.8 | 0.83 | 0.6 |
| H4 | 405 | 2.5 | 55 | M | 4.0 | 2.6 | 0.7 | 1.6 | 1.2 | 0.58 | 0.7 |
| H5 | 190 | 10.0 | 52 | M | 4.7 | 3.5 | 0.6 | 1.1 | 1.1 | 0.55 | 0.8 |
| H6 | 414 | 2.2 | 50 | M | 4.0 | 2.3 | 0.7 | 2.0 | 1.3 | 0.54 | 0.7 |
|
| |||||||||||
| Mean±SD | 461±198 | 3.9±4.3 | 53±3 | 4.5±0.8 | 2.9±0.6 | 1.0±0.5 | 1.4±0.5 | 1.4±0.4* | 0.68±0.15 | 0.7±0.1 | |
|
| |||||||||||
| ΔNefHIV | |||||||||||
| C49 | 1006 | 0 | 44 | F | 3.4 | 2.1 | 0.7 | 1.8 | 1.2 | 0.58 | 0.8 |
| C54V | 1054 | 0.04 | 71 | M | 3.2 | 1.4 | 1.5 | 0.6 | 1.4 | 1.07 | 0.4 |
| C64 | 850 | 0 | 73 | F | 5.7 | 3.1 | 2.0 | 1.3 | 1.9 | 1.05 | 0.9 |
| C98 | 435 | 0.02 | 61 | M | 5.0 | 3.1 | 1.2 | 1.4 | 1.5 | 0.80 | 1.1 |
| C135 | 480 | 0 | 53 | M | 4.2 | 2.6 | 1.1 | 0.8 | 1.2 | 0.92 | 0.8 |
| D36 | ND | ND | 40 | M | 4.0 | 2.4 | 0.8 | 1.8 | 1.3 | 0.62 | 0.8 |
|
| |||||||||||
| Mean±SD | 765±291# | 0.01±0.02# | 57±14 | 4.2±1.0 | 2.5±0.6 | 1.2±0.5 | 1.3±0.5 | 1.4±0.3* | 0.84±0.21 | 0.8±0.2 | |
The ΔNefHIV-infected patients were matched with patients infected with Nef-positive (WT) HIV-1 strain. Patients infected with Nef-positive strain of the virus (n=6) were selected from a cohort of treatment-naïve HIV patients described in our previous study [13]. All subjects were males. Reference values for HIV-negative subjects were obtained by analyzing plasma samples of six HIV negative subjects (all males) selected from a group of healthy volunteers from the blood bank of the Baker IDI Heart and Diabetes Institute.
All subjects were not undergoing any lipid-lowering therapy and did not have a history of cardiovascular disease. All samples were stored at −80°C and were analyzed retrospectively.
Plasma samples of both groups of HIV-infected patients used in this study were from the previous studies [9, 13]; original human ethics approvals permitted for the extension of analysis of the collected samples.
Lipoprotein profile
Total cholesterol (TC), low density lipoprotein cholesterol (LDL-C), high density lipoprotein cholesterol (HDL-C) and triglycerides (TG) were analyzed using Cobas blood analyser. Apolipoprotein A-I and apolipoprotein B concentrations were analyzed using ELISA kits (Mabtech, Sweden).
2.2. HDL size
Distribution of apoA-I among HDL subfractions was analyzed by non-denaturing PAGE followed by immunoblotting using antibody against human apoA-I as described previously [14]. The following definitions of HDL subfractions were used: HDL3c, 7.2–7.8 nm; HDL3b, 7.8–8.2 nm; HDL3a, 8.2–8.8 nm; HDL2a, 8.8–9.7 nm; HDL2b, 9.7–12 nm.
2.3. Cholesterol efflux
Cholesterol efflux assay was performed using THP-1 human macrophages activated with LXR agonist as described previously [15]. 1% plasma or 1.1% apoB-depleted plasma (obtained as described previously [15]) were used as an acceptor.
2.4. Lipidomic analysis
Lipidomic analysis was performed as described previously [16]. In brief, lipids from 10 μl of plasma were extracted using a modified, single phase Folch method; the analysis was done in triplicates. The analysis was performed by liquid chromatography electrospray ionization-tandem mass spectrometry (LC ESI-MS/MS) using a Agilent 1200 liquid chromatography system, and Applied Biosystems API 4000 Q/TRAP mass spectrometer with a turbo-ion spray source (350 °C) and Analyst 1.5 and MultiQuant data systems using a Zorbax C18, 1.8 μm, 50 × 2.1-mm column (Agilent Technologies). Lipid concentrations were calculated by relating the peak area of each species to the peak area of the corresponding internal standard.
2.5. Small RNA deep sequencing and bioinformatics analysis
RNA from plasma was extracted using the miRNeasy Mini Kit (Qiagen, Australia). The manufacturers’ protocol was followed with a slight modification involving the use of Trizol LS (Life Technologies, Australia). The small RNA yield, composition and quality was analyzed using the Agilent 2100 Bioanalyser with the Small RNA kit (Agilent Technologies). Sequencing adapters were ligated onto the small RNA sample followed by conversion into cDNA libraries using the Ion Total RNA-Seq Kit V2 (Life Technologies, Australia) and prepared for deep sequencing on the Ion Torrent Personal Genome Machine (PGM™). Pooled libraries with unique barcodes were loaded on 318™ sequencing chips and run on the Ion Torrent PGM (Life Technologies, Australia) using the Ion PGM™ 200 Sequencing Kit (Life Technologies). The Torrent Suite 4.2.1 was used to manage the Ion Torrent PGM™ to process raw signals and perform base calling. The sequences were then assessed for quality, and primer-adapter sequences were trimmed by the Torrent Suite software, followed by alignment to the human reference genome (HG19) using bowtie 2 followed by a second pass through TMAP using Partek Flow. The trimmed and aligned data was mapped to known miRNA using miRBase V.20. The number of reads for each miRNA was adjusted to reads per million (RPM) and normalized using the lognormal linear method across all samples. Samples containing less than 5 reads per million were removed. The data was then transferred to Partek Genomics Suite for statistical analysis, hierarchical clustering and to identify unique miRNA in each sample type. TargetScan, mirDB and TarBase were interrogated to predict targets for the selected miRNAs. Raw sequencing files in .BAM format for each sample sequenced can be downloaded from http://www.ebi.ac.uk/ena/data/view/PRJEB7984
3. Results
3.1. Plasma lipoproteins
Patient plasma lipoprotein and apolipoprotein levels are shown in Table 1. Total cholesterol, triglyceride and apolipoprotein B levels were similar between the groups, while LDL-C levels trended higher in WT HIV group. In comparison to a reference group of HIV-negative subjects level of HDL-C was reduced by 25% in WT HIV infected patients, but only by 8% in ΔNefHIV infected subjects, however, due to limited power of the study these substantial differences did not reach statistical significance. However, when we defined hypoalphalipoproteinemia (HALP) as HDL-C below 1 mmol/l, the proportion of subjects with HALP was significantly lower among patients infected with ΔNefHIV compared to those in WT HIV group (chi-square, P=0.05). Levels of apoA-I were similar in both HIV-infected groups, consequently, the HDL-C/apoA-I ratio, which may be considered a surrogate marker of HDL lipidation, was 20% higher in ΔNefHIV patients, but again, the difference was not statistically significant.
3.2. HDL structure and functionality
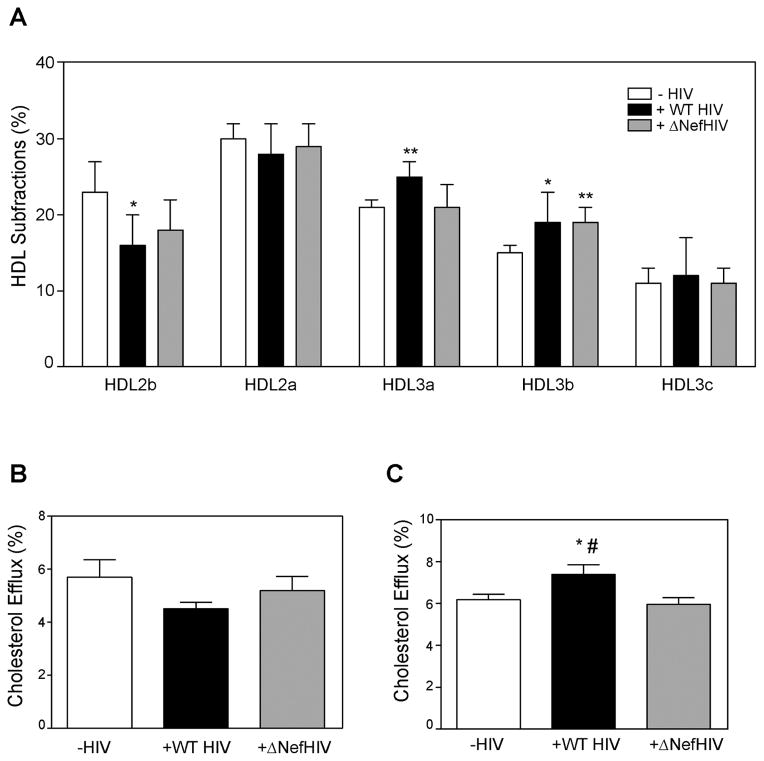
We previously demonstrated that Nef reduces abundance and functionality of ABCA1 [5, 6, 17]; the most likely consequence of this effect at systemic level would be changes in lipidation of HDL, which, in turn, may affect HDL functionality. An indication of this was a higher ratio of HDL-C/apoA-I in ΔNefHIV patients compared to HIV patients (Table 1). We therefore analyzed the distribution of HDL among particles of different sizes. In patients infected with WT HIV the proportion of large HDL2b particles (9.7–12 nm) was reduced relative to that in the reference group of uninfected subjects, whereas this effect was partially eliminated in patients infected with ΔNefHIV (Fig. 1A). The proportion of smaller HDL3a particles (8.2–8.8 nm) was increased in plasma of WT HIV infected patients, but not in patients infected with ΔNefHIV. Proportion of HDL3b particles (7.8–8.2 nm) was increased in plasma of patients from both WT HIV and ΔNefHIV groups, while the proportion of smallest HDL3c particles (7.2–7.8 nm) was not affected by HIV status. Thus, HIV infection reduced the proportion of large fully lipidated HDL particles and increased the proportion of smaller under-lipidated HDL, this effect was partially eliminated in patients infected with ΔNefHIV.
Figure 1. The effect of HIV and ΔNefHIV infection on the structure (A) and function (B, C) of plasma high density lipoprotein.
A – Analysis of distribution of HDL among different subfractions in plasma of HIV-negative subjects and patients infected with WT HIV or ΔNefHIV. *p<0.05; **p<0.01 versus HIV negative subjects. B- Cholesterol efflux from human THP-1 monocyte-macrophages to 1% plasma from HIV-negative subjects and patients infected with WT HIV or ΔNefHIV. C- Cholesterol efflux from human THP-1 monocyte-macrophages to 1.1% apoB-depleted plasma from HIV-negative subjects and patients infected with WT HIV or ΔNefHIV. *p<0.05 versus HIV-negative; #p<0.05 versus ΔNefHIV.
To investigate the effect of these changes in HDL structure on HDL functionality, the ability of whole plasma and of HDL to support cholesterol efflux from human macrophages was tested. Macrophages were activated with LXR agonist to ensure that most of the efflux represents specific ABCA1/G1-dependent cholesterol efflux. When the efflux to whole plasma was tested, we found no statistically significant difference between the groups in the capacity of plasma to support cholesterol efflux (Fig. 1B). Next, we measured the efflux to plasma depleted of apoB-containing lipoproteins (i.e. plasma where HDL is the only available lipoprotein). We found that while there was no difference between the efflux to apoB-depleted plasma from HIV-negative subjects and subjects infected with ΔNefHIV, cholesterol efflux to plasma from WT HIV group was higher (Fig. 1C). We hypothesized that increased cholesterol efflux to HDL from WT HIV-infected subjects was due to increased proportion of small HDL3a particles, and indeed when data for all three groups were combined, there was a correlation between the capacity of HDL to support cholesterol efflux and proportion of HDL3a particles (r=0.50; p<0.05).
3.3. Lipidomic profiling
We have previously reported the effects of HIV infection on plasma lipidomic profile [16]. Using a similar approach, we compared lipidomic profile of patients infected with WT or Nef HIV. The full lipidomic profile (330 lipid species) is provided in the accompanying Data in Brief article [18], and lipid species significantly different between the groups are shown in Table 2. Compared to the reference group of uninfected subjects, three species of phosphatidylserine (PS) and PS as a group were reduced in WT HIV group, but not in ΔNefHIV group. Lipid species affected in ΔNefHIV group but not affected in WT HIV group, include individual species of dihydroceramide (dhCer), ceramide, alkylphosphatidylethanolamine, phosphatidylethanolamine (PE) and two species of sphingomyelin. Individual species of phosphatidylinositol and dhCer were increased in both HIV infected groups. One species each of dihexosylceramide, PE and PS were only increased in ΔNefHIV group.
Table 2.
Lipidomic analysis
| Lipid Species | HIV Negative | WT HIV | ΔNef-HIV |
|---|---|---|---|
| PS 36:1 | 1418.5 ± 935.8 | 259.5 ± 259.9** | 7733.5 ± 9425.3# |
| PS 38:3 | 299.8 ± 158.4 | 53.8 ± 91.9* | 1559.4 ± 1791.7# |
| PS 40:5 | 123.0 ± 65.5 | 13.6 ± 33.4** | 590.9 ± 659.4# |
| PS (total) | 2161.5 ± 1317.7 | 334.2 ± 121.4* | 10,772.2 ± 12,619.5# |
| dhCer 20:0 | 20.8 ± 5.6 | 23.4 ± 9.1 | 115.7 ± 125.8* |
| Cer 18:0 | 80.4 ± 16 | 102.9 ± 26.6 | 140.7 ± 76.9* |
| SM 32:0 | 423.0 ± 84.8 | 512.0 ± 136.4 | 666.2 ± 176.0* |
| SM 34:0 | 4,369 ± 686.2 | 5,402.1 ± 1,442.6 | 6,326 ± 803.1** |
| PE(O-36:6) | 34.3 ± 10.4 | 71.7 ± 44.9 | 95.4 ± 32.5** |
| dhCer (total) | 519.0 ± 77.7 | 571.9 ± 254.3 | 1439.2 ± 1033.8* |
| PI 34:0 | 153.5 ± 58.0 | 362.2 ± 298.8* | 342.8 ± 287.7 |
| dhCer 16:0 | 46.3 ± 6.7 | 71.5 ± 15.7* | 120.7 ± 70.5** |
| dhCer 18:0 | 46.1 ± 11.4 | 79.6 ± 22.0* | 194.1 ± 184.3* |
| DHC 20:0 | 111.3 ± 38.4 | 85.0 ± 38.6 | 256.2 ± 249.5# |
| PE(22:6/0:0) | 1378.0 ± 402.6 | 1723.5 ± 836.7 | 1042.8 ± 168.4# |
| PS 40:6 | 128.4 ± 53.9 | 36.6 ± 49.7 | 572.1 ± 619.6# |
Abbreviations: dhCer, dihydroceramide; Cer, ceramide; DHC, dihexosylceramide; PE, phosphatidylethanolamine; PE(O), alkylphosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin;
p < 0.05 vs HIV Negative;
p < 0.01 vs HIV Negative;
p < 0.05 vs WT HIV (ANOVA)
When data for all three groups were combined, there was positive correlation between the proportion of HDL2a particles (which was similar in the three groups) and plasma abundance of phosphatidylcholine (r=0.69, p<0.002), cholesterol (r=0.54, p<0.05), alkenylphosphatidylcholine (r=0.47, p<0.05) and phosphatidylethanolamine (r=0.50, p<0.05); all these lipid species are major constituents of large lipidated HDL particles. There was also negative correlation between cholesterol efflux to whole plasma and the abundance of cholesteryl esters (r=−0.50, p<0.05) and ceramide (r=−0.45, p<0.05).
3.4. miRNA profiling
Abundance of small RNAs extracted from plasma samples was analyzed by deep sequencing. RNA from two patients infected with ΔNefHIV did not pass quality control testing and were not sequenced. Unsupervised hierarchical clustering was performed on significantly differentially expressed miRNA using Euclidean average linkage by miRNA. The full dataset is presented in Supplemental Dataset 1, and differentially expressed miRNAs are shown in Fig. 2. Twenty-four miRNAs were found to be significantly deregulated in patients infected with WT HIV compared to the ΔNefHIV groups. These 24 miRNAs were observed to show similar or recovered expression levels within ΔNefHIV and uninfected subject groups but were significantly deregulated in HIV patients. There are two major nodes of the dendrogram. Node 1 contains 4 miRNAs which were found to be up-regulated in WT HIV group. Node 2 contains 20 miRNA which were found to be down-regulated in WT HIV group. Predicted and validated targets for these miRNAs that are related to lipid metabolism and/or HIV infection are listed in Table 3.
Figure 2. Hierarchical clustering of differentially regulated miRNA in WT HIV and ΔNefHIV patients.
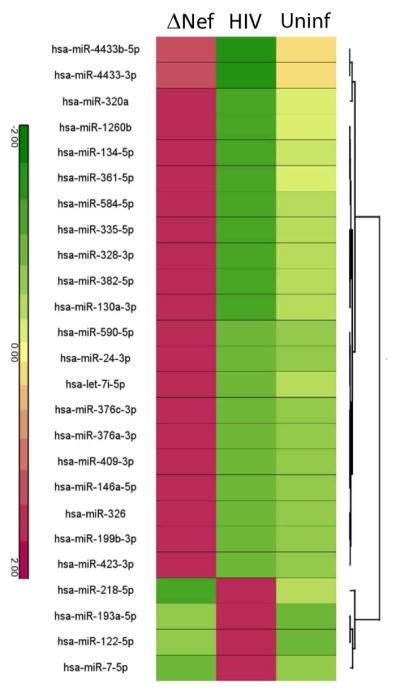
Abundance of miRNAs that show similar or recovered expression levels within ΔNefHIV patients and uninfected patients however significantly deregulated in HIV patients. Lines connect pairs with p<0.01.
Table 3.
Selected predicted targets of miRNAs depended on presence of Nef
| microRNA | Predicted targets | Validated targets | Gene name | Reference to HIV |
|---|---|---|---|---|
| hsa-miR-199b-3p | ABCA1 | ATP-binding cassette transporter A1 | Many | |
| ABCA13 | ATP binding cassette, transporter A13 | |||
| LRP2 | LDL receptor-related protein 2 | |||
| hsa-miR-409-3p | ABCA12 | ATP-binding cassette, transporter A12 | Many | |
| ABCB10 | ATP-binding cassette, transporter B10 | |||
| LRP | LDL receptor-related protein | PMID:22412921 | ||
| hsa-miR-320a | CANX | Calnexin | PMID: 25170080 | |
| XPO1/CRM1 | Exportin-1 | PMID: 22318151 | ||
| hsa-miR-24-3p | ABCB9 | ABCB9 | ATP-binding cassette transporter B9 | |
| hsa-miR-382-5p | LRP12 | LDL receptor-related protein 12 | ||
| hsa-miR-130a-3p | LRP8 | LDL receptor-related protein 8 | ||
| LDLR | LDLR | LDL Receptor | PMID:24191141 | |
| ACSL4 | acyl-CoA synthetase long-chain family member 4 | |||
| PLCB1 | phospholipase C, beta 1 | PMID:23555755 | ||
| CD69 | CD69 | PMID:25340508 | ||
| hsa-miR-4433b-5p | VLDLR | VLDL Receptor | PMID:25394062 | |
| SMPD3 | Neutral sphingomyelinase II | |||
| hsa-miR-7-5p | APOA2 | Apolipoprotein A2 | PMID:21978398 | |
| IRS2 | IRS2 | Insulin receptor substrate 2 | PMID:15590971 | |
| ACSL4 | ACSL4 | acyl-CoA synthetase long-chain family member 4 | ||
| IDE | IDE | insulin-degrading enzyme | PMID:17026490 | |
| hsa-miR-218-5p | ACSL1 | acyl-CoA synthetase long-chain family member 1 | ||
| hsa-miR-376c-3p | VLDLR | VLDL receptor | PMID:25394062 | |
| ACSL1 | acyl-CoA synthetase long-chain family member 1 | |||
| ALCAM | activated leukocyte cell adhesion molecule | PMID25420915 | ||
| hsa-miR-590-5p | IL12A | Interleukin 12A | PMID: 12444143 | |
| hsa-miR-326 | SLC2A1/GLUT1 | MFS transporter, solute carrier family 2 | PMID: 24335483 | |
| hsa-miR-193a-5p | SRF | Serum response factor | PMID: 12120892 | |
| hsa-miR-146a-5p | CCL8/MCP-2 | CCL8/MCP-2 | Chemokine (C-C motif) ligand 8 | PMID: 20181935 |
| hsa-let-7i-5p | TBP | TBP | TATA Box motif | PMID: 25336585 |
| hsa-miR-122-5p | HMOX1 | HMOX1 | Heme oxygenase (decycling) 1 | PMID: 24752012 |
| BACH1 | BACH1 | BTB and CNC homology 1 | PMID: 24752012 | |
| hsa-miR-134-5p | SHP | SHP | Src homology-2 domain-containing protein tyrosine phosphatase | PMID: 18776219 PMID: 23029125 |
4. Discussion
Metabolic complications are an important part of HIV disease and include impairment of cellular and systemic lipid metabolism and enhanced development of atherosclerosis. We have demonstrated the key role of HIV protein Nef in disturbances of cellular cholesterol metabolism caused by HIV infection [5]. Nef is an HIV accessory protein expressed early in infection; it is located on the plasma membrane of infected cells (for review see [19]) and is also released from infected cells. The documented effects of released Nef on uninfected cells include apoptosis [20], endothelial dysfunction [21], impairment of immune response [22], and impairment of cholesterol metabolism. The effects of extracellular Nef on lipid metabolism in bystander cells include down regulation of ABCA1 and inhibition of cholesterol efflux in vitro and hypoalphalipoproteinemia and hyperetrigliceridemia in vivo [6, 7]. Furthermore, Nef affects the spectrum of miRNAs released from infected cells in exosomes, including several miRNAs with established role in cholesterol metabolism, such as miR-33a*, miR-16b, miR-145, miR-144* [23]. Collectively, these findings suggest that Nef released from infected cells may be an important factor in pathogenesis of metabolic complications of HIV disease, but this concept was never tested in a clinical setting. Patients infected with ΔNefHIV provided a unique opportunity to close this gap.
The main finding of this study is a lower prevalence of hypoalphalipoprotenemia in patients infected with ΔNefHIV compared to patients infected with WT HIV. Furthermore, the proportion of smaller, presumably immature, HDL was increased in WT HIV infected patients similar to what was documented for SIV infection [6]. This effect was not observed in patients infected with ΔNefHIV. The HDL-C/apoA-I ratio trended to be higher in ΔNefHIV infected patients, also suggesting that HDL particles in these patients are more lipidated. These findings are consistent with our previous observations implicating Nef in reducing liver ABCA1 and impairing generation and/or maturation of HDL particles [6, 7]. There was no effect of HIV, with or without Nef, on the ability of whole plasma to support cholesterol efflux, which is consistent with our previous findings [3, 13]. However, when the efflux to HDL was measured, HDL from WT HIV group, but not from ΔNefHIV group, had higher functionality in cholesterol efflux assay. The most likely explanation of this finding is higher proportion of small, presumably under-lipidated HDL particles in plasma of WT HIV infected patients compared to two other groups; small HDL particles are better acceptors of cholesterol compared to larger fully lipidated HDL particles [24]. This finding is inconsistent with findings of another recent study where the efflux to apoB-depleted plasma from WT HIV infected patients was reduced [25]; however, patient demographics in this study was different. Our hypothesis, however, is that the primary effect of extracellular Nef is in reducing abundance of ABCA1in both hepatic and extra-hepatic cells. In hepatocytes, reduction of ABCA1 would reduce plasma levels of HDL. In extra-hepatic cells, reduction of ABCA1 would reduce capacity of the cells for cholesterol efflux; contribution of the inhibition of ABCA1 in macrophages used in this study was not tested as cells were neither infected with HIV nor treated with Nef.
Lipidomic analysis of patient plasma showed a significant difference between WT HIV and uninfected subjects as well as between WT HIV and ΔNefHIV groups in several species of phosphatidylserine. The lower level of phosphatidylserine in plasma from WT HIV group relative to ΔNefHIV groups may reflect different circulating microparticle levels in these groups. Phosphatidylserine is a major lipid in the platelet plasma membrane and in platelet derived microparticles, thus the lower level observed in plasma of WT HIV group may reflect a role of Nef in suppressing platelet activation and/or microparticle production. Also of interest is the lipid species that show a significant difference between the uninfected group and the ΔNefHIV group, but no difference between uninfected and WT HIV groups. These lipids include primarily sphingolipids (dihexosylceramide, ceramide and sphingomyelin) which were elevated in the ΔNefHIV infected patients. We have previously reported that ceramides, while not associated with HIV itself, are positively associated with risk of future cardiovascular events in HIV infected individuals, and that sphingomyelin, which was negatively associated with WT HIV, also showed a positive association with future cardiovascular events [16].
Profiling of plasma miRNA showed that the abundance of 24 miRNAs was likely dependent on the presence of Nef. Predicted targets of these miRNAs included ABCA1 itself (hsa-miR-199b-3p) and two factors directly involved in regulation of ABCA1 abundance, ABCA12 (hsa-miR-409-3p) [26] and calnexin (hsa-miR-320a) [27]. MicroRNAs apparently regulated by Nef were also predicted to target a number of other ABC transporters (ABCA10, ABCA13, ABCB9), lipoprotein receptors (LDL receptor, VLDL receptor, several members of LDL receptor related protein family) and ligands (apolipoprotein A-II) and enzymes involved in lipid biosynthesis (members of long-chain acyl-CoA synthetase family, neutral sphingomyelinase). Fifteen of these miRNAs, or their close homologs, were also found to be regulated by Nef in exosomes released from Nef-transfected cells [23], although the direction of change was not always the same. Despite relatively large changes in the abundance of these miRNAs, this did not cause profound changes in plasma lipoprotein profile. However, the target cells of these miRNA are not necessarily the cells involved in regulation of plasma lipoprotein metabolism, but may be cells where changes in intracellular cholesterol metabolism play a role in pathogenesis of common complications of HIV, such as cells of vessel wall involved in atherosclerosis or β-cells involved in diabetes. Thus, within a limitation of analysis restricted to circulating miRNAs and the fact that not all targets were validated, our findings are consistent with a hypothesis that alteration of expression and/or release of miRNAs may be an important mechanism by which HIV Nef dysregulates lipid metabolism in HIV disease.
5. Limitations
This study has several important limitations. First, the group size of ΔNefHIV infected patients was small. Unfortunately, the cohort of these patients is unique and no more patients infected with ΔNefHIV are known to us, therefore, this cohort could not be expanded. A change in the treatment guidelines shifting initiation of treatment to primary care also prevented expansion of the matching treatment-naïve WT HIV group. The ΔNefHIV group included two female patients, but both were of postmenopausal age, therefore, this should not have greatly affected the results. Plasma samples from patients of ΔNefHIV group were collected at a different time and by different operators and were stored for longer time compared to samples in WT HIV group, potentially contributing to the observed variability. Finally, infection with Nef-deficient HIV resulted in much lower viral loads and in a milder presentation of several elements of immunologic dysfunction compared to patients infected with WT HIV. This makes it difficult to distinguish between the direct effects of the deficiency of Nef and indirect effects of milder impairment of immunologic status and lower viral load. This compromise is difficult to avoid as viral loads similar to those in patients infected with ΔNefHIV are only achievable after effective treatment with HAART, which would represent an even bigger confounding factor. Despite these limitations, the findings of this study support the hypothesis that Nef secreted from HIV infected cells plays, directly or indirectly, a major role in pathogenesis of metabolic complications of HIV disease.
Supplementary Material
Highlights.
Patients infected with Nef-deficient strain of HIV(ΔNefHIV) have reduced prevalence of hypoalphalipoproteinema
Several lipid classes in plasma lipidomic profile are different in patients infected with WT HIV and ΔNefHIV
HIV induced changes in profile of lipid metabolism related microRNA are attenuated in ΔNefHIV patients
Findings are consistent with Nef playing a significant role in lipid abnormalities in HIV patients
Acknowledgments
Funding sources
This study was supported by grants from the National Health and Medical Research Council of Australia (GNT1019847) and NIH (HL101274) to DS and MB and in part by the Victorian Government’s OIS Program. DS, PM and AH are Fellows of the National Health and Medical Research Council of Australia.
Footnotes
Conflict of interests
The authors have no conflict of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sudano I, Spieker LE, Noll G, Corti R, Weber R, Luscher TF. Cardiovascular disease in HIV infection. Am Heart J. 2006;151:1147–1155. doi: 10.1016/j.ahj.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 2.Glesby MJ. Coronary heart disease in HIV-infected patients, Curr. HIV/AIDS Rep. 2005;2:68–73. doi: 10.1007/s11904-005-0021-7. [DOI] [PubMed] [Google Scholar]
- 3.Rose H, Hoy J, Woolley I, Tchoua U, Bukrinsky M, Dart A, Sviridov D. HIV infection and high density lipoprotein metabolism. Atherosclerosis. 2008;199:79–86. doi: 10.1016/j.atherosclerosis.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guaraldi G, Zona S, Orlando G, Carli F, Ligabue G, Fiocchi F, Rossi R, Modena MG, Raggi P. Progression of coronary artery calcium in men affected by human immunodeficiency virus infection. Int J Cardiovasc Imaging. 2012;28:935–941. doi: 10.1007/s10554-011-9898-y. [DOI] [PubMed] [Google Scholar]
- 5.Mujawar Z, Rose H, Morrow MP, Pushkarsky T, Dubrovsky L, Mukhamedova N, Fu Y, Dart A, Orenstein JM, Bobryshev YV, Bukrinsky M, Sviridov D. Human Immunodeficiency Virus Impairs Reverse Cholesterol Transport from Macrophages. PLoS Biol. 2006;4:e365. doi: 10.1371/journal.pbio.0040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asztalos BF, Mujawar Z, Morrow MP, Grant A, Pushkarsky T, Wanke C, Shannon R, Geyer M, Kirchhoff F, Sviridov D, Fitzgerald ML, Bukrinsky M, Mansfield KG. Circulating Nef induces dyslipidemia in simian immunodeficiency virus-infected macaques by suppressing cholesterol efflux. J Infect Dis. 2010;202:614–623. doi: 10.1086/654817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui HL, Ditiatkovski M, Kesani R, Bobryshev YV, Liu Y, Geyer M, Mukhamedova N, Bukrinsky M, Sviridov D. HIV protein Nef causes dyslipidemia and formation of foam cells in mouse models of atherosclerosis. FASEB J. 2014;28:2828–2839. doi: 10.1096/fj.13-246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verity EE, Zotos D, Wilson K, Chatfield C, Lawson VA, Dwyer DE, Cunningham A, Learmont J, Dyer W, Sullivan J, Churchill M, Wesselingh SL, Gabuzda D, Gorry PR, McPhee DA. Viral phenotypes and antibody responses in long-term survivors infected with attenuated human immunodeficiency virus type 1 containing deletions in the nef and long terminal repeat regions. J Virol. 2007;81:9268–9278. doi: 10.1128/JVI.00650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaunders JJ, Geczy AF, Dyer WB, McIntyre LB, Cooley MA, Ashton LJ, Raynes-Greenow CH, Learmont J, Cooper DA, Sullivan JS. Effect of long-term infection with nef-defective attenuated HIV type 1 on CD4+ and CD8+ T lymphocytes: increased CD45RO+CD4+ T lymphocytes and limited activation of CD8+ T lymphocytes, AIDS Res. Hum Retroviruses. 1999;15:1519–1527. doi: 10.1089/088922299309801. [DOI] [PubMed] [Google Scholar]
- 10.Learmont JC, Geczy AF, Mills J, Ashton LJ, Raynes-Greenow CH, Garsia RJ, Dyer WB, McIntyre L, Oelrichs RB, Rhodes DI, Deacon NJ, Sullivan JS. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. [DOI] [PubMed] [Google Scholar]
- 11.Gorry PR, Churchill M, Learmont J, Cherry C, Dyer WB, Wesselingh SL, Sullivan JS. Replication-dependent pathogenicity of attenuated nef-deleted HIV-1 in vivo. J Acquir Immune Defic Syndr. 2007;46:390–394. doi: 10.1097/QAI.0b013e31815aba08. [DOI] [PubMed] [Google Scholar]
- 12.Gorry P, McPhee D, Verity E, Dyer W, Wesselingh S, Learmont J, Sullivan J, Roche M, Zaunders J, Gabuzda D, Crowe S, Mills J, Lewin S, Brew B, Cunningham A, Churchill M. Pathogenicity and immunogenicity of attenuated, nef-deleted HIV-1 strains in vivo. Retrovirology. 2007;4:66. doi: 10.1186/1742-4690-4-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose H, Low H, Dewar E, Bukrinsky M, Hoy J, Dart A, Sviridov D. The effect of HIV infection on atherosclerosis and lipoprotein metabolism: A one year prospective study. Atherosclerosis. 2013;229:206–211. doi: 10.1016/j.atherosclerosis.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sviridov D, Chin-Dusting J, Nestel P, Kingwell B, Hoang A, Olchawa B, Starr J, Dart A. Elevated HDL Cholesterol is Functionally Ineffective in Cardiac Transplant Recipients: Evidence for Impaired Reverse Cholesterol Transport. Transplantation. 2006;81:361–366. doi: 10.1097/01.tp.0000197556.83675.a6. [DOI] [PubMed] [Google Scholar]
- 15.Hoang A, Drew BG, Low H, Remaley AT, Nestel P, Kingwell BA, Sviridov D. Mechanism of cholesterol efflux in humans after infusion of reconstituted high-density lipoprotein. Eur Heart J. 2012;33:657–665. doi: 10.1093/eurheartj/ehr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong G, Trevillyan JM, Fatou B, Cinel M, Weir JM, Hoy JF, Meikle PJ. Plasma lipidomic profiling of treated HIV-positive individuals and the implications for cardiovascular risk prediction. PLoS One. 2014;9:e94810. doi: 10.1371/journal.pone.0094810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui HL, Grant A, Mukhamedova N, Pushkarsky T, Jennelle L, Dubrovsky L, Gaus K, Fitzgerald ML, Sviridov D, Bukrinsky M. HIV-1 Nef mobilizes lipid rafts in macrophages through a pathway that competes with ABCA1-dependent cholesterol efflux. J Lipid Res. 2012;53:696–708. doi: 10.1194/jlr.M023119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meikle P, Low H, Churchill MJ, Bukrinsky M, Sviridov D. Lipidomic profile of plasma from patients infected with wild type and nef-deficient HIV-1 strain. Data in Brief. 2015 doi: 10.1016/j.dib.2015.11.067. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arora VK, Fredericksen BL, Garcia JV. Nef: agent of cell subversion, Microbes. Infect. 2002;4:189–199. doi: 10.1016/s1286-4579(01)01527-1. [DOI] [PubMed] [Google Scholar]
- 20.Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic. 2010;11:110–122. doi: 10.1111/j.1600-0854.2009.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, Green LA, Gupta SK, Kim C, Wang L, Almodovar S, Flores SC, Prudovsky IA, Jolicoeur P, Liu Z, Clauss M. Transfer of Intracellular HIV Nef to Endothelium Causes Endothelial Dysfunction. PLoS One. 2014;9:e91063. doi: 10.1371/journal.pone.0091063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiao X, He B, Chiu A, Knowles DM, Chadburn A, Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells, Nat. Immunol. 2006;7:302–310. doi: 10.1038/ni1302. [DOI] [PubMed] [Google Scholar]
- 23.Aqil M, Naqvi AR, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J Extracell Vesicles. 2014;3:23129–23140. doi: 10.3402/jev.v3.23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du XM, Kim MJ, Hou L, Le Goff W, Chapman MJ, Van Eck M, Curtiss LK, Burnett JR, Cartland SP, Quinn CM, Kockx M, Kontush A, Rye KA, Kritharides L, Jessup W. HDL Particle Size Is a Critical Determinant of ABCA1-Mediated Macrophage Cellular Cholesterol Export, Circ. Res. 2015;116:1133–1142. doi: 10.1161/CIRCRESAHA.116.305485. [DOI] [PubMed] [Google Scholar]
- 25.Siegel MO, Borkowska AG, Dubrovsky L, Roth M, Welti R, Roberts AD, Parenti DM, Simon GL, Sviridov D, Simmens S, Bukrinsky M, Fitzgerald ML. HIV infection induces structural and functional changes in high density lipoproteins. Atherosclerosis. 2015;243:19–29. doi: 10.1016/j.atherosclerosis.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, Mukhamedova N, Ip S, D’Souza W, Henley Katya J, DiTommaso T, Kesani R, Ditiatkovski M, Jones L, Lane Rachael M, Jennings G, Smyth Ian M, Kile Benjamin T, Sviridov D. ABCA12 Regulates ABCA1-Dependent Cholesterol Efflux from Macrophages and the Development of Atherosclerosis. Cell Metabol. 2013;18:225–238. doi: 10.1016/j.cmet.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Jennelle L, Hunegnaw R, Dubrovsky L, Pushkarsky T, Fitzgerald ML, Sviridov D, Popratiloff A, Brichacek B, Bukrinsky M. HIV-1 Protein Nef Inhibits Activity of ATP-binding Cassette Transporter A1 by Targeting Endoplasmic Reticulum Chaperone Calnexin. J Biol Chem. 2014;289:28870–28884. doi: 10.1074/jbc.M114.583591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.