Full-body 56Fe-irradiation showed the greatest level of gene modulation and key transcriptional regulatory nodes were identified. Validation of networks highlighted a unique and long-lasting molecular response in cardiomyocytes. Our findings infer potential novel biomarkers for prediction of long-term cardiovascular risks in radiotherapy patients and astronauts during exploration-type space missions.
Keywords: cardiac physiology, cardiomyocyte, radiation biology, gene expression molecular signaling
Abstract
There are 160,000 cancer patients worldwide treated with particle radiotherapy (RT). With the advent of proton, and high (H) charge (Z) and energy (E) HZE ionizing particle RT, the cardiovascular diseases risk estimates are uncertain. In addition, future deep space exploratory-type missions will expose humans to unknown but low doses of particle irradiation (IR). We examined molecular responses using transcriptome profiling in left ventricular murine cardiomyocytes isolated from mice that were exposed to 90 cGy, 1 GeV proton (1H) and 15 cGy, 1 GeV/nucleon iron (56Fe) over 28 days after exposure. Unsupervised clustering analysis of gene expression segregated samples according to the IR response and time after exposure, with 56Fe-IR showing the greatest level of gene modulation. 1H-IR showed little differential transcript modulation. Network analysis categorized the major differentially expressed genes into cell cycle, oxidative responses, and transcriptional regulation functional groups. Transcriptional networks identified key nodes regulating expression. Validation of the signal transduction network by protein analysis and gel shift assay showed that particle IR clearly regulates a long-lived signaling mechanism for ERK1/2, p38 MAPK signaling and identified NFATc4, GATA4, STAT3, and NF-κB as regulators of the response at specific time points. These data suggest that the molecular responses and gene expression to 56Fe-IR in cardiomyocytes are unique and long-lasting. Our study may have significant implications for the efforts of National Aeronautics and Space Administration to develop heart disease risk estimates for astronauts and for patients receiving conventional and particle RT via identification of specific HZE-IR molecular markers.
NEW & NOTEWORTHY
Full-body 56Fe-irradiation showed the greatest level of gene modulation and key transcriptional regulatory nodes were identified. Validation of networks highlighted a unique and long-lasting molecular response in cardiomyocytes. Our findings infer potential novel biomarkers for prediction of long-term cardiovascular risks in radiotherapy patients and astronauts during exploration-type space missions.
it is estimated that stroke and heart disease account for one-third the number of ionizing radiation-associated excess deaths in atomic bomb (A-bomb) survivors compared with cancer (48). There was a significant increase in noncancer-associated diseases decades after exposure of a single acute dose in A-bomb survivors that includes heart disease, stroke, digestive diseases, and respiratory diseases (59). Even moderate doses of 0.5–2 Gy have been indicated to increase the incidence of heart disease in A-bomb survivors. However, it is unclear if low-dose radiation poses long-term degenerative risks, as other studies have shown no increases in cardiovascular (CV) disease risk with significant association that can be attributed to radiation exposure in the A-bomb studies (77). The potential association of increased CV risk at low-dose exposure must be investigated (3). Furthermore, the time-dependent latency of significant biological effects of high charge and energy (HZE) radiation on cardiac tissue requires further exploration to understand the risk of human exposure to low doses of space-type ionizing particle radiation.
The effect of cosmic radiation exposure to HZE nuclei in humans is largely unknown and is based on data extrapolated from low-linear energy transfer (LET) radiation. An individual in interplanetary space may be exposed to ∼1–2 mSv of radiation per day from a mixed field of cosmic rays (16). While intracellular effects such as DNA damage and repair and cell cytotoxicity have been studied, a complete understanding of HZE particles on the heart and other specialized cell types, as well as various organ tissues in the body, is lacking. Additional exposure studies are also needed since HZE particles are now being used as part of treatment regimens in cancer radiotherapy (RT) patients. Thus a more extensive study of HZE particles and their relationship to CV function and disease is warranted.
It is well recognized that irradiation (IR)-induced CV disease is a side effect of RT (1, 4, 9, 11, 14, 26, 34, 36, 39, 44, 51, 56, 63, 65, 69, 71, 72, 74). A more recent epidemiological study of 2,168 women who underwent RT for breast cancer between 1958 and 2001 in Europe has shown that the rates of major coronary events increase linearly with the mean dose to the heart by 7.4% per Gy, with no apparent lower or upper threshold (overall average of the mean doses to the whole heart was 4.9 Gy, range 0.03–27.72 Gy) (17). The absence of no apparent lower or upper threshold is a clear indication that IR dose is not the only defining factor for the harmful effects of IR, and the responses are unlikely to be linear.
Multiple factors may be involved in the development of CV diseases following IR exposure, which may be compounded by other factors during spaceflight. For instance, MRI measurements obtained from four astronauts following a 10-day space mission revealed an ∼12% decrease in left ventricular (LV) mass (10, 15), most likely due to effects of zero or near zero gravity. Changes in gravity could also lead to transient ischemia that may affect heart function and produce major changes in the heart perfusion (61). This means that major cell types in the heart may play a role in tissue remodeling and IR responses that are critical in maintaining the heart function.
Cardiomyocytes (CMs) are the basic contractile cells within the heart, whose function directly influences the pathogenesis of heart disease and the development of heart failure (75). Examination of the molecular response in CMs after insults caused by exposure to IR, such as particle radiation (proton and iron) at low doses, is important for our understanding of CV system function during and after exploration-type space missions. It is well known that, at both low and high doses, the activation of multiple biological pathways by IR is dose dependent (2, 76). Previous studies have characterized high-dose effects in CMs, which are functionally IR resistant to acute exposures (35, 58). Gene expression in CMs at high doses of IR has shown that the primary response is to upregulate gene transcription (7). There is little information available on how low doses of particle radiation may affect CMs or what regulatory mechanisms are responsive.
The objective of the present study is to characterize the effects of whole body radiation exposure to low-dose, high-energy proton (90 cGy, 1 GeV) and low-dose HZE particle iron [15 cGy, 1 GeV/nucleon (n)] on the gene expression and regulatory pathways in isolated LV CMs using a murine model with sampling at 1, 3, 7, 14, and 28 days after exposure.
MATERIALS AND METHODS
IR and Dosimetry
Whole body IR of adult (8–9 mo old) male C57Bl/6NT (Taconic) mice was performed at the NASA Space Radiation Laboratory (NSRL) at Brookhaven National Laboratory (Upton, NY). Using dose rates between 0.05 and 0.17 Gy/min, iron and proton, respectively, 0.15 Gy was delivered using 1 GeV/n iron ions (LET ∼151 keV/μm) or 0.9 Gy at 1 GeV energy protons (LET ∼0.22 keV/μm). All mice were handled in accordance with the guidelines set, reviewed, and approved by the International Animal Care and Use Committees (IACUC) at both GeneSys Research Institute and Brookhaven National Labs.
Cardiac Cell Isolation
Primary LV cardiac cells from 8–9 mo old C57Bl/6NT mice were isolated using standard preparation protocol that included cannulation of the aorta and collagenase digestion, followed by a Ca2+ gradient selection (25, 32, 33, 42). This procedure yielded >90% of Ca2+-tolerant CMs. The resulting cell suspension could contain a small population of inflammatory and/or endothelial cells. Following isolation of CMs, RNA was extracted via TRIzol (Life Technologies, Grand Island, NY), according to the manufacturer's instructions. RNA quality and quantity were assessed using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Gene Expression Microarrays and Data Analysis
Expression arrays were chosen to delineate differences between controls (sham IR) and IR samples (protons or iron particle IR vs. sham IR samples). The arrays were also selected to compare proton vs. iron IR samples across same time points. For genomewide expression profiling, the Affymetrix GeneChip 1.0ST array system was used at the Boston University School of Medicine Microarray Core Facility. For iron IR samples, there were a total of 15 microarrays that were from individual mouse samples separated into three per group for the non-IR controls, day 1 as well as day 3 after IR (3 replicate arrays), or two per group (2 replicate arrays) at days 7, 14, or 28 after IR. For proton samples, a total of 14 microarrays included three mice per group for non-IR controls and day 3 after IR (3 replicate arrays), or two mice per group at days 1, 7, 14, or 28 after IR (2 replicate arrays). The log2 transformation data were normalized and background corrected. Statistical analysis was performed using a one-way ANOVA. A cutoff P value <0.05 resulted in 5,220 genes for the iron and 1,651 genes for the proton datasets. Adjustment with the Benjamini-Hochberg correction method for a false discovery rate (FDR) <10% resulted in 1,538 genes in the iron dataset (5). The data were then filtered for all samples with an average signal above background noise using a minimum cutoff value of 20 and at least one sample group with a >1.5-fold change up- or downregulated compared with the non-IR control, resulting in 400 unique genes. There were, however, no genes in the proton dataset that satisfied the FDR cutoff <10%. Raising this threshold to FDR <33% resulted in only 133 genes, but additional filtering for an average signal >20 for all samples and greater than ±1.5-fold-change reduced this gene list to only 21 genes. Analysis for iron vs. proton also used the log2 transformation of data for robust means analysis normalized and background corrected microarray signals. Statistical analysis focused on filtering data with >1.5-fold up- or downregulated and a FDR = 5% to identify significant genes for pathway analysis, as described below. All microarray data were uploaded to the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE68876.
Biological Pathway Analysis
Functional analysis and predicted upstream regulators were performed through the use of Ingenuity Pathway Analysis (IPA) core analysis software (Ingenuity Systems, http://www.ingenuity.com) and Database for Annotation, Visualization and Integrated Discovery (DAVID; at https://david.ncifcrf.gov) (29). Limited pathway analysis was performed on the proton dataset due to a small number of genes meeting the statistical significance cutoff. The 400 significant genes list from the iron dataset were uploaded and resulted in a total of 381 annotated genes. The core analysis performed for each time point resulted in 164, 215, 193, 83, and 162 gene lists for days 1, 3, 7, 14, or 28, respectively. The TFactS website (http://www.tfacts.org) (23) was used to predict which transcription factors (TFs) were regulated based on the total lists of upregulated and downregulated iron-responsive genes generated across the microarray experiments. Significant TFs and potential genes regulated were identified based on a multiple parameter P < 0.05. Functional categories or individual upstream regulators with a P < 0.05 and a z-score above +2.0 (activated) or below −2.0 (inhibited) were considered significant within IPA.
Western Blot Analysis
A portion of the LV (3 × 3 mm) was collected from non-IR/control and iron-IR mice on days 7, 14, and 28 and was lysed using RIPA lysis buffer (Fisher Scientific, Pittsburgh, PA) to prepare whole tissue lysates. Total protein was quantified using Bradford assay analysis so as to load equal amounts of 50 μg total protein for each sample. Lysates were then assayed by Western blotting after SDS-PAGE analysis using phosphorylation-specific NFATc4 (nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 4) (Ser168/170, Santa Cruz Biotechnology, Santa Cruz, CA), p38 (Thr180/Tyr182), JNK (Thr182/Tyr185), and ERK1/2 (p44/42, all antibodies from Cell Signaling, Danvers, MA), GATA4 (Ser105, from SAB Signalway Antibody, College Park, MD), and STAT3 (Tyr705, NeoBiolab, Woburn, MA) primary antibodies. We also assayed for specific total protein levels using antibodies against NFATc4 (Santa Cruz Biotechnology), p38, JNK, and ERK1/2 (all antibodies from Cell Signaling), and GATA4 and STAT3 (both antibodies from Bethyl Laboratories, Montgomery, TX). Proteins were visualized using a goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (Cell Signaling) and ECL enhanced chemiluminescence (Amersham).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) analysis was performed using the method previously described (52, 81). Nuclear extracts were prepared from heart tissue of iron-IR or sham-IR mice. Tissue pieces were homogenized in lysis buffer containing 10 mM HEPES, pH 8.0, 10 mM KCl, 2 mM MgCl2, 1 mM dithiothrietol, 0.1 mM EDTA, 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 10 μl of protease inhibitor (Sigma-Aldrich), and 0.2% Nonidet P-40. The total protein concentration in each sample was measured using the bicinchoninic acid method following the manufacturer's protocol (Pierce, Rockford, IL). The EMSA was performed using a double-stranded oligonucleotides of nuclear factor (NF)-κB, STAT3, and GATA4 (NF-κB: 5′-AGT TGA GGG GAC TTT CCC ACG C-3′; STAT3: 5′-GAT CCT TCT GGG AAT TCC TAG ATC-3′, and GATA4: 5′-CAC TTG ATA ACA GAA AGT GAT AAC TCT-3′) end-labeled with T4 polynucleotide kinase. Super shift assays were performed using rabbit polyclonal antibodies raised against 1) a peptide mapping the COOH-terminus of NF-κB p65 (Santa Cruz Biotechnology); 2) amino acid residues of 675–725 of full-length STAT3 (Bethyl Laboratories, Montgomery, TX); and 3) amino acid residues of 392–442 of full-length GATA4 (Bethyl Laboratories, Montgomery, TX). The nuclear extract of heart samples from mice that are either sham-IR or exposed to iron-IR was preincubated with 1 μg of antibody against either NF-κB, GATA4, or STAT3 for 20 min at room temperature before the addition of γ-32P-labeled consensus oligonucleotides of respective TF. For competition assay, the nuclear extracts (20 μg) were preincubated with homologous unlabeled NF-κB, STAT3, or GATA4 oligonucleotide (in excess of 50–100 times of the labeled probe concentration) for 5 min on ice, followed by addition of γ-32P-labeled probes of respective TFs. All samples were electrophoresed at 100 V through 6% polyacrylamide gels in Tris-glycine buffer (25 mM Tris, 190 mM glycine, and 1 mM EDTA). DNA binding activity of TFs was estimated by subjecting the dried gel to phosphor imaging using a Bio-Rad Phosphor Imaging System (GS-525; Bio-Rad, Hercules, CA) or to expose to X-ray film and semiquantitated using National Institutes of Health (NIH) Image J integrated density program.
Cardiac Physiology Studies
Echocardiography.
Animals were lightly anesthetized with isoflurane vaporized in O2 (1.5–2%) at the rate of 1 l/min using a nose cone on a warming pad post-body weight measurement. Two-dimensional guided M-mode echocardiography was performed with a 15-MHz (15-6L) pediatric open heart surgery transducer (Agilent Technologies, Santa Clara, CA), as detailed earlier (19). At least five sequential beats were analyzed (n = 6–8/group). Heart rate, LV end-diastolic diameter (EDD) and end-systolic diameter (ESD), and LV wall thickness were measured. LV ejection fraction percent (EF%) and fractional shortening percent (FS%) were calculated using standard formulas EF% = (EDV − ESV)/EDV × 100 and FS% = (EDD − ESD)/EDD × 100.
Hemodynamic measurements.
For hemodynamic analysis post-IR, in vivo LV pressure measurements were performed by direct LV catheterization using Millar Mikro-Tip Blood Pressure System (1.2F, SciSense, Ithaca, NY) (n = 6–8/group), as described previously (19). Heart rate, LV systolic pressure, LV end-diastolic pressure, maximum change in pressure over time, and minimum change in pressure over time were also recorded.
Cardiac fibrosis (Masson's Trichrome staining).
To determine the effects of low-dose 56Fe-IR on IR-induced cardiac fibrosis 1 mo post-IR, serial 10-μm sections of cardiac tissue were processed for Masson's Trichrome staining (Electron Microscopy Sciences, Hatfield, PA), and random regions of the heart were imaged at ×200 (n = 20 images/sample per group) to be analyzed using Image-J program (version 1.40, NIH) to quantify for percentage of fibrosis (blue pixels).
Immunofluorescence Staining and Analysis
Ventricular tissue cross sections (6–8 μm) of OCT embedded heart tissue from sham-IR and 56Fe ion-IR mice, collected at 1, 7, 14, and 28 days post-IR, were fixed in 4% paraformaldehyde for 15 min at room temperature and washed with 1× phosphate-buffered saline for 5 min. Sections were permeabilized with 0.1% Triton X-100 (Sigma, St. Louis, MO) for 20 min at room temperature and washed three times in 1× phosphate-buffered saline for 5 min. To evaluate inflammatory infiltration, the expression of CD68, a glycoprotein normally expressed on macrophages, also known in mice as macrosialin, was quantified. Heart tissue sections were stained with rat anti-mouse CD68 monoclonal antibody (AbD Serotec, Raleigh, NC), along with Alexa-488 goat anti-rat secondary antibody (Life Technologies). Topro-3 was used to visualize nuclei (Life Technologies). The immunostained sections were examined using laser scanning confocal microscope (LSM 510 Meta, ZEISS, Thornwood, NY) at ×400 magnification. Data was obtained from three replicate samples for sham-IR and 56Fe ion-IR cardiac tissues at each time point. Using a computer-assisted image analysis algorithm based on pixel and color distribution for CD68, staining was evaluated and graphs were plotted.
Quantitative RT-PCR Analysis
Total RNA isolated from CMs, as detailed in the section above, were converted to cDNA using the TaqMan Reverse Transcription Kit (Life Technologies). The quantitative RT (qRT)-PCR was performed on five target genes [insulin-like growth factor-binding protein 6 (IGFBP6), chemokine (C-C motif) receptor 9 (CCR9), a disintegrin and metalloprotease 19 (ADAM19), D site-binding protein (DBP), and GAPDH] to confirm our findings in gene expression microarray analysis for CMs isolated from 15 cGy full-body 56Fe-IR mice at various time points postexposure. The samples were analyzed using Applied Biosystems 7300 Real-Time PCR machine and software.
Statistical Analysis
Data are expressed as means ± SE. Significant differences were evaluated using the Student t-test, one-way ANOVA, right-tailed Fisher's exact test, and χ2 when appropriate.
RESULTS
Significant Changes in Transcriptional Activity over 1 mo after Low-Dose High-Energy Particle Radiation Exposure
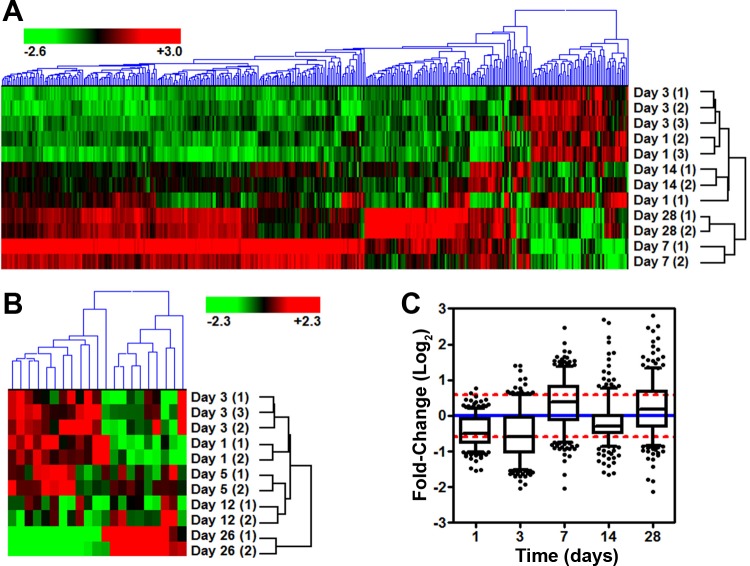
Primary ventricular CMs were isolated to examine the temporal response following whole body in vivo exposure of adult 8- to 9-mo-old C57Bl/6NT mice to 15 cGy of 1 GeV/n iron ions or 90 cGy of 1 GeV protons. A total of 29 microarrays were used to analyze global gene expression changes. Following standard preprocessing methods, P values were corrected for multiple hypothesis testing (FDR < 10%), and genes were chosen that displayed significant expression above background as well as displayed at least a ±1.5-fold change. This resulted in 400 significantly expressed genes for the iron dataset (Fig. 1A, ANOVA P value < 7.22 × 10−3; FDR < 10%). The majority of these genes displayed a clear increase on days 7 and 28 that was absent on days 1, 3, and 14 (Fig. 1, A and C). No genes were significantly expressed in the proton dataset with the same filtering criteria used for iron exposure. In fact, raising the FDR cutoff (FDR < 33%) resulted in only 21 genes for proton dataset (Fig. 1B). Furthermore, there were only four genes in common (Car14, Dbp, Lcn2, and Trim7) between the iron's 400 gene set (FDR < 10%) and the proton's 21 gene set (FDR < 33%). This data highlights the very distinct biological response to low-dose (15 cGy, 1 GeV/n) iron particle IR.
Fig. 1.
Gene microarray results for proton and iron irradiations (IRs). Hierarchical clustering using an Euclidean algorithm for gene lists with a fold-change greater than ±1.5 from iron-IR samples with a false discovery rate (FDR) < 0.1 (A), or proton samples with an FDR < 0.33 (B) are shown. Relative fold-change to the non-IR control is shown. C: plot of the average fold-change distribution by day with the 400-gene list from the iron-IR. Whiskers show the 5th to 95th percentile with outliers (solid circles). Red dashed lines show ±1.5 fold-change, and blue dash line shows no change compared with non-IR control samples.
Delayed Increase in Inflammatory Responses after 56Fe-IR over 1 mo
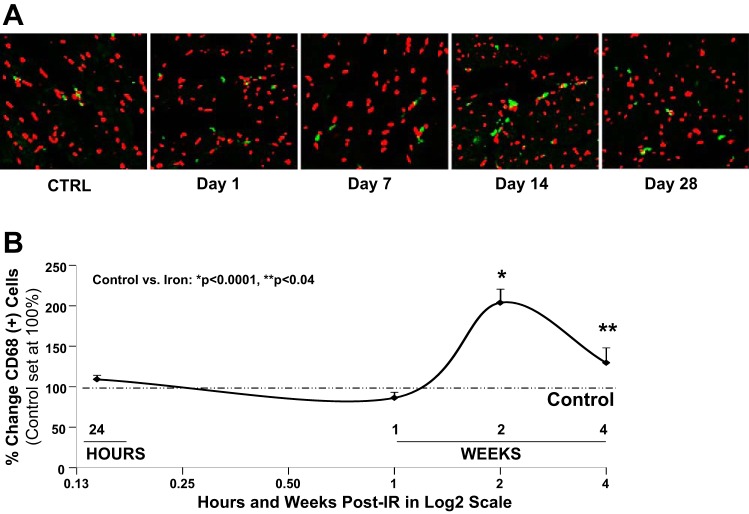
CD68 analysis for macrophage infiltration in heart tissue of mice exposed to full-body, low-dose 56Fe-IR demonstrated a delayed yet significant increase in inflammatory response by days 14 and 28 post-IR (Fig. 2). By day 14, there was a twofold increase in inflammatory cells compared with control, followed by a slight decrease by day 28, which was still ∼35% higher than control tissue (Fig. 2). The increase in inflammatory responses weeks after initial exposure may lead to release of various cytokines, superoxide, nitric oxide, and other signaling molecules by immune cells (i.e., macrophages), which are capable of causing oxidative tissue damage (50).
Fig. 2.
Inflammatory response in heart tissue after low-dose, full-body 56Fe-IR. A: representative ×400 confocal microscopy images for immunostaining with macrophage marker CD68 (green) and Topro-3 stained nuclei (red) in heart tissue after a single, low-dose, full-body 56Fe-IR in mice on days 1, 7, 14, and 28 post-IR, along with respective non-IR controls (CTRL). Note, when CD68-green cytoplasmic staining is overlaid with nuclei red staining, it appears as yellow/green staining. B: graphic representation of %change in CD68 positive (+) cells in the heart tissue of non-IR control and 56Fe-IR mice at 24 h and up to 28 days post-IR. CD68 (+) cells in control hearts were set at 100% (dash-dotted line). Values are means ± SE; n = 6–8 animals per time point/group for sham-IR controls and 56Fe-IR groups. Statistical significance was assigned when P < 0.05. *P < 0.0001 and **P < 0.04 for control vs. 56Fe-IR. [These data are a revised interpretation of previously published work (78), with permission.]
Functional Analysis Reveals Time-Dependent Responses
To better understand the biological implications of the gene expression results, IPA's functional annotation library was used to analyze the 381 annotated genes of the 400 differential iron gene set. The analysis identified categories that were most statistically significant to the data set using a P value threshold and predicted activation or inhibition of that category by z-score (Table 1). As might be expected with radiation exposure, there was an increase in cell death, necrosis, and apoptosis appearing 7 days after IR that was reversed at days 14 and 28, when apoptotic-related genes were decreased and survival was increased. Free radical scavenging production was also found to be increased on days 7 and 28, while earlier on day 3, synthesis and production were decreased. There were also a significant number of genes associated with inflammation found in these categories. On days 1 and 3, there was an inhibition in genes with a functional association with arthritis, rheumatic disease, and inflammation. This was in contrast to the increase in this category seen on day 7 after IR.
Table 1.
IPA analysis identified significant functional categories modulated by iron irradiation
| Time | Functional Category (Function) | P Value | Predicted Activation | Activation z-Score |
|---|---|---|---|---|
| Day 1 | Inflammatory response (arthritis, rheumatic disease & inflammation) | 2.86E-08 | Decreased | −2.52 |
| cellular growth and proliferation (proliferation) | 2.50E-03 | Increased | 2.07 | |
| Day 3 | Cardiovascular disease (vascular disease) | 7.14E-07 | Decreased | −2.33 |
| Cellular development (differentiation) | 1.39E-07 | Decreased | −2.27 | |
| Free-radical scavenging (synthesis & production) | 4.90E-05 | Decreased | −2.70 | |
| Inflammatory response (arthritis, rheumatic disease & inflammation) | 1.67E-06 | Decreased | −2.15 | |
| Day 7 | Infectious disease (infection) | 1.75E-11 | Decreased | −2.72 |
| Cell death and survival (cell death, necrosis & apoptosis) | 5.18E-04 | Increased | 2.00 | |
| Cell-to-cell signaling and interaction (binding) | 8.08E-05 | Increased | 2.74 | |
| Free-radical scavenging (production) | 6.98E-04 | Increased | 2.002 | |
| Hematological system development and function, cellular development & tissue morphology (differentiation & quantity) | 9.27E-06 | Increased | 2.07 | |
| Inflammatory response (arthritis, rheumatic disease, inflammation & immune response) | 3.20E-05 | Increased | 2.10 | |
| Molecular transport (quantity) | 1.10E-03 | Increased | 2.28 | |
| Day 14 | Cell death and survival (apoptosis) | 9.04E-05 | Decreased | −2.21 |
| Cell-to-cell signaling and interaction (activation) | 1.50E-02 | Decreased | −2.59 | |
| Inflammatory response (immune response) | 3.09E-04 | Decreased | −2.39 | |
| Tissue development & cellular assembly and organization (formation & fibrogenesis) | 1.25E-03 | Decreased | −2.01 | |
| Day 28 | Inflammatory response (activation) | 2.18E-03 | Decreased | −2.41 |
| Hematological system development and function & tissue morphology (quantity) | 2.24E-03 | Decreased | −2.00 | |
| Cell death and survival (apoptosis) | 1.05E-02 | Decreased | −2.00 | |
| Cell death and survival (survival) | 1.76E-03 | Increased | 2.08 | |
| Cardiovascular system development and function | 8.27E-05 | Increased | 2.13 | |
| Embryonic, organismal, and organ development, & tissue development (development) | 3.02E-03 | Increased | 2.32 | |
| Free-radical scavenging (production) | 8.41E-03 | Increased | 2.15 | |
| Hematological system development and function, humoral immune response & tissue morphology (quantity) | 1.19E-04 | Increased | 2.33 |
Functional categories were examined by day and considered significant with a z-score > 2.0, activated, or below less than −2.0, inhibited. Right-tailed Fisher's exact test was used to calculate a P value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
Additionally, hematological system development and function, cell-to-cell binding, and molecular transport were also activated on day 7 (Table 1). By day 14 following exposure, there was a general lack of significantly expressed genes with predicted inhibition in functional categories, including inflammation and cell-to-cell activation. At the last time point, 28 days after IR, there were again a number of functional categories with increased as well as inhibited activity that was reminiscent of a wavelike response to high doses of low-LET gamma-ray exposures (47). Network diagrams of functional categories for cell death and survival, free radical scavenging, and inflammation were created (data not shown). The genes within these networks again emphasized early decreases in gene expression on days 1 and 3 that were increased on day 7.
To help explain the cause of the changes in gene expression, predicted upstream regulator activity was analyzed based on changes in downstream target genes identified in the 400 differential gene set. There were two notable types of upstream regulators, cytokines and transcriptional regulators, that may regulate the genes we identified. Table 2 highlights the global transcription binding factors that were indicated as playing a role in up- or downregulation after iron exposure, regardless of time after exposure. TFs were selected using http://www.tfacts.org based on inputting a gene list. Significant regulators were selected based on P < 0.05 and FDR < 10%. Table 2 identifies the number of genes within the microarray dataset, as well as the number of potential known target genes. A total of 12 major TFs were identified as significant for upregulation, while 14 factors played a role in downregulation. TFs such as Notch1, ETS1, SP1, CREB1, and RBPJ were identified on both lists, indicating a significant role in up- and downregulation, which may be associated with the cyclic nature of iron IR-associated signaling in CM over time.
Table 2.
Identification of significant transcription factors involved in iron irradiation based on global gene expression analysis
| Transcription Factor | P Value | FDR Control | Microarray, no. | Target Genes, no. |
|---|---|---|---|---|
| Upregulation | ||||
| ETS1 | 0.00E+00 | 5.26E-04 | 14 | 136 |
| CEBPA | 1.00E-05 | 1.05E-03 | 8 | 119 |
| NOTCH1 | 2.00E-05 | 1.58E-03 | 5 | 40 |
| SP1 | 7.00E-05 | 2.11E-03 | 13 | 428 |
| CREB1 | 8.00E-05 | 2.63E-03 | 9 | 211 |
| FLI1 | 9.00E-05 | 3.16E-03 | 4 | 28 |
| ELF1 | 2.30E-04 | 3.68E-03 | 2 | 3 |
| RBPJ | 2.80E-04 | 4.21E-03 | 6 | 104 |
| PPARG | 4.30E-04 | 4.74E-03 | 4 | 41 |
| RELB | 4.50E-04 | 5.26E-03 | 2 | 4 |
| ATF4 | 4.80E-04 | 5.79E-03 | 3 | 18 |
| EBF1 | 4.80E-04 | 6.32E-03 | 3 | 18 |
| Downregulation | ||||
| ETV4 | 0.00E +00 | 5.10E-04 | 6 | 31 |
| ETS1 | 0.00E +00 | 1.02E-03 | 13 | 136 |
| NOTCH1 | 1.00E-05 | 1.53E-03 | 6 | 40 |
| SP1 | 1.00E-05 | 2.04E-03 | 8 | 93 |
| POU1F1 | 2.00E-05 | 2.55E-03 | 7 | 74 |
| EBF1 | 4.00E-05 | 3.06E-03 | 4 | 18 |
| CTNNB1 | 4.00E-05 | 3.57E-03 | 13 | 306 |
| POU2F2 | 7.00E-05 | 4.08E-03 | 4 | 20 |
| CREB1 | 1.30E-04 | 4.59E-03 | 10 | 211 |
| RBPJ | 1.70E-04 | 5.10E-03 | 7 | 104 |
| FOS | 2.00E-04 | 5.61E-03 | 5 | 48 |
| ETS2 | 3.50E-04 | 6.12E-03 | 4 | 30 |
| RELA | 3.70E-04 | 6.63E-03 | 6 | 84 |
| GLI1 | 5.00E-04 | 7.14E-03 | 7 | 124 |
Iron-modulated genes are shown based on analysis within http://www.tfacts.org that identify significant transcription factors (P < 0.05) based on genes profiled on the microarray compared with possible regulated target genes.
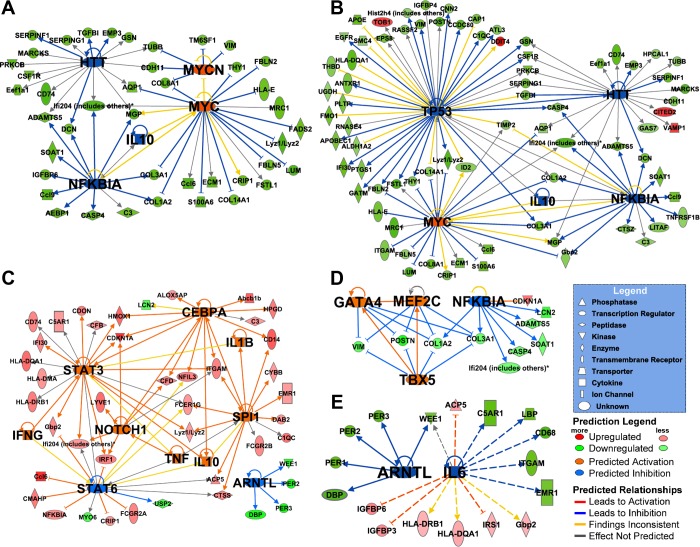
Table 3 highlights cytokines and TFs that were also indicated as playing a role in up- or downregulation after iron exposure as a function of days post-IR. Networks by day for the predicted upstream regulators and downstream targets were created (Fig. 3). The networks with a large number of affected downstream gene expression activity were found on days 1, 3, and 7 (Fig. 3, A–C). The same upstream regulators were predicted to be inhibited on days 1 and 3, such as NFKB1A (day 1, P < 4.76E-05 and day 3, P < 9.72E-06), interleukin (IL)-10 (day 1, P < 1.75E-05 and day 3, P < 3.67E-07), and HTT (day 1, P < 6.54E-05 and day 3, P < 1.44E-05), whereas MYC (day 1, P < 4.87E-06 and day 3, P < 1.73E-05) was activated on both days. The effect on downstream gene targets on days 1 and 3 clearly depicted a strong decrease in gene expression activity as an early response to iron IR. However, on day 7, a different set of upstream regulators (IFN-γ, IL-1β, TNF-α, IL-10) were predicted, most of which were activated and may induce the general increase in gene expression activity noted earlier (Fig. 3C).
Table 3.
Predicted upstream cytokine and transcriptional regulators based on dataset overlap with Ingenuity Pathway Analysis database
| z-Score (No. of Target Genes) |
||||||
|---|---|---|---|---|---|---|
| Type | Upstream Regulator | Day 1 | Day 3 | Day 7 | Day 14 | Day 28 |
| Transcriptional regulator | MYCN | +2.22 (5) | ||||
| Transcriptional regulator | TP53 | −2.05 (44) | ||||
| Transcriptional regulator | MYC | +2.85 (20) | +2.69 (22) | |||
| Transcriptional regulator | HTT | −2.00 (17) | −2.00 (21) | |||
| Transcriptional regulator | NFKBIA | −2.47 (13) | −3.21 (16) | −2.81 (8) | ||
| Cytokine | IL10 | −2.36 (12) | −2.65 (16) | +2.59 (14) | ||
| Transcriptional regulator | CEBPA | +2.57 (12) | ||||
| Cytokine | IFNG | +4.14 (48) | ||||
| Cytokine | IL1B | +3.28 (32) | ||||
| Transcriptional regulator | NOTCH1 | +2.17 (6) | ||||
| Transcriptional regulator | SFPI1 | +3.09 (12) | ||||
| Cytokine | TNF | +3.26 (52) | ||||
| Transcriptional regulator | STAT3 | +2.06 (16) | ||||
| Transcriptional regulator | STAT6 | −2.21 (14) | ||||
| Transcriptional regulator | GATA4 | +2.78 (4) | ||||
| Transcriptional regulator | MEF2C | +2.76 (4) | ||||
| Transcriptional regulator | TBX5 | +2.80 (4) | ||||
| Transcriptional regulator | ARNTL | −2.51 (4) | −2.15 (5) | |||
| Cytokine | IL6 | −2.19 (13) | ||||
A significant P value <0.05 and z-score (more than +2.0, activated, or less than −2.0, inhibited) were chosen.
Fig. 3.
Downstream gene targets of predicted cytokine and transcriptional regulators. Networks were generated using Ingenuity Pathway Analysis (IPA) for gene expression relative to non-IR control for day 1 (A), day 3 (B), day 7 (C), day 14 (D), or day 28 (E) after iron-IR. Gene expression signal is colored red (upregulated) or green (downregulated), while predicted upstream regulators have a larger font size and are colored orange (activated) or blue (inhibited).
There was a time-dependent change in gene expression that results in an overall increase in the activity of several TFs (upregulated: CEBPA, Notch1, SFPI1, STAT3; and downregulated: STAT6 and ARNTL) by day 7 post-IR (Table 3). On day 14, several developmental TFs (TBX5, GATA4, MEF2C) that are required for the maintenance of cardiac morphogenesis, myogenesis, vascular development, and protection from pressure overload-induced heart failure were activated (6, 30, 55).
To identify time-independent changes in biological pathways, we also analyzed differences between iron and proton arrays to identified 1,387 transcripts (P = 0.05 and FDR = 5%). These were further analyzed in DAVID to identify global KEGG pathways in IPA and NIH DAVID. The top five significant pathways identified were Parkinson's disease, Alzheimer's, oxidative phosphorylation, cardiac muscle contraction, and Huntington's disease, followed by hypertrophic cardiomyopathy and dilated cardiomyopathy (Table 4). These top five significant pathways shared the majority of transcripts that were involved in mitochondrial and oxidative phosphorylation function (specified in italic font in Table 4), illustrating the interrelationship between the same oxidative phosphorylation genes that play a role in neurodegenerative and CV disorders/diseases.
Table 4.
Major biological and disease-related pathways associated with iron exposures
| KEGG Pathway | Genes, No. | P Value | Genes | Benjamini and Hochberg | FDR, % |
|---|---|---|---|---|---|
| Parkinson's disease | 28 | 4.576E-13 | NDUFB7, ATP5B, COX7B, UBE2G2, CYC1, COX7C, NDUFS6,CASP9, NDUFS1, ATP5J, ATP6, ND1, COX7A1, ND2, ND3, NDUFA1, PARK7, COX6C, VDAC1, SDHA, ND4L, COX3, UQCRH,COX2, COX1, NDUFV2, COX6A2, ATP5C1 | 6.544E-11 | 5.420E-10 |
| Alzheimer's disease | 28 | 1.486E-10 | NDUFB7, ATP5B, COX7B, CYC1, COX7C, NDUFS6, CASP9, PPP3CB, GAPDH, NDUFS1, ATP5J, ATP6, LPL, COX7A1, NDUFA1, NAE1, COX6C, SDHA, ATP2A2, PSEN1,COX3, UQCRH, COX2, COX1, NDUFV2, ATP2A1, COX6A2, ATP5C1 | 1.062E-08 | 1.760E-07 |
| Oxidative phosphorylation | 25 | 2.214E-10 | NDUFB7, ATP5B, COX7B, CYC1, COX7C, ATP6V0B, NDUFS6, NDUFS1, ATP5J, ATP6, ND1, COX7A1, ND2, ND3, NDUFA1, COX6C, SDHA, ND4L, COX3, UQCRH,COX2, COX1, NDUFV2, ATP5C1, COX6A2 | 1.055E-08 | 2.622E-07 |
| Cardiac muscle contraction | 18 | 1.600E-08 | ACTC1, CACNA2D1, COX7A1, MYL2, MYL3, COX7B, CYC1, COX7C, MYH6, CACNB4, COX6C, COX3, ATP2A2, COX2, UQCRH,COX1, COX6A2, ATP6 | 5.720E-07 | 1.895E-05 |
| Huntington's disease | 24 | 5.967E-07 | POLR2E, COX7A1, NDUFB7, ATP5B, COX7B, CYC1, COX7C, NDUFA1, DCTN1, COX6C, VDAC1, SDHA, NDUFS6, COX3, CASP9,COX2, UQCRH, COX1, NDUFV2, COX6A2, ATP5C1, NDUFS1, ATP6, ATP5J | 1.707E-05 | 7.067E-04 |
| HCM | 9 | 0.013 | ACTC1, CACNA2D1, ATP2A2, MYL2, MYL3, ITGB5, MYH6, CACNB4, PRKAA2 | 0.269 | 14.438 |
| Dilated cardiomyopathy | 9 | 0.020 | ACTC1, CACNA2D1, ATP2A2, MYL2, MYL3, PLN, ITGB5, MYH6, CACNB4 | 0.343 | 21.585 |
| Wnt signaling pathway | 12 | 0.024 | PSEN1, PPP2R5B, PPP2R5C, CAMK2D, SMAD4, PPP3CB, FRAT1, FZD1, CXXC4, FBXW11, RBX1, AXIN1 | 0.354 | 25.128 |
| Glycerolipid metabolism | 6 | 0.025 | GLYCTK, LPL, DGKB, PPAP2A, ALDH3A2, AGPAT2 | 0.327 | 25.579 |
| Ubiquitin mediated proteolysis | 11 | 0.030 | RFWD2, CUL3, UBE2D3, UBE2Z, UBE3B, UBE3A, WWP1, UBE2G2, ANAPC11, FBXW11, RBX1 | 0.358 | 30.691 |
| Fatty acid elongation in mitochondria | 3 | 0.033 | PPT2, HADHA, HADHB | 0.355 | 32.954 |
| Glycerophospholipid metabolism | 7 | 0.040 | PGS1, DGKB, PLA2G12B, LYPLA2, PPAP2A, AGPAT2, PTDSS2 | 0.382 | 38.015 |
HCM, hypertrophic cardiomyopathy.
Genes listed were among the top 200 most differentially expressed genes [false discovery rate (FDR) <0.05] between all noncontrol iron microarrays and all noncontrol proton microarrays. P values are Benjamini and Hochberg (Ref. 5) P values. Genes with overrepresentation FDR < 0.05 are listed in bold.
Quantitative Real-Time PCR Confirmed MICROARRAY-Based Time-Dependent Gene Expression Responses up to 28 Days after Low-Dose High-Energy Particle Radiation Exposure in CMs
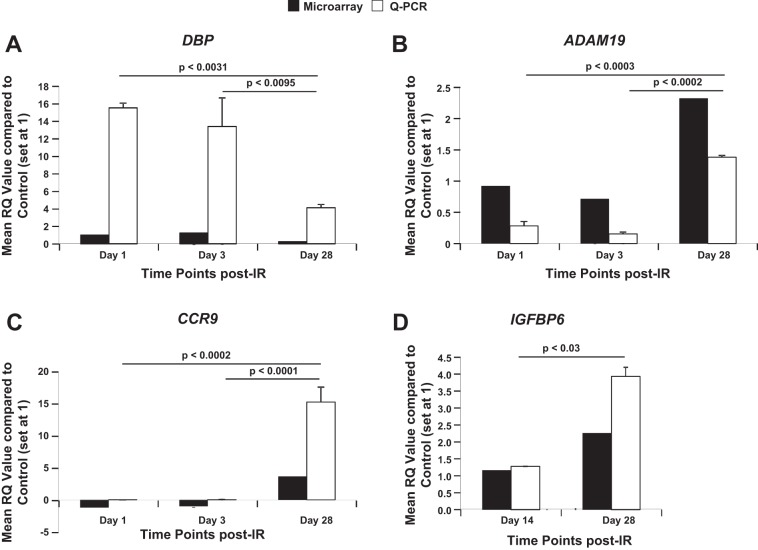
To determine the effects of full-body 56Fe-IR on expression of few selected genes, total RNA from 56Fe-IR CMs were processed for qRT-PCR. We examined the gene expression in our 56Fe-IR CM samples at 1 and 3 days, as well as 14 and 28 days, post-IR for comparison to the microarray analysis. The qRT-PCR data for DBP demonstrated an ∼3.6-fold and ∼4.4-fold decrease in expression by 28 days compared with day 1 and 3 time points post-IR, respectively (Fig. 4A, open bars). This was consistent with the trend observed in microarray analysis, which showed an ∼3.8-fold and ∼3.2-fold decrease by 28 days compared with 1- and 3-day time points, respectively (Fig. 4A, solid bars). This decrease in gene expression at 28 days by qRT-PCR analysis was significant compared with both earlier time points, 1 and 3 days. To further confirm our microarray findings, two other genes, ADAM19 and CCR9, that demonstrated an ∼2.5–3.3-fold and ∼4.1–4.7-fold increase respectively, in expression by 28 days, compared with 1- and 3-day time points (Fig. 4, B and C, solid bars), were also analyzed for qRT-PCR. As expected, the qRT-PCR data showed a similar trend in modulation of gene expression, where we observed an ∼4.8- to 8.9-fold and >100- to 150-fold increase in ADAM19 and CCR9, respectively, by 28 days compared with 1- and 3-day time points (Fig. 4, B and C, open bars). This increase in gene expression at 28 days by qRT-PCR analysis was significant compared with both earlier time points, 1 and 3 days. We also looked at the later time point gene expression for another gene, IGFBP6, which demonstrated a significant ∼3.1-fold increase by 28 days compared with 14 days (Fig. 4D, open bars). This trend for gene expression was consistent with our observations using microarrays for IGFPB6, where there was a significant approximately twofold increase by 28 days vs. 14 days (Fig. 4D, solid bars).
Fig. 4.
Quantitative real-time PCR (Q-PCR) confirmed microarray-based time-dependent gene expression responses up to 28 days post-IR. Graphic representation of Q-PCR analysis, mean relative quantification (RQ) values (open bars) compared with control (set at 1), and respective microarray data (solid bars) of cardiomyocytes from full-body 15 cGy 56Fe-IR mice at 1, 3, and 28 days post-IR for D site-binding protein (DBP; A), a disintegrin and metalloprotease 19 (ADAM19; B), and chemokine (C-C motif) receptor 9 (CCR9; C), and at 14 and 28 days for insulin-like growth factor-binding protein 6 (IGFBP6; D). Values in all graphs represent Q-PCR data as means ± SE of the pooled data from n = 3 animals per time point/group for non-IR control and 56Fe-IR groups. Statistical significance was assigned when P < 0.05.
Activation of TFs Play a Role in Gene Regulation after Iron Exposure
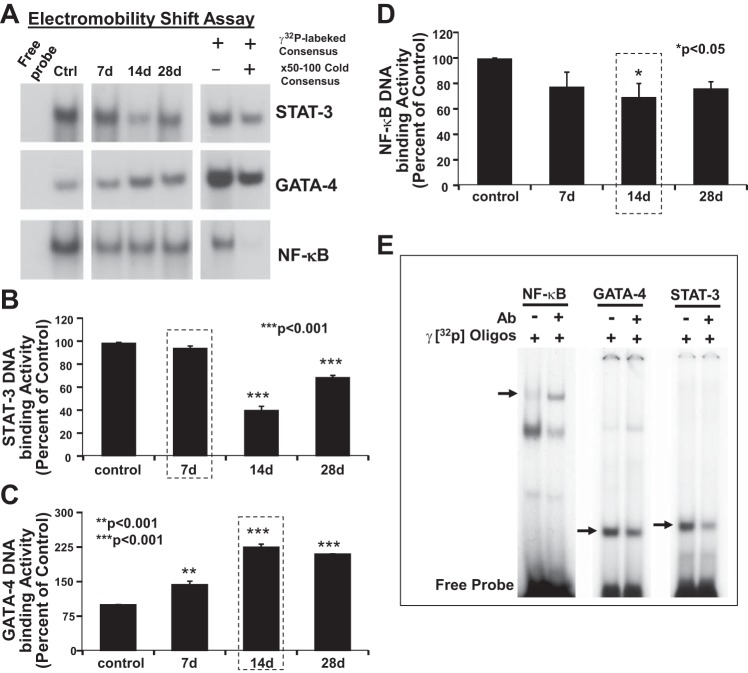
EMSA was used to detect and validate selected TFs modulated in response to iron-IR. We chose to validate three key TFs identified that play a major role in CM maintenance and function (22, 57, 66) and by microarray analysis (Table 3), namely STAT3, GATA4, and NF-κB using EMSA (Fig. 5). Heart tissues were sampled before (controls), and 7, 14, and 28 days after iron exposure. Induction of STAT3 activation showed a cyclic response with increased detection of DNA binding activity in control samples. The activation level at 7 days after exposure was similar to that of control (Fig. 5, A and B). The levels of STAT3 activation were, however, decreased at 14 days post-IR and then increased again at 28 days. The TF GATA4, on the other hand, had a consistent upregulation at 7, 14, and 28 days compared with sham-IR controls. Compared with controls, the levels of GATA4 DNA-binding activity were 1.5-, 3.2-, and 3-fold increased at 7, 14, and 28 days post-IR, respectively (Fig. 5, A and C). NF-κB DNA binding activity was at the detectable level in the control samples. Compared with control in IR-exposed samples, NF-κB showed suppression at 7, 14, and 28 days after iron exposure (Fig. 5, A and D), suggesting that this could be a compensatory response to a development of pressure overload-induced cardiac dysfunction (66). To confirm that the DNA-binding activity was due to specific binding of STAT3, GATA4, and NF-κB to their sequence-specific oligonucleotide, a competition binding assay was performed. Nuclear extracts obtained from heart tissue samples were preincubated in the presence or absence of homologous unlabeled (cold) oligonucleotide competitor identical to the respective TF oligonucleotide-specific probes. As shown (Fig. 5A, left), the DNA binding activity was competitively reduced by the addition of homologous unlabeled oligonucleotides. The competitive inhibition of the DNA binding activity confirmed STAT3, GATA4, and NF-κB-specific binding. Super shift assays were used to further confirm the validity of these results (Fig. 5E), which links key transcriptional regulatory nodes identified by gene expression to functional protein-DNA complexes being modulated over 28 days after iron exposure.
Fig. 5.
Activation of transcription factors STAT3, GATA4, and NF-κB after iron radiation. A: autoradiogram showing a time-dependent activation of STAT3 and GATA4, and inhibition of NF-κB DNA-binding activity after iron-IR. Left: EMSA was performed using 20 μg of nuclear protein isolated from heart tissue of control mice and respective free probe. Middle: EMSA was performed using 20 μg of nuclear protein isolated from heart tissue of iron-IR mice at 7, 14, and 28 days post-IR. Please note that data in the left and middle were derived from the same autoradiograph, and original blots were spliced to remove the 24-h time point due to no significant relevance for validation of IPA prediction. Right: specificity of binding activity of iron IR-induced TF activation is confirmed by competition assay. Significant decrease in STAT3, GATA4, and NF-κB binding activity in the presence of respective competitor (lane 2) confirmed the specificity. B–D: graphic representation of the percent change in STAT3, GATA4, and NF-κB DNA binding activity, respectively, on days 7, 14, and 28, compared with control samples. Values are means ± SE of the pooled data from n = 2–3 animals per time point/group for non-IR control and iron-IR groups. Statistical significance was assigned when P < 0.05. E: super shift assay performed with factor-specific antibodies (Ab) for NF-κB, GATA4, and STAT3. Note that NF-κB factor-specific Ab showed a significant super shift (left), whereas GATA4 and STAT3 Ab revealed a competitor effect. Decreases in the GATA4 and STAT3 band intensities are indicated by arrows (middle and right).
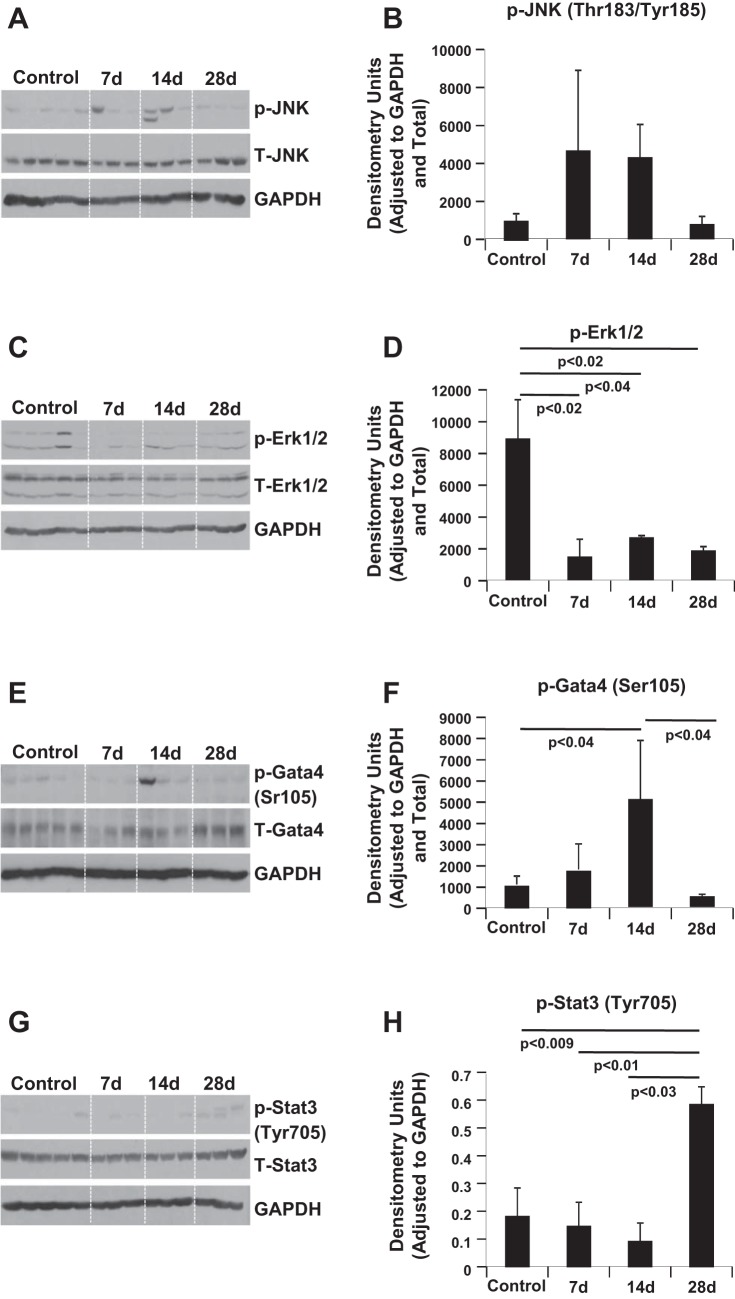
JNK, ERK1/2, GATA4, and STAT3 are Modulated by Iron IR
Phosphorylation on residues threonine-183 and tyrosine-185 is required for activation of JNK (37). Western blot analysis was performed to compare JNK activation over time at 7, 14, and 28 days after 56Fe-IR. Compared with control, 56Fe-IR resulted in nonsignificant yet similar elevated activation levels of JNK at days 7 and 14 post-IR, which normalized to control levels by day 28 (Fig. 6, A and B). At the same time, 56Fe-IR caused a significant approximately fourfold decrease (P < 0.04) in the phosphorylation levels of ERK1/2 at all examined time points post-IR compared with control (Fig. 6, C and D). We also examined the phosphorylation levels of GATA4 at serine-105 and STAT3 at tyrosine-705, and intensity of the bands was normalized to that of GAPDH on the respective membranes. Time course analysis revealed that GATA4 levels at day 7 were similar to those of control, but were significantly elevated by day 14 after 56Fe-IR (Fig. 6, E and F). However, this approximately fivefold increase at day 14 (compared with control) was followed by a significant (∼10-fold) decrease by day 28 compared with day 14 and were comparable to control levels (Fig. 6, E and F). STAT3 phosphorylation levels remained comparable to nonexposed control mice hearts at 7 and 14 days post-IR (Fig. 6, G and H). This was followed by a significant approximately three- to sixfold increase at day 28 compared with control, day 7, and day 14 (Fig. 6, G and H).
Fig. 6.
Western blotting validates differential regulation and signaling in response to iron radiation. A small portion of the left ventricle (3 × 3 mm) from 3 animals per treatment condition was homogenized, and the total protein was isolated and then processed for Western blot analyses. Representative Western blot scans are shown of heart tissue homogenates from controls and iron-IR mice at days 7, 14, and 28 post-IR. Bands represent phosphorylated (p) and total (T) levels for JNK and GAPDH (A), GATA4 and GAPDH (C), ERK1/2 and GAPDH (E), and STAT3 and GAPDH (G). Quantification and graphic representation are shown of total protein levels and phosphorylation using densitometric analysis of phospho-band intensities after adjusting for corresponding loading controls and total levels for p-JNK (Thr183/Tyr185; B), p-ERK1/2 (D), p-GATA4 (Ser105; F), and p-STAT3 (Tyr705; H). Results in graphs are means ± SE of the pooled data from n = 3–5 animals per time point/group for non-IR control and iron-IR mice at days 7, 14, and day 28 post-IR. Statistical significance was assigned when P < 0.05.
Cardiac Physiology and Fibrosis
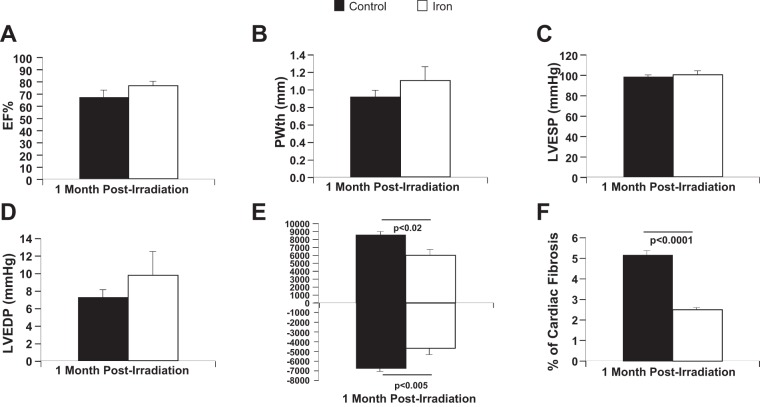
Postradiation with low-dose 1H-IR and 56Fe-IR, mice in all groups, including sham controls, did not demonstrate any signs of stress, loss of body weight, reduction in food intake, or loss of physical activity, with average body weights of mice for all study groups in the range of 29.46–34 g. No considerable change was observed in EF% at 1 mo post-IR (Fig. 7A). However, hemodynamic and echocardiography analysis showed that, in 56Fe-IR hearts, LV posterior wall thickness was slightly increased at 1 mo (Fig. 7B), which was associated with slightly higher LV end-diastolic pressure (Fig. 7D), as well as significant decreases in LV maximum change in pressure over time and LV minimum change in pressure over time (Fig. 7E), suggesting that 56Fe-IR hearts developed systolic and diastolic dysfunction, indicative of possible hear failure in development, as early as 1 mo post-IR. Average whole heart weight measurements at 1 mo post-IR performed during hemodynamic analysis were in the range of 0.133–0.14 g for all groups.
Fig. 7.
Echocardiography and hemodynamic measurements of cardiac functions and evaluation of cardiac fibrosis at 1 mo post-IR. Echocardiography analysis is shown of cardiac function in the hearts of full-body 56Fe-IR and non-IR control mice 1 mo post-IR for ejection fraction percent (EF%; A) and posterior wall thickness [PWth (mm); B]. Hemodynamic measurements and analysis are shown of cardiac function in the hearts of full-body 56Fe-IR and non-IR control mice 1 mo post-IR for left ventricular (LV) end-systolic pressure [LVESP (mmHg); C], LV end-diastolic pressure [LVEDP (mmHg); D], and LV maximum and minimum pressure change (mmHg/s; E). F: graphic representation of %fibrosis in the hearts of full-body 56Fe-IR and non-IR control mice 1 mo post-IR evaluated with Masson's trichrome staining. Values in all graphs are means ± SE; n = 6–8 animals per time point/group for non-IR control (solid bars) and 56Fe-IR (open bars). Statistical significance was assigned when P < 0.05. [These data are a revised interpretation of previously published work (78), with permission.]
In contrast to the common expectation that ionizing radiation, especially at moderate and high doses, increases cardiac fibrosis months after exposure (67), we found a significant twofold decrease in cardiac fibrosis at 1 mo after low-dose, full-body 56Fe-IR (Fig. 7F). These findings may suggest different mechanisms of IR-induced injury for low and high doses, as well as after terrestrial vs. particle radiation, and preclude the simple extrapolation of previously known effects of IR in biological tissues, including heart (28).
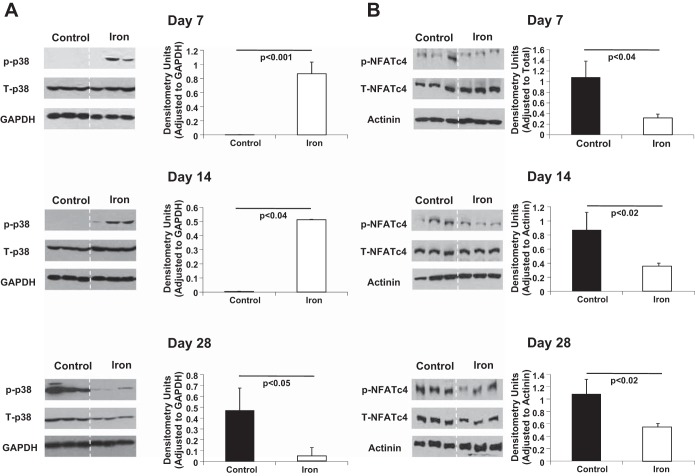
Iron Heavy Particles Activate p38 MAPK
The activation of p38 requires phosphorylation on residues threonine-180 and tyrosine-182 (60). Therefore, we used phospho-specific anti-p38 antibody and performed Western blot analysis to compare p38 kinase activation in response to iron-IR. Phosphorylated and total p38 proteins were measured. Time course analysis revealed that p38 was phosphorylated after iron exposure compared with the control (Fig. 8A). Iron-IR showed increased p38 phosphorylation (activation) in day 7 samples (Fig. 8A, top). A similar signal intensity response was observed in day 14 hearts (Fig. 8A, middle) after iron exposure, indicating continued activation. In contrast, by day 28 (Fig. 8A, bottom), the iron modulated signal was significantly decreased compared with control.
Fig. 8.
Western blotting validates differential regulation and signaling in response to iron radiation. A small portion of the left ventricle (3 × 3 mm) from 3 animals per treatment condition was homogenized, and the total protein was isolated and then processed for Western blot analyses. A: representative Western blot scans of heart tissue homogenates from controls and iron-IR mice at days 7, 14, and 28 post-IR. Bands represent p- and T-p38, and GAPDH was used as loading control. Quantification and graphic representation is shown of total protein levels and phosphorylation using densitometric analysis of phospho-band intensities after adjusting for corresponding GAPDH and total p38 band intensities. B: representative Western blot scans of heart tissue homogenates from controls and iron-IR mice at days 7, 14, and 28 post-IR. Bands represent p- and T-NFATc4 (nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 4), and actinin was used as loading control. Quantification and graphic representation is shown of total protein levels and phosphorylation using densitometric analysis of phospho-band intensities after adjusting for corresponding actinin and total NFATc4 band intensities. Results are means ± SE of the pooled data from n = 3 animals per time point/group for non-IR control (solid bars) and iron-IR (open bars) mice at days 7 14, and 28 post-IR. Statistical significance was assigned when P < 0.05.
NFATc4 under basal unstimulated condition remains highly phosphorylated and inactive (79), and dephosphorylation of NFATc4 results in its activation. NFATc4 is associated with initiation of multiple biological processes, including cytokine gene expression, adipocyte differentiation, and adult cardiac hypertrophy (12, 24). Therefore, we used phospho-specific antibodies against the NFATc4 homology domain to perform Western blot analysis to compare NFATc4 activation in response to iron-IR. Time course analysis revealed that NFATc4 was significantly dephosphorylated after iron exposure compared with the controls (Fig. 8B). NFATc4 dephosphorylation levels remained lower than in nonexposed cells at 7, 14, and 28 days, suggesting activation of cardiac hypertrophy signaling in iron-IR mice hearts. However, by day 28 (Fig. 8B, bottom) compared with non-IR/control samples, the iron-exposed samples started to show increased levels of NFATc4 phosphorylation (<2-fold difference at day 28 vs. 3.5- and 2.5-fold at days 7 and 14, respectively), indicating a possible p38 regulation, i.e., the decrease in the p38 phosphorylation by day 28 (Fig. 8A, bottom).
DISCUSSION
Full-body, low-dose space IR-induced responses in CMs may be radiation-type dependent and have long-lasting effects. Determining alterations on the systems level in CM gene expression and function is paramount to our understanding of degenerative CV risks associated with space IR. Our studies revealed an immense complexity of RNA transcription, regulation of biological pathways, including, but not limited to, inflammation, immune cell trafficking, DNA damage, and repair and free-radical scavenging following iron IR (15 cGy, 1 GeV/n) over 28 days after exposure.
Whole body iron-IR exposure has previously been shown to increase in vivo aortic stiffness and ex vivo aortic tension as long as 6–8 mo after a single 1-Gy but not 0.5-Gy exposure (62). Aortic lesions were also found to be increased 13 wk following targeted 2- to 5-Gy iron-IR in apolipoprotein E-deficient mice (82). Using gene expression microarrays, we profiled the transcriptional changes in isolated CMs from whole body iron- and proton-IR mice from days to weeks after exposure. Proton-IR at 90 cGy and 1 GeV energy failed to elicit a robust alteration in significant gene expression compared with non-IR controls. However, iron exposure at 15 cGy and 1 GeV/n resulted in significant alterations in many genes that were similarly downregulated after exposure at 1 and 3 days but fluctuated at 7, 14, and 28 days. The use of qRT-PCR validated differential transcript responses over these same time periods. Although the fold changes were not identical, the direction of the transcript modulations was similar for the different time points.
It is well known that the biological effectiveness of ionizing radiation for cell cytotoxicity, mutagenesis, and carcinogenesis can differ with the type of IR exposure. However, the molecular mechanisms for the different responses are not clear. A number of studies have suggested that the biological and molecular response to very low doses and dose rates of gamma-IR may be different from that of high doses (2, 76, 80), and that the response to very high, ablative, doses of IR may also be unique. Whether there are unique responses based on particle quality is also unknown. Several studies have looked at comparisons between different qualities of particles over limited time points or in different cells or model systems. One of the major findings is that common genes changed as a function of particle energy (20). It has been identified that cytokines and TFs, such as NF-κB can be modulated by HZE particles (21). However, many of these aforementioned comparative studies focused on other model systems, such as the rat hippocampus or human bronchial epithelial cell line (HEBC3 KT). This study demonstrates that CMs are also differentially responsive to low doses of iron HZE particles.
Given the excess risk modeled for lung cancer for HZE particle exposure in the human bronchial epithelial cell line (HEBC3 KT), a nononcogenically immortalized cell line that does not form tumors in immune-compromised mice was used (20). To provide a comprehensive picture of cellular responses at the molecular level, the global transcriptome changes in HEBC3 KT at three LETs, the very low LET of γ-rays (0.2 keV/μm), an intermediate LET (40 keV/μm) of 1 GeV/n of 28Si, and that of 1 GeV/n 56Fe (150 keV/μm, an LET that is at or near the maximum relative biological effectiveness for a number of endpoints), were examined (20). As others have reported, cell survival was LET dependent (20, 21, 64).
The studies in this paper identified different genes as modulated compared with the aforementioned studies using other cell lines such as HEBC3 KT cells (20, 21). There are multiple reasons for these differences in our iron exposure data, which include the cell type (CMs), dose (15 cGy), and time (up to 28 days) after whole body exposure in mice. Despite the low level of gene concordance, there were similarities between this work and the two different published studies, including increased pathway responses elicited by iron particle exposures. In the Ding et al. 2013 study (20) of the five major pathways, they identified that iron-IR always showed the greatest level of differential responses as measured by significance analysis. However, unlike our findings, the Ding et al. papers did not include protons for comparison (20). Pathway analysis also showed similar major themes to include cellular growth and proliferation, cytokine regulation, and cell cycle. The similarities also included pathways associated with p53, the DNA response pathway, as well as pathways associated with apoptosis (Fig. 3). All of these pathways generally appear to be regulated by IR responses or other genotoxic responses (2, 6, 20, 21, 76). Our finding demonstrates that these pathways normally associated with higher dose responses extend down to 15 cGy iron-IR exposures over a period of ∼1 mo. The fact that genes may differ between studies may not be as important as identifying their linkage with major biological pathways that are shared across multiple studies and serve similar functions across cells and tissue systems, which may drive cell fate.
Alterations in functional cardiac physiology from low dose of iron-IR may be directly linked to molecular mechanisms that are identified as iron responsive in CMs. Major biological pathway analysis illustrates that multiple disease-related pathways, such as cardiomyopathies and neurological disorders, are affected by iron-IR, and these common pathways are linked by molecular mechanisms associated with mitochondrial function, as well as oxidative phosphorylation (Table 4). The top five significant pathways identified were Parkinson's disease, Alzheimer's, oxidative phosphorylation, cardiac muscle contraction, and Huntington's, and all share many of the same energy-related transcripts. These five pathways were followed by hypertrophic and dilated cardiomyopathy pathways (Table 4). The significant transcript overlap between neurodegenerative disease pathways with cardiac muscle-specific pathways was surprising, given that we focused only on CMs. However, these findings may substantiate a possible common underlying molecular mechanism for IR-induced neurodegenerative and cardio-degenerative diseases.
Our gene profile studies were predictive for activation of specific TFs and signaling pathways that controlled validated transcripts identified within this study. We focused on validating our microarray finding by characterizing STAT3, GATA4, and NF-κB TFs using EMSAs. The EMSA findings were validated over a period of 28 days after iron exposure, and our findings indicated that the pathway analysis was predictive for these TF protein activation responses.
Various cytokines and growth factors via tyrosine phosphorylation are known to activate STATs (43), and phosphorylation of tyrosine-705 site results in activation of STAT3, which induces dimerization, followed by nuclear translocation and DNA binding (18, 31). This increase in STAT3 (Tyr705) phosphorylation levels at day 28 compared with day 14 confirms our findings from EMSA analysis, wherein we detected a similar trend. Similar levels of STAT3 activation on day 7 compared with control, followed by a decrease on day 14, concurred with our findings of nuclear translocation and DNA binding activity of STAT3.
Cardiac hypertrophy is known to occur in response to stimulants, such as injury or stress (27), and involves multiple signaling molecules culminating in activation of TFs and subsequent gene expression (41, 70). The major contributors responsible for regulation of cardiac hypertrophy gene regulation involves GATA4 through its phosphorylation of serine 105 (Ser105), which in turn activates DNA binding and transcription of GATA4 (13, 41). Since JNK, ERK1/2, and p38 MAPK have been known to regulate the phosphorylation of GATA4 at Ser105 in cultured CMs (13, 41, 68), we evaluated the expression levels of these signaling molecules as viable upstream candidates across all time points post-IR. Increased levels of GATA4 phosphorylation on day 14 were confirmed by the higher expression levels for both JNK and p38, thus suggesting an increased DNA binding activity. Even though ERK1/2 levels post-IR were lower compared with control, day 14 still showed a slightly higher phosphorylation compared with days 7 and 28, and this subtle change in ERK1/2 phosphorylation might enhance GATA4 DNA binding activity (41) when combined with increased phosphorylation of JNK and p38 on day 14 (Fig. 6, A and B, and Fig. 8A, middle).
We also studied p38 MAPK phosphorylation in our samples. Increased phosphorylation of p38 indicates activation of p38 MAPK, and active p38 regulates CM survival/apoptosis (45, 73), cardiac hypertrophy (8, 38, 54), and transition from cardiac hypertrophy to heart failure via activation of the JAK/STAT3 pathway (38, 53). In addition, activation of the p38 MAPK pathway through phosphorylation of p38 also results in protection against heart failure by aiding in cardiac restoration (46). Phosphorylation of p38 MAPK was significantly increased in iron-IR hearts at 7 and 14 days and showed reduced phosphorylation at 28 days. Increased p38 MAPK phosphorylation on days 7 and 14 after iron-IR suggests activation of compensatory/protective mechanism, whereas reduced p38 MAPK phosphorylation on day 28 may be an early indication of cardiac decompensation in development. Decreases in activation of p38 MAPK signaling in 56Fe ion-IR hearts could further negate a possible cardiac protective mechanism(s).
One of the major mechanisms of cardiac compensation is hypertrophy of the heart, and the TF, NFATc4, is one of the well-studied regulators of cardiac hypertrophy (12, 40). MAPK p38 have been shown to regulate NFATc4 phosphorylation and nuclear translocation (79). This activation of p38 affects dephosphorylated NFATc4 by rephosphorylation that antagonizes the Ca2+-mediated dephosphorylation and nuclear translocation of NFATc4, which results in NFATc4 being exported out of the nucleus and termination of NFATc4-mediated transcription (40, 49, 79), hence cardiac hypertrophy signaling. There was activation of NFATc4 in 56Fe ion-IR hearts at days 7, 14, and 28, which may indicate activation of cardiac hypertrophy signaling.
Further studies will be needed to determine whether these changes are shared across different cell and organ types in the body. Other factors associated with immune cell invasion and timing should also be studied. Additional characterization will also be needed to determine whether other HZE particles modulate transcripts and proteins at similar dose levels for the aforementioned biological pathways. This work provides new insights for thinking about biological pathways and molecular mechanisms that may be important for heart risk estimates for radiation therapy, astronaut safety, and medical diagnostics.
In summary, our data present several new findings: 1) at 90 cGy, 1 GeV dose of 1H-IR, gene expression in CMs is not significantly affected over 28 days; 2) at 15 cGy, 1 GeV/n dose of 56Fe-IR, CMs exhibit a time-dependent change in gene expression that results in an overall increase in the activity of inflammatory, free-radical scavenging and CV development and function pathways, 7 and 28 days post-IR; 3) our differential genes were predictive for modulation of proteins such as TFs (STAT3, GATA4, NF-κB, NFATc4) and p38 MAPK signaling; 4) there was a significant transcript overlap between neurodegenerative disease and cardiac muscle disorder-specific pathways that may support a possibility of common underlying molecular mechanism for radiation-induced neurodegenerative and cardio-degenerative diseases/disorders; and 5) 14 days after a single 56Fe-IR, the activation of several developmental TFs, such as TBX5, GATA4, and MEF2C, which are required for the maintenance of cardiac homeostasis, strongly suggests activation of cardio-protective and regeneration responses in 56Fe-IR-exposed hearts. It is very important to emphasize here that cardio-protective genes were activated in CMs before any detectable clinical symptoms in cardiac function. Using the same experimental model, our group has shown that the first detectable changes in cardiac function were observed only 1 mo after 56Fe-IR (Fig. 7) (78). These findings may infer potential novel biomarkers that can augment known CV disease biomarkers currently in the clinic for specific use in deep space missions.
GRANTS
This work was supported by NASA under Grant NNX11AD22G and American Heart Association (AHA) Grant 14GRNT18860032 to D. A. Goukassian. This work was also supported in part by the US Department of Energy (DOE) under contract no. DE-AC52-07NA27344, with funding from the US DOE Low Dose Radiation Research Program Grant KP110202 to M. A. Coleman, and by AHA Grant 10GRNT4710003 and National Heart, Lung, and Blood Institute Grant HL-106098 to X. Yan. This work was also supported by National Space Biomedical Research Institute (NSBRI) Grant CA02802 through NASA NCC 9–58-298 to M. Natarajan.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.A.C., S.P.S., M.N., K.M., L.E.P., and Y.A. analyzed data; M.A.C., S.P.S., M.N., L.E.P., and D.A.G. interpreted results of experiments; M.A.C., S.P.S., M.N., and K.M. prepared figures; M.A.C., S.P.S., and D.A.G. drafted manuscript; M.A.C., S.P.S., M.N., S.M., L.E.P., X.Y., and D.A.G. edited and revised manuscript; S.P.S., J.O., M.N., K.M., J.S., and Y.A. performed experiments; D.A.G. conception and design of research; D.A.G. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank members of National Aeronautics and Space Administration (NASA) Space Radiation Laboratory (NSRL) and Biological Environmental and Climate Sciences Department at Brookhaven National Laboratory, Drs. Adam Rusek and Peter Guida and their teams, for the help and support of our research studies. We also thank Dr. J. T. McDonald for initial bioinformatics processing and initial analysis of microarray data.
REFERENCES
- 1.Adams MJ, Hardenbergh PH, Constine LS, Lipshultz SE. Radiation-associated cardiovascular disease. Crit Rev Oncol Hematol 45: 55–75, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, Fornace AJ Jr. Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res 1: 445–452, 2003. [PubMed] [Google Scholar]
- 3.Baker JE, Moulder JE, Hopewell JW. Radiation as a risk factor for cardiovascular disease. Antioxid Redox Signal 15: 1945–1956, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basavaraju SR, Easterly CE. Pathophysiological effects of radiation on atherosclerosis development and progression, and the incidence of cardiovascular complications. Med Phys 29: 2391–2403, 2002. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300, 1995. [Google Scholar]
- 6.Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, Rajagopal S, Son JK, Ma Q, Springer Z, Kang PM, Izumo S, Pu WT. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci U S A 103: 14471–14476, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerma M, van der Wees CG, Vrieling H, Svensson JP, Wondergem J, van der Laarse A, Mullenders LH, van Zeeland AA. Microarray analysis of gene expression profiles of cardiac myocytes and fibroblasts after mechanical stress, ionising or ultraviolet radiation. BMC Genomics 6: 6, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braz JC, Bueno OF, Liang Q, Wilkins BJ, Dai YS, Parsons S, Braunwart J, Glascock BJ, Klevitsky R, Kimball TF, Hewett TE, Molkentin JD. Targeted inhibition of p38 MAPK promotes hypertrophic cardiomyopathy through upregulation of calcineurin-NFAT signaling. J Clin Invest 111: 1475–1486, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brosius FC 3rd, Waller BF, Roberts WC. Radiation heart disease. Analysis of 16 young (aged 15 to 33 years) necropsy patients who received over 3,500 rads to the heart. Am J Med 70: 519–530, 1981. [DOI] [PubMed] [Google Scholar]
- 10.Bungo MW, Goldwater DJ, Popp RL, Sandler H. Echocardiographic evaluation of space shuttle crewmembers. J Appl Physiol 62: 278–283, 1987. [DOI] [PubMed] [Google Scholar]
- 11.Burns RJ, Bar-Shlomo BZ, Druck MN, Herman JG, Gilbert BW, Perrault DJ, McLaughlin PR. Detection of radiation cardiomyopathy by gated radionuclide angiography. Am J Med 74: 297–302, 1983. [DOI] [PubMed] [Google Scholar]
- 12.Bushdid PB, Osinska H, Waclaw RR, Molkentin JD, Yutzey KE. NFATc3 and NFATc4 are required for cardiac development and mitochondrial function. Circ Res 92: 1305–1313, 2003. [DOI] [PubMed] [Google Scholar]
- 13.Charron F, Tsimiklis G, Arcand M, Robitaille L, Liang Q, Molkentin JD, Meloche S, Nemer M. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev 15: 2702–2719, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen SI, Bharati S, Glass J, Lev M. Radiotherapy as a cause of complete atrioventricular block in Hodgkin's disease. An electrophysiological-pathological correlation. Arch Intern Med 141: 676–679, 1981. [PubMed] [Google Scholar]
- 15.Convertino VA, Cooke WH, Lurie KG. Inspiratory resistance as a potential treatment for orthostatic intolerance and hemorrhagic shock. Aviat Space Environ Med 76: 319–325, 2005. [PubMed] [Google Scholar]
- 16.Cucinotta FA, Durante M. Cancer risk from exposure to galactic cosmic rays: implications for space exploration by human beings. Lancet Oncol 7: 431–435, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Darby SC, Ewertz M, McGale P, Bennet AM, Blom-Goldman U, Bronnum D, Correa C, Cutter D, Gagliardi G, Gigante B, Jensen MB, Nisbet A, Peto R, Rahimi K, Taylor C, Hall P. Risk of ischemic heart disease in women after radiotherapy for breast cancer. N Engl J Med 368: 987–998, 2013. [DOI] [PubMed] [Google Scholar]
- 18.Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421, 1994. [DOI] [PubMed] [Google Scholar]
- 19.Ding B, Price RL, Goldsmith EC, Borg TK, Yan X, Douglas PS, Weinberg EO, Bartunek J, Thielen T, Didenko VV, Lorell BH. Left ventricular hypertrophy in ascending aortic stenosis mice: anoikis and the progression to early failure. Circulation 101: 2854–2862, 2000. [DOI] [PubMed] [Google Scholar]
- 20.Ding LH, Park S, Peyton M, Girard L, Xie Y, Minna JD, Story MD. Distinct transcriptome profiles identified in normal human bronchial epithelial cells after exposure to gamma-rays and different elemental particles of high Z and energy. BMC Genomics 14: 372, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding LH, Shingyoji M, Chen F, Chatterjee A, Kasai KE, Chen DJ. Gene expression changes in normal human skin fibroblasts induced by HZE-particle radiation. Radiat Res 164: 523–526, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Enomoto D, Obana M, Miyawaki A, Maeda M, Nakayama H, Fujio Y. Cardiac-specific ablation of the STAT3 gene in the subacute phase of myocardial infarction exacerbated cardiac remodeling. Am J Physiol Heart Circ Physiol 309: H471–H480, 2015. [DOI] [PubMed] [Google Scholar]
- 23.Essaghir A, Toffalini F, Knoops L, Kallin A, van Helden J, Demoulin JB. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res 38: e120, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Facundo HT, Brainard RE, Watson LJ, Ngoh GA, Hamid T, Prabhu SD, Jones SP. O-GlcNAc signaling is essential for NFAT-mediated transcriptional reprogramming during cardiomyocyte hypertrophy. Am J Physiol Heart Circ Physiol 302: H2122–H2130, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham EL, Balla C, Franchino H, Melman Y, del Monte F, Das S. Isolation, culture, and functional characterization of adult mouse cardiomyoctyes. Journal of visualized experiments : JoVE: e50289, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatoum OA, Otterson MF, Kopelman D, Miura H, Sukhotnik I, Larsen BT, Selle RM, Moulder JE, Gutterman DD. Radiation induces endothelial dysfunction in murine intestinal arterioles via enhanced production of reactive oxygen species. Arterioscler Thromb Vasc Biol 26: 287–294, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 7: 589–600, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Hendry JH. Threshold doses and circulatory disease risks. Ann ICRP 44: 69–75, 2015. [DOI] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y, Wright CD, Kobayashi S, Healy CL, Elgethun M, Cypher A, Liang Q, O′Connell TD. GATA4 is a survival factor in adult cardiac myocytes but is not required for alpha1A-adrenergic receptor survival signaling. Am J Physiol Heart Circ Physiol 295: H699–H707, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ihle JN. Cytokine receptor signalling. Nature 377: 591–594, 1995. [DOI] [PubMed] [Google Scholar]
- 32.Ito K, Yan X, Feng X, Manning WJ, Dillmann WH, Lorell BH. Transgenic expression of sarcoplasmic reticulum Ca(2+) ATPase modifies the transition from hypertrophy to early heart failure. Circ Res 89: 422–429, 2001. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, Yan X, Tajima M, Su Z, Barry WH, Lorell BH. Contractile reserve and intracellular calcium regulation in mouse myocytes from normal and hypertrophied failing hearts. Circ Res 87: 588–595, 2000. [DOI] [PubMed] [Google Scholar]
- 34.La Vecchia L. Physiologic dual chamber pacing in radiation-induced atrioventricular block. Chest 110: 580–581, 1996. [DOI] [PubMed] [Google Scholar]
- 35.Lampidis TJ, Weichselbaum RR, Little JB. Letter: Gamma-irradiation of mammalian beating heart cells in vitro. Effects on cellular function. Int J Radiat Biol Relat Stud Phys Chem Med 28: 99–102, 1975. [DOI] [PubMed] [Google Scholar]
- 36.Larsen RL, Jakacki RI, Vetter VL, Meadows AT, Silber JH, Barber G. Electrocardiographic changes and arrhythmias after cancer therapy in children and young adults. Am J Cardiol 70: 73–77, 1992. [DOI] [PubMed] [Google Scholar]
- 37.Lee SA, Dritschilo A, Jung M. Impaired ionizing radiation-induced activation of a nuclear signal essential for phosphorylation of c-Jun by dually phosphorylated c-Jun amino-terminal kinases in ataxia telangiectasia fibroblasts. J Biol Chem 273: 32889–32894, 1998. [DOI] [PubMed] [Google Scholar]
- 38.Lemke LE, Bloem LJ, Fouts R, Esterman M, Sandusky G, Vlahos CJ. Decreased p38 MAPK activity in end-stage failing human myocardium: p38 MAPK alpha is the predominant isoform expressed in human heart. J Mol Cell Cardiol 33: 1527–1540, 2001. [DOI] [PubMed] [Google Scholar]
- 39.Letsas KP, Korantzopoulos P, Evangelou D, Pappas LK, Kardaras F. Acute myocardial infarction with normal coronary arteries in a patient with Hodgkin's disease: a late complication of irradiation and chemotherapy. Tex Heart Inst J 33: 512–514, 2006. [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Li J, Cai X, Sun H, Jiao J, Bai T, Zhou XW, Chen X, Gill DL, Tang XD. Protein kinase D3 is a pivotal activator of pathological cardiac hypertrophy by selectively increasing the expression of hypertrophic transcription factors. J Biol Chem 286: 40782–40791, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang Q, Wiese RJ, Bueno OF, Dai YS, Markham BE, Molkentin JD. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol Cell Biol 21: 7460–7469, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao R, Jain M. Isolation, culture, and functional analysis of adult mouse cardiomyocytes. Methods Mol Med 139: 251–262, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Lim CP, Cao X. Serine phosphorylation and negative regulation of Stat3 by JNK. J Biol Chem 274: 31055–31061, 1999. [DOI] [PubMed] [Google Scholar]
- 44.Lipshultz SE, Sallan SE. Cardiovascular abnormalities in long-term survivors of childhood malignancy. J Clin Oncol 11: 1199–1203, 1993. [DOI] [PubMed] [Google Scholar]
- 45.Ma XL, Kumar S, Gao F, Louden CS, Lopez BL, Christopher TA, Wang C, Lee JC, Feuerstein GZ, Yue TL. Inhibition of p38 mitogen-activated protein kinase decreases cardiomyocyte apoptosis and improves cardiac function after myocardial ischemia and reperfusion. Circulation 99: 1685–1691, 1999. [DOI] [PubMed] [Google Scholar]
- 46.Marber MS, Rose B, Wang Y. The p38 mitogen-activated protein kinase pathway–a potential target for intervention in infarction, hypertrophy, and heart failure. J Mol Cell Cardiol 51: 485–490, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride WH, Chiang CS, Olson JL, Wang CC, Hong JH, Pajonk F, Dougherty GJ, Iwamoto KS, Pervan M, Liao YP. A sense of danger from radiation. Radiat Res 162: 1–19, 2004. [DOI] [PubMed] [Google Scholar]
- 48.McDougall JA, Sakata R, Sugiyama H, Grant E, Davis S, Nishi N, Soda M, Shimizu Y, Tatsukawa Y, Kasagi F, Suyama A, Ross P, Kopecky KJ, Li CI. Timing of menarche and first birth in relation to risk of breast cancer in A-bomb survivors. Cancer Epidemiol Biomarkers Prev 19: 1746–1754, 2010. [DOI] [PubMed] [Google Scholar]
- 49.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mungrue IN, Gros R, You X, Pirani A, Azad A, Csont T, Schulz R, Butany J, Stewart DJ, Husain M. Cardiomyocyte overexpression of iNOS in mice results in peroxynitrite generation, heart block, and sudden death. J Clin Invest 109: 735–743, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakao T, Kanaya H, Namura M, Ohsato K, Araki T, Ohka T, Sugihara N, Takeda R. Complete atrioventricular block following radiation therapy for malignant thymoma. Jpn J Med 29: 104–110, 1990. [DOI] [PubMed] [Google Scholar]
- 52.Natarajan M, Aravindan N, Meltz ML, Herman TS. Post-translational modification of I-kappa B alpha activates NF-kappa B in human monocytes exposed to 56Fe ions. Radiat Environ Biophys 41: 139–144, 2002. [DOI] [PubMed] [Google Scholar]
- 53.Negoro S, Oh H, Tone E, Kunisada K, Fujio Y, Walsh K, Kishimoto T, Yamauchi-Takihara K. Glycoprotein 130 regulates cardiac myocyte survival in doxorubicin-induced apoptosis through phosphatidylinositol 3-kinase/Akt phosphorylation and Bcl-xL/caspase-3 interaction. Circulation 103: 555–561, 2001. [DOI] [PubMed] [Google Scholar]
- 54.Nemoto S, Sheng Z, Lin A. Opposing effects of Jun kinase and p38 mitogen-activated protein kinases on cardiomyocyte hypertrophy. Mol Cell Biol 18: 3518–3526, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholas SB, Philipson KD. Cardiac expression of the Na+/Ca2+ exchanger NCX1 is GATA factor dependent. Am J Physiol Heart Circ Physiol 277: H324–H330, 1999. [DOI] [PubMed] [Google Scholar]
- 56.Orzan F, Brusca A, Conte MR, Presbitero P, Figliomeni MC. Severe coronary artery disease after radiation therapy of the chest and mediastinum: clinical presentation and treatment. Br Heart J 69: 496–500, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paulin R, Meloche J, Jacob MH, Bisserier M, Courboulin A, Bonnet S. Dehydroepiandrosterone inhibits the Src/STAT3 constitutive activation in pulmonary arterial hypertension. Am J Physiol Heart Circ Physiol 301: H1798–H1809, 2011. [DOI] [PubMed] [Google Scholar]
- 58.Petrovic D, Brown SM, Yatvin MB. Effects of adriamycin and irradiation on beating of rat heart muscle cells in culture. Int J Radiat Oncol Biol Phys 2: 505–513, 1977. [DOI] [PubMed] [Google Scholar]
- 59.Preston DL, Shimizu Y, Pierce DA, Suyama A, Mabuchi K. Studies of mortality of atomic bomb survivors. Report 13: Solid cancer and noncancer disease mortality: 1950–1997. Radiat Res 160: 381–407, 2003. [DOI] [PubMed] [Google Scholar]
- 60.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem 270: 7420–7426, 1995. [DOI] [PubMed] [Google Scholar]
- 61.Sanford GL, Harris-Hooker S, Lui J, Melhado-Gardner C, Pink Y, Wallace T, Bosah FN. Influence of changes in gravity on the response of lung and vascular cells to ischemia/reperfusion in vitro. J Gravit Physiol 6: P27–P28, 1999. [PubMed] [Google Scholar]
- 62.Soucy KG, Lim HK, Kim JH, Oh Y, Attarzadeh DO, Sevinc B, Kuo MM, Shoukas AA, Vazquez ME, Berkowitz DE. HZE (5)(6)Fe-ion irradiation induces endothelial dysfunction in rat aorta: role of xanthine oxidase. Radiat Res 176: 474–485, 2011. [DOI] [PubMed] [Google Scholar]
- 63.Stewart FA, Heeneman S, Te Poele J, Kruse J, Russell NS, Gijbels M, Daemen M. Ionizing radiation accelerates the development of atherosclerotic lesions in ApoE−/− mice and predisposes to an inflammatory plaque phenotype prone to hemorrhage. Am J Pathol 168: 649–658, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stewart J, Ko YH, Kennedy AR. Protective effects of l-selenomethionine on space radiation induced changes in gene expression. Radiat Environ Biophys 46: 161–165, 2007. [DOI] [PubMed] [Google Scholar]
- 65.Stewart JR, Fajardo LF, Gillette SM, Constine LS. Radiation injury to the heart. Int J Radiat Oncol Biol Phys 31: 1205–1211, 1995. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka T, Ogawa M, Suzuki J, Sekinishi A, Itai A, Hirata Y, Nagai R, Isobe M. Inhibition of IkappaB phosphorylation prevents load-induced cardiac dysfunction in mice. Am J Physiol Heart Circ Physiol 303: H1435–H1445, 2012. [DOI] [PubMed] [Google Scholar]
- 67.Taunk NK, Haffty BG, Kostis JB, Goyal S. Radiation-induced heart disease: pathologic abnormalities and putative mechanisms. Front Oncol 5: 39, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tenhunen O, Sarman B, Kerkela R, Szokodi I, Papp L, Toth M, Ruskoaho H. Mitogen-activated protein kinases p38 and ERK 1/2 mediate the wall stress-induced activation of GATA-4 binding in adult heart. J Biol Chem 279: 24852–24860, 2004. [DOI] [PubMed] [Google Scholar]
- 69.Tribble DL, Barcellos-Hoff MH, Chu BM, Gong EL. Ionizing radiation accelerates aortic lesion formation in fat-fed mice via SOD-inhibitable processes. Arterioscler Thromb Vasc Biol 19: 1387–1392, 1999. [DOI] [PubMed] [Google Scholar]
- 70.van Berlo JH, Elrod JW, Aronow BJ, Pu WT, Molkentin JD. Serine 105 phosphorylation of transcription factor GATA4 is necessary for stress-induced cardiac hypertrophy in vivo. Proc Natl Acad Sci U S A 108: 12331–12336, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veinot JP, Edwards WD. Pathology of radiation-induced heart disease: a surgical and autopsy study of 27 cases. Hum Pathol 27: 766–773, 1996. [DOI] [PubMed] [Google Scholar]
- 72.Virmani R, Farb A, Carter AJ, Jones RM. Pathology of radiation-induced coronary artery disease in human and pig. Cardiovasc Radiat Med 1: 98–101, 1999. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Huang S, Sah VP, Ross J Jr, Brown JH, Han J, Chien KR. Cardiac muscle cell hypertrophy and apoptosis induced by distinct members of the p38 mitogen-activated protein kinase family. J Biol Chem 273: 2161–2168, 1998. [DOI] [PubMed] [Google Scholar]
- 74.Weintraub NL, Jones WK, Manka D. Understanding radiation-induced vascular disease. J Am Coll Cardiol 55: 1237–1239, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woodcock EA, Matkovich SJ. Cardiomyocytes structure, function and associated pathologies. Int J Biochem Cell Biol 37: 1746–1751, 2005. [DOI] [PubMed] [Google Scholar]
- 76.Wyrobek AJ, Manohar CF, Krishnan VV, Nelson DO, Furtado MR, Bhattacharya MS, Marchetti F, Coleman MA. Low dose radiation response curves, networks and pathways in human lymphoblastoid cells exposed from 1 to 10 cGy of acute gamma radiation. Mutat Res 722: 119–130, 2011. [DOI] [PubMed] [Google Scholar]
- 77.Yamada M, Wong FL, Fujiwara S, Akahoshi M, Suzuki G. Noncancer disease incidence in atomic bomb survivors, 1958–1998. Radiat Res 161: 622–632, 2004. [DOI] [PubMed] [Google Scholar]
- 78.Yan X, Sasi SP, Gee H, Lee J, Yang Y, Mehrzad R, Onufrak J, Song J, Enderling H, Agarwal A, Rahimi L, Morgan J, Wilson PF, Carrozza J, Walsh K, Kishore R, Goukassian DA. Cardiovascular risks associated with low dose ionizing particle radiation. PLoS One 9: e110269, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang TT, Xiong Q, Enslen H, Davis RJ, Chow CW. Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Mol Cell Biol 22: 3892–3904, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin E, Nelson DO, Coleman MA, Peterson LE, Wyrobek AJ. Gene expression changes in mouse brain after exposure to low-dose ionizing radiation. Int J Radiat Biol 79: 759–775, 2003. [DOI] [PubMed] [Google Scholar]
- 81.Yu H, Mohan S, Natarajan M. Radiation-triggered NF-kappaB activation is responsible for the angiogenic signaling pathway and neovascularization for breast cancer cell proliferation and growth. Breast Cancer (Auckl) 6: 125–135, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu T, Parks BW, Yu S, Srivastava R, Gupta KW, X, Khaled S, Chang PY, Kabarowski JH, Kucik DF. Iron Ion (56Fe) Radiation Increases the Size of Pre-Existing Atherosclerotic Lesions in ApoE−/− mice. Gravit Space Biol 25: 57–59, 2011. [Google Scholar]