Abstract
WDR5 is a core component of SET1-family complexes that achieve transcriptional activation via methylation of histone H3 on Nζ of Lys4 (H3K4). The role of WDR5 in the MLL1 complex has recently been described as specific recognition of dimethyl-K4 in the context of a histone H3 amino terminus; WDR5 is essential for vertebrate development, Hox gene activation and global H3K4 trimethylation. We report the high-resolution X-ray structures of WDR5 in the unliganded form and complexed with histone H3 peptides having unmodified and mono-, di- and trimethylated K4, which together provide the first comprehensive analysis of methylated histone recognition by the ubiquitous WD40-repeat fold. Contrary to predictions, the structures reveal that WDR5 does not read out the methylation state of K4 directly, but instead serves to present the K4 side chain for further methylation by SET1-family complexes.
The structure and dynamics of chromatin are increasingly recognized to be essential for mediating both global and local regulation of the genome1,2. Vital to maintaining and remodeling the various chromatin states are a collection of post-translational modifications of histones that are introduced in a highly regulated fashion and cause specific functional consequences3. In particular, post-translational introduction of methyl groups onto lysine side chains of histone proteins greatly affects chromatin function in complex and often opposing ways, resulting in either transcriptional repression or activation, contingent upon the precise location and degree of methylation4–7. For example, trimethylation of histone H3 at K9 or K27 is implicated in silencing of the underlying DNA6,8, whereas methylation at other sites, notably H3K4, is coupled to transcriptional activation4,7. (Throughout this manuscript, single-letter amino acid abbreviations are used to refer to histone residues, whereas three-letter abbreviations are used for WDR5 residues.) K4 is trimethylated in nucleosomes proximal to promoters and coding regions of actively transcribed genes9.
SET domain proteins are the major catalytic components of a number of histone methyltransferase (HMTase) complexes that effect lysine methylation10–15. The sole yeast enzyme capable of catalyzing H3K4 methylation is the SET domain protein Set1, which was identified as the catalytic component of the Set1 complex (also called the COMPASS)11,13,15. In higher eukaryotes, the situation is more complex: there are a number of yeast Set1 orthologs (SET1a and SET1b) and paralogs (MLLs and Ash1) collectively referred to as the SET1 family16. Among these proteins, the most extensively studied is mixed lineage leukemia-1 (MLL1), a proto-oncogene whose fusion with a number of partners causes a variety of human leukemias17. The SET1 family of transcription-activating K4 HMTase complexes (MLL1, MLL2 and SET1 in humans) share a common set of core elements, including Ash2L, two WD40-repeat proteins (WDR5 and RbBP5) and a SET domain polypeptide18. Whereas WDR5 knockdown produces a genome-wide deficiency in H3K4 trimethylation19, MLL1 knockdown does not (Y. Dou and R.G. Roeder, Rockefeller University, personal communication). WDR5 is essential to vertebrate development and hematopoiesis19, and deficiency phenotypes are analogous, at least in part, to those observed in Mll−/− and MllΔSET mice20,21, probably owing to a common Hox gene regulation defect. Thus, WDR5's function is not restricted to the MLL1 complex, although participation in MLL1 complexes seems to be important in organismal viability.
The molecular mechanisms of WDR5-dependent gene activation have only recently begun to be elucidated. Although it has been demonstrated that WDR5 preferentially associates with histone H3 dimethylated at K4 (H3K4me2)19, the mechanism by which this binding preference results in global K4 trimethylation remains obscure (Fig. 1a). To understand how WDR5 recognizes its substrates and to begin to describe a more precise molecular mechanism for the function of this subunit in the MLL1 complex and in a variety of other SET1-family complexes, we undertook the structural and biochemical characterization of human WDR5 and histone H3 peptides. A very recent report of WDR5 in complex with an H3K4me2 peptide has provided initial characterization of one of the five complexes reported here22. Nevertheless, only the complete set of structures and quantitative binding measurements presented herein permit insight into a more complex mechanism of K4 methyl-form discrimination by WDR5 than has previously been suggested22. Our exhaustive set of structures also suggests a previously uncharacterized role for WDR5 in MLL1 and related methylation complexes: WDR5 binding to histone H3 appears to present the K4 side chain for further methylation.
Figure 1.
The overall structure of WDR5 complexes with histone H3. (a) The set of possible methylation states of H3K4. In general, nucleosomes di- and trimethylated at K4 of H3 are associated with transcriptional activation. (b) The overall structure of WDR5 in complex with H3K4me2. WDR5 is a canonical seven-bladed β-propeller (red); the histone H3 peptide binds across the top face of the protein (green, with amino (N′) and carboxyl (C′) termini and R2 labeled). (c) Electrostatic surface of WDR5 contoured from −8 to +8 kT using ABPS52 shows a series of acidic patches on the upper face of WDR5 (arrows 1–4), whereas the remainder of the protein is highly positively charged. H3K4me2 peptide is shown in green; potential aromatic cage residues are situated around arrow 5. Remaining arrows indicate patches of positive electrostatic potential discussed in the text. Cutaway view of this surface along the pseudo seven-fold symmetry axis (top) demonstrates how deeply buried R2 is in the cavity of the β-propeller fold's torus.
RESULTS
Structures of WDR5–histone H3 complexes
N-terminally truncated WDR5 was crystallized alone and in complex with a series of histone H3 peptides bearing each of the possible methylation states at K4 (Fig. 1a). The five structures of the unliganded and peptide-bound WDR5 were solved to 2.2- to 1.5-Å resolution by molecular replacement, using the C-terminal domain of Tup1 as the search model23. All six crystal forms have two nearly identical WDR5 protomers per asymmetric unit (Supplementary Fig. 1 online), with the exception of our additional H3K4me2 crystal complex (form II), which contains only one, and therefore below we depict only one of each protomer pair. There is also extensive similarity shared by the unliganded protein and the series of four peptide-liganded structures (Supplementary Fig. 1); this is discussed in further detail below. The use of histone peptides of differing lengths yielded crystals of varying quality, with different space groups and unit cell dimensions (data not shown). Neither different packing environs for the surface-bound peptide nor differing lengths of bound peptide produced any discernable changes in the bound peptide conformation.
WDR5 has the expected canonical β-propeller fold made up of seven WD40 repeats (Fig. 1b). The β-propeller has an overall shape of a cone truncated at the apex, with a water-filled cavity running through the center along the seven-fold symmetry axis (Fig. 1b,c). The majority of the protein surface is positively charged, owing to the presence of numerous lysine and arginine residues, but a few patches of negative charge are found on the upper face of the protein in the orientation shown in Figure 1c.
For all five liganded crystal forms, omit maps calculated from high-resolution datasets revealed unambiguously interpretable electron density for the histone H3 peptides situated across the narrow end of the β-propeller cone (Fig. 1b,c). The N-terminal 6–8 amino acids of the peptide ligand could be easily modeled into the excellent density, whereas the C-terminal 1–3 residues were less ordered and therefore were not modeled (Fig. 2a). Indeed, the diffraction data were of sufficiently high quality to enable direct observation of the methyl groups on K4 in the various methylated forms, and even to distinguish the trigonal geometry about the Nε of K4 in the dimethyl form from the planar geometry of the similarly sized guanidinium group of R2 (Supplementary Fig. 2 online).
Figure 2.
Bound histone H3 peptide electron density and conformation. (a) Stereo view of the H3K4me2 peptide model in a simulated annealing omit map contoured at 1.0 σ about the peptide, calculated from the protein model and the 1.5-Å data set (form I). (b) Main chain superposition of all of the WDR5-bound peptides. Nitrogen atoms are colored in blue, oxygen atoms in red, and carbon atoms are colored differently for each peptide as indicated in key. Points of conformational divergence are localized to the K4 and Q5 side chains.
Histone H3 peptide recognition
Histone H3 peptides are bound in a nearly identical manner throughout the entire series of liganded structures, with the minor differences being localized to the K4 side chain conformation (discussed below) and to the C-terminal end of the peptide, where the interpretable electron density extended furthest in the structures having the highest resolution. Main chain superpositions of the peptides alone (Fig. 2b) closely resemble superpositions generated using the entire WDR5 complex (Supplementary Fig. 1), indicating that the overall binding mode of the peptide to WDR5 is not substantially affected by the methylation state of K4. This being the case, we will first detail the conserved features of peptide recognition and then discuss the differences.
The first three residues of the peptide N terminus (A1, R2, T3) adopt a backbone conformation best described as a truncated helix, with dihedral Φ and Ψ angles characteristic of α- and 310-helices, but interrupted at the C-terminal end so that only a single intramolecular i → i + 4 hydrogen bond is observed (that between the A1 carbonyl oxygen and the K4 amide proton, Fig. 3a). An ordered water molecule is involved in a bridging intrapeptide contact between the Nα and the Oγ-hydroxyl of T3. The A1 carbonyl also participates in a hydrogen bond to another ordered water molecule that seems to serve as the hub of a hydrogen-bonding network conserved throughout the liganded structures; this water also spans the Oγ-hydroxyl of T6 and the K4 carbonyl (Fig. 3a). This set of interactions serves to orient the main chain of K4, enforcing the projection of the side chain outward from the body of WDR5.
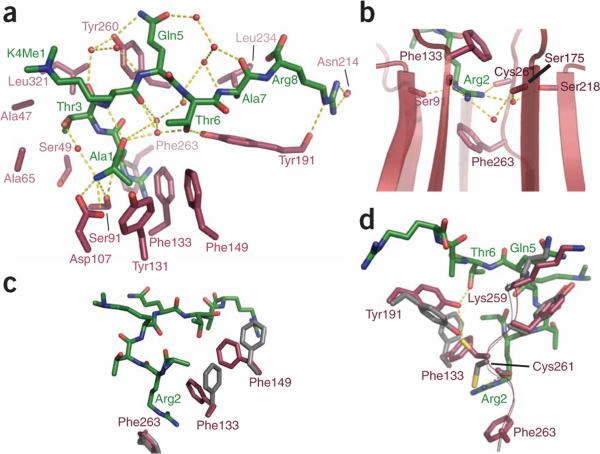
Figure 3.
Peptide recognition by WDR5 and conformational changes upon peptide binding. (a) The peptide is recognized by an elaborate series of direct and indirect contacts. Orientation of the peptide–WDR5 complex is the same as in the lower panel of Figure 1c. The majority of direct contacts from WDR5 are made to the N terminus and the first three residues. These residues adopt an approximately helical main chain conformation, with one hydrogen bond between the A1 and K4 backbone. Water-mediated contacts are important in recognition of the C-terminal residues of the peptide, as all waters shown (red spheres) are conserved among the peptide-bound structures. Tyr191 apparently acts as a central platform in this peptide-bound water network. (b) Phe133 and Phe263 form an aromatic sandwich about the R2 guanidinium moiety, equatorially flanked by a number of backbone carbonyl–mediated hydrogen bonds. These tight hydrogen bonds are thought to impart specificity for arginine over dimethyllysine, particularly the one from Nε of R2 to the Ser91 carbonyl. (c) Apparent coordinated movement of Phe133 and Phe149 to form the top of the aromatic sandwich recognition element when peptide is bound. The relevant apostructure side chains are depicted in gray and a representative liganded structure (H3K4me2 complex I) is in crimson. (d) Retraction of the loop bearing Lys259 causes a reorganization of the residues lining the central cavity, which permits tight R2 coordination. Coloring is as in c. This movement may be driven by a steric clash between this lysine and the incoming peptide Q5 side chain.
The histone H3 N terminus appears to be specifically recognized by WDR5 through a series of hydrogen bonds from the α-amino group of A1 to the Asp107 and Ser91 side chains (Fig. 3a). The hydrophobic side chain methyl group of A1 appears to be specifically recognized through van der Waals interactions with the side chains of Tyr131 and Phe149. This methyl-recognition pocket appears to be large enough to accept a methyl group, but too small to accommodate any larger side chain without engendering a severe steric clash. The guanidine-bearing side chain of R2 extends deep into the acidic end of the WDR5 central cavity and creates multiple specific intermolecular interactions. The top and bottom faces of the R2 guanidinium moiety are framed in a sandwich-like configuration of cation-π and π-π stacking interactions by Phe133 and Phe263 (Fig. 3b). A further layer of arginine-specific recognition is contributed by interactions with the edge of R2: main chain carbonyls contributed by the N-terminal strand (A-strand) of five of the seven β-propellers directly or indirectly hydrogen-bond to every nitrogen in the R2 guanidinium functional group. The indirect hydrogen bonds are mediated by two structurally conserved water molecules (Fig. 3b). This convergence of backbone amides on a single moiety of a bound ligand is reminiscent of the oxyanion hole of serine proteases, but in such proteases, it is the opposite ends of the amide dipoles that converge on the developing anion24. T3 is locked in place by a short water-mediated contact with the peptide N terminus, while the γ-methyl group projects into a hemispherical hydrophobic pocket comprised of the side chains of Ala47, Ala65 and Leu321 and the aliphatic methylene of Ser49 (Fig. 3a). This extensive array of interactions with the N-terminal end of the H3 protein provides an ample basis for specific recognition of that sequence over other N-terminal peptide sequences present in the cell.
Apparent in our structures, but absent in the recently reported WDR5–H3K4me2 complex22, are the apparently specific contacts that give rise to recognition of the backbone and side chains of Q5, T6 and R9. We attribute this additional detail to the enhanced resolution of the datasets. Though the newer structures suggest that the first three histone H3 residues are more specifically recognized than those that follow C-terminally, there are nonetheless sufficient numbers and types of contacts to residues 4–9 to impart additional binding energy and specificity. Indeed, the structure suggests that WDR5 recognizes a longer stretch of bound peptide than most other histone modification–recognition modules.
Many of the interactions between the peptide and protein consist of waters conserved throughout our liganded structure series that mediate H3 peptide-protein backbone contacts via bridging hydrogen bonds. As these waters are present in each of the peptide-bound structures and largely absent or substantially shifted in the apoprotein structure, we consider them to be an important part of peptide recognition (Fig. 3a). Notably, these water-promoted contacts are almost exclusively restricted to recognition of the peptide backbone and oxygen-bearing side chains; in particular, the backbone of peptide residues 4–7 forms a central spine of hydrated peptide. Specificity for the latter portion of this sequence is imparted by direct and indirect contacts to Q5 and T6 side chains by Tyr260 and Tyr191, respectively (Fig. 3a). By contrast, the A7 side chain is recognized by a shallow hydrophobic cleft composed of Leu234 and an aromatic face of Tyr191, analogous to the A1 side chain recognition mode (Fig. 3a). R8 forms a bidentate hydrogen bond with the backbone carbonyl of Asn214, whereas the Ne contributes a hydrogen bond to the carbonyl of Tyr191. A large patch of acidic residues is present in the vicinity of this side chain, as evident in the surface electrostatic potential renderings (Fig. 1c, arrow 3), and the acidic patch presumably forms favorable coulombic interactions with the R8 guanidinium group. Partial occupancy at this site is still apparent in the structures in which this residue is less ordered, although another conformer that positions the R8 and K9 near a different area of negatively charged surface also exists, confounding unambiguous interpretation (Fig. 1c, arrow 2).
Conformational changes upon binding
The unliganded structure is quite similar overall to the four peptide-bound species, with an average main chain r.m.s. deviation of 0.269Å that is comparable to the respective coordinate errors of each model (Supplementary Fig. 1). That stated, a few differences in WDR5 conformation between the bound and free structures are worth noting, and all of these are localized to the peptide-binding interface. In the bound structures, two aromatic residues flip down into the cavity relative to the apo form: Phe133 forms the top of the aromatic sandwich involved in the recognition of R2, and Phe149 stacks edge-to-face with Phe133 (quadrapole-to-quadrapole stacking) to stabilize the sandwich (Fig. 3c). Consistent with this important role in R2 recognition and their putative coordinated function, alanine substitutions at either position produce similar H3 peptide binding deficiencies, with mutation of Phe133 most severely attenuating binding (Fig. 4c). A loop comprising residues Lys259 to Cys261 shifts distal to the seven-fold symmetry axis, retracting Lys259 from a potential steric clash with the incoming peptide Q5 side chain and concomitantly freeing the water-filled central cavity for R2 guanidinium binding. This loop movement also shifts the carbonyl of Cys261 0.51Å closer to the Nη1 of R2, presumably strengthening this hydrogen-bonding interaction in the complex (Fig. 3d). However, this is the only portion of the R2 recognition pocket aside from Phe133 that shows appreciable movement, suggesting that this structural element is largely static. When the R2 pocket is vacated, a new side chain rotamer of Cys269 is established that forms a hydrogen bond with Tyr191's hydroxyl (Fig. 3d). The subtle withdrawal of Tyr191 from the pocket upon peptide binding permits a direct hydrogen bond to the T6 side chain in addition to stabilizing the conserved water network found in the liganded structures (Fig. 3). Mutation of this residue leads to an approximately ten-fold loss of binding energy for the H3K4me2 species (Supplementary Fig. 3 online), consistent with an important but nonessential role in peptide binding.
Figure 4.
Peptide binding by WDR5. (a) Peptide injections over immobilized WDR5-biotin. Reference and blank traces were subtracted and traces from each functionalized flow cell (three traces per concentration) were averaged for ease of viewing. Injection concentrations of peptides (residues 1–14): H3 unmodified and H3K4me3, 0.5, 0.75, 1.0, 2.0, 4.0, 6.0, 10.0 and 12.0 μM; H3K4me1, 1.0, 2.0, 4.0, 6.0, 10.0 and 12.0 μM; H3K4me2, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.75, 0.8, 0.9, 1.0, 2.0, 4.0 and 5.0 μM. The association/injection is from 0 to 200 s and the dissociation/buffer flow is from 200 to 460 s. The more gradual association and dissociation of H3K4me2 indicates both a slower on and slower off rate than those of the other peptides. Similar experiments with H31–9-derived peptide had slightly weaker binding but recapitulated the methyl-form binding trend shown. (b) Equilibrium Kd analysis. Equilibrium Kds were determined by fitting response saturation fractions calculated by the Langmuir binding isotherm (see Methods). H3 unmodified, Kd 3.3 ± 0.2 μM; H3K4me1, Kd = 8.7 ± 0.3 μM; H3K4me2, Kd = 1.02 ± 0.05 μM; H3K4me, Kd = 7.8 ± 0.2 μM. (c) Single-protein injections of wild-type WDR5 and mutants, all at 750 nM, over immobilized H31–20-biotin peptides. Reference and blank traces were subtracted. Consistent with the importance of interactions with the N terminus of the H3 peptide, mutation of Asp107 to alanine caused the greatest decrease in peptide binding among all the mutant proteins examined. Mutation of Tyr131 to alanine leads to slightly enhanced binding of the H3 peptide, suggesting that even a methyl group is slightly larger than optimal for the A1-binding pocket. By contrast, the F149A mutant showed greatly decreased H3 peptide binding, perhaps owing to Phe149's additional role in stabilizing the R2-binding pocket.
Although most protein modules that bind the H3 N terminus impart a fairly extended peptide conformation proximal to K4, with Ψ-Φ angles within the β-strand region of Ramachandran space25–28, H3 peptides bound to WDR5 are quite deformed. Notably, the peptide conformation is similar to those found in complexes with SET domains29,30. However, a shallow binding cleft in the MLL1 SET domain would be required to accommodate the composite surface of a WDR5-presented peptide without steric infractions upon complex formation.
Measurement of peptide binding
To understand the energetic contribution made by various contacts between WDR5 and the H3 peptide and the dependence of binding on the methylation state at K4, we performed detailed binding analyses using surface plasmon resonance, formatted using chip preparations having either WDR5 or the H3 peptide immobilized. With immobilized full-length protein, equilibrium analysis and kinetic-parameter fitting of the association and dissociation rates furnished similar Kd values for the series of peptides binding to the wild-type protein, validating the measurements (Fig. 4a,b). The assays with immobilized peptides were performed with wild-type and mutant proteins to assess the roles of residues deemed crucial for peptide binding on the basis of the structure (Fig. 4c). In these experiments, exact numerical analysis is confounded by moderate nonspecific adhesion of WDR5 to the dextran-streptavidin matrix; nevertheless, qualitative comparison of affinities can be made on the basis of the data.
Our binding studies show that the initially reported specificity of WDR5 for H3K4me2 is not as pronounced as that measured by peptide pull-down assays19. However, the general trend of binding is recapitulated: H3K4me2 binds WDR5 most strongly, with a measured Kd of 1.02 ± 0.05 μM, whereas the analogous monomethylated (H3K4me1) and trimethylated (H3K4me3) peptides bind more weakly (Kd = 8.7 ± 0.3 μM and 7.8 ± 0.2 μM, respectively). However, substantial binding to unmodified H3 peptide is detected, with a Kd of 3.3 ± 0.2 μM. The minor disparity from the previously reported trend19 is not surprising, as peptide pull-down experiments are qualitative measurements dominated by the koff term (due to resin-washing steps), and therefore they may amplify small differences in apparent affinity. Overall, the affinities measured here are similar: the difference between the most tightly and the most weakly binding peptide is only about eight-fold, indicative of very modest binding-affinity differences, on the order of 1.2 kcal mol−1.
Differences in the kinetics of binding suggest that recognition of the K4 methylation state and decomposition of the bound complex may proceed through some transient intermediate before achieving the final bound form observed in the crystal structures. The rates of association and dissociation for H3K3me2 binding to WDR5 are discernibly unique: relative to the other peptides, H3K3me2 has a much slower kon, manifested as a slower approach to equilibrium, and a slower koff, apparent in the sluggish decay of bound peptide signal in the dissociation phase (Fig. 4a).
Discrimination of histone H3 methylation states
Among the complex structures in our series, it is difficult to discern any specific interactions arising as a consequence of differential methylation at K4 that appear to be important for peptide discrimination. This is surprising, given that all other methylated lysine–binding modules that have been studied use an aromatic cage, presumably as a consequence of convergent evolution25–28. The K4 side chain adopts two general conformers divergent at χ3 (the torsion angle between Cγ and Cδ), in a manner apparently dependent on the methylation status of K4 (Fig. 5). Perhaps this is a consequence of crystal packing, although the entire methylated series was crystallized under the same conditions, in the same space group, with nearly identical unit cells (Fig. 5). Furthermore, we obtained structures of several other H3K4me2-bound WDR5 complexes crystallized in different space groups that were not appreciably different from the corresponding complex with the 9-residue peptide (complex II and data not shown). Moreover, the conformation of K4me2 in the recently reported structures of this complex22 appears to be identical to that reported here.
Figure 5.
Differences in K4 conformation in the different methylation states. (a–d) The crystal-packing interface relevant to K4 conformation is depicted for the H3K4me3 complex (C2 space group, a), H3K4me2 complex I (C2, b), H3K4me1 complex (C2, c) and unmodified H3 complex (P21, d). Peptides are colored as in Figure 2b; the principal WDR5 protomer is red; and the symmetry-related protomer at the peptide interface is gray. Note the rotation about χ3 moving from the tri- and dimethylated species to the monomethylated and unmodified species. In H3K4me2 complex I, the distances between the ζ-methyl carbons and the Glu322 carboxylate Oδ1 are 3.27 Å and 3.37 Å for the closest methyl group in each of the two complexes per asymmetric unit, whereas these distances are 3.83 Å and 3.87 Å for the more distant methyl group. For comparison, the previously reported shorter distances for these measurements were 3.15 Å and 3.42 Å22.
The recent report on WDR5 in complex with H3K4me2 peptides suggested unconventional hydrogen bonds between the ζ-methyl groups and Glu322 as the source of discrimination among the methylation states of K4 (ref. 22). The distances measured in this 1.9-Å structure between the methyl carbons and the Glu322 carboxylate oxygen fell slightly within the sum of atomic van der Waals radii, and these groups were therefore assumed to be repulsive in the absence of the putative hydrogen bonds. In our 1.5-Å H3K4me2 structure, these distances are longer than those previously measured (Fig. 5b). Furthermore, the interceding proton between the postulated donor and acceptor atoms would be 2.49–2.57Å away from the Oδ2, a distance longer than the average H···O hydrogen bond distance31. According to a calculation with the AMBER force field (ignoring the attractive force of a putative CH···O hydrogen bond), there is still an attractive potential resulting from a NH·m=.·O hydrogen bond and salt bridge that outweighs van der Waals repulsion at the distances in our structure32. Thus, even in this intimate distance regime, there is still modest attraction, suggesting that unconventional hydrogen-bonding is not required to explain the observed distances. Regardless, the K4 side chain of H3K4me2 forms more extensive intracomplex interactions than that of any other peptide, generating a hydrogen-bonding network between the ammonium-proximal waters and Glu322 (Fig. 5b).
In the H3K4me1-bound WDR5 structure, the K4 conformer in the crystal has a different χ3 angle from that found in the other two methylated peptide structures, although there is ~ 15% occupancy of the conformer observed for K4me2 and K4me3. Consequently, there is no substantial interaction between the K4me1 side chain and Glu322 in this structure, despite predictions that one unconventional hydrogen bond should exist (Fig. 5c)22. Rather, there are contacts from the K4 side chain to Glu151 and a backbone carbonyl of a symmetry-related protomer in the crystal (Fig. 5c). The unmodified peptide is found in a nearly identical conformation in a different space group, although this packing arrangement produces a similar set of contacts (Fig. 5d).
With H3K4me3, the Glu322 side chain retracts from what would be a very repulsive Cζ-Oδ1 distance of 2.76 Å between this side chain and the third methyl group. This conformation of the Glu322 side chain places the carboxylate so that the shortest distance between Oδ2 and the two H3K4 ζ-methyl groups is 3.75 Å (Fig. 5a), longer than the putative range of CH···O hydrogen bonds 31. Thus, the interactions predicted by ref. 22 appear not to exist in the H3K4me3 structure. Rather, the distances observed are consistent with a solvent-exposed salt bridge.
DISCUSSION
The role of WDR5 in SET1-family complexes
A paradox is apparent in the previous WDR5 literature: WDR5 binds preferentially to histone H3 dimethylated at K4, yet somehow affects global trimethylation at the very same residue19. This paradox is resolved, at least in part, by the present data coupled with the report of Dou et al.33 The most noteworthy result of structural analysis is that K4 of the peptide appears to be presented for recognition by another component of the MLL1 complex, and the binding data confirm that dimethylation at this position is modestly recognized. This structure-derived hypothesis is further validated biochemically in the paper of Dou et al.33, which shows that WDR5 mutations that attenuate H3-binding activity, devised by inspection of the present structures, decrease the HMTase activity of the MLL1 core complex when incorporated in place of the native WDR5.
The significance of the modest differences in H3 peptide binding to isolated WDR5 remains unclear; however, in the context of histone methylation complexes such as MLL1, WDR5 clearly associates with nucleosomes bearing di- and trimethylation at H3K4 (ref. 19). This strong preferential association is the result of either additional binding affinity imparted by some other complex module or processive turnover by the SET domain acting on the presented K4. There is precedent for SET domains having preferences for the methylation state of their lysine substrates manifested as Km effects34 and kcat effects29,35 such that further discrimination of the methylation form could be accomplished by the SET domain in its binding and turnover. The recombinant MLL1 SET domain (like several other isolated SET proteins36) has very poor HMTase activity, particularly in performing the di-to-trimethylation reaction12. Yet the addition of WDR5 does not enhance the HMTase activity of recombinant MLL (data not shown). Thus, other complex components must be important both in modulating catalysis and in amplifying the modest methyl-form discrimination in WDR5's intrinsic binding activity.
Global H3K4 trimethylation is lost upon WDR5 knockdown in animals19, yet the MLL1 complex may only be present at a fraction of actively transcribed promoters37. Thus, it is likely that WDR5 has a ‘peptide presentation’ role in most if not all HMT complexes involved in H3K4 trimethylation.
Methyl-form discrimination by WDR5
Although we cannot rule out the possibility that unconventional CH···O hydrogen bonds have some role in determining methyl-form specificity22, this explanation does not satisfactorily explain the observed trend of binding affinity and is inconsistent with our mutagenesis studies. If noncanonical hydrogen bonds (from the methyl groups of the more highly methylated forms of K4 to Glu322) were the key determinant of the observed binding specificity, a loss of methyl-form preference would be expected upon mutation of this residue. Although the E322A mutant binds all forms of the peptide more weakly than wild-type WDR5, identical preferences for the different methylated states are retained (Fig. 4c). Moreover, according to the above hypothesis, more highly methylated forms of K4 should bind more tightly than the unmodified form22, so that affinity decreases monotonically with methylation. Our binding data do not show such a simple trend.
In the absence of a convincing structural basis for the established preference for dimethylation at K4, this binding preference may be attributed to a modest kinetic discrimination for H3K4me2, manifested as a slower koff relative to other methylated forms of the peptide (Fig. 5a). This modest preference may be imparted by a transient binding intermediate that collapses to the thermodynamic minima under saturating peptide concentrations on a crystallographic time-scale, so that the crystallized structures do not show this transient intermediate state. Perhaps the H3K4me2 preference results from binding of the methylated lysine by an aromatic cage–like structure, analogous to recognition motifs observed in all other methyllysine-binding modules25–28. It is tempting to speculate that the aromatic residues Tyr131, Phe133 and Phe149 near the final resting place of the K4 side chain have this role (Fig. 1c, arrow 5), but this is not clear from the mutagenesis, given the additional functions of these residues in R2 recognition. The presence of negative surface potential patches proximal to the observed peptide-binding groove also could represent intermediate association sites for this polycationic peptide (Fig. 1c, arrows 1–4). Furthermore, the positions of waters in liganded structures relative to the apo form indicates that appreciable solvent rearrangement must occur between these two limiting states. Perhaps differential water rearrangement or differences in the entropy of freeing waters contribute to the overall modest preference for H3K4me2. Any of these factors could produce a ~ 1.2 kcal mol−1 difference in affinity without apparent structural basis.
All known SET domains perform methyltransferase chemistry within a deep cavity29,30. Thus, WDR5 would not be expected to interrogate the methylation state of the K4 side chain and simultaneously present this same moiety for SET-mediated methylation. Regardless of how the modest kinetic selection is actually achieved, it enables the dual functions of H3K4 recognition and presentation by WDR5 to occur sequentially. Moreover, the somewhat promiscuous binding with regard to K4 methylation also permits, albeit to a lesser extent, the presentation of less highly methylated and unmodified forms of H3 for methylation. Thus, we envision the WDR5 presentation of H3K4 and its subsequent methylation to be sensitive to, yet not wholly dependent upon, preexisting K4 methylation. It is noteworthy that a slight reduction in monomethylation of K4 accompanies the abrogation of global K4 trimethylation upon WDR5 knockdown19, consistent with the measured peptide affinity trend.
WD40-repeat proteins in histone modification
WD40-repeat proteins that are coupled with SET domain proteins may represent a common mechanism for substrate selection and presentation preserved among HMTase complexes that methylate other histone positions, even those that promote the lysine methylation events that cause DNA silencing. EZH2, the archetypal SET domain protein involved in silencing, does not show HMTase activity outside of a complex that also contains two WD40-repeat proteins, EED and RbBP4 (also called RbAp48), the latter of which has been shown to bind histones38,39.
There is good reason to anticipate that peptide recognition and presentation by WD40-repeat proteins is a more general phenomenon extending to other post-translational modification complexes. There are a number of WD-repeat domain proteins that physically and genetically interact with histone-modification catalytic proteins. Examples of such WD40-repeat proteins include Groucho proteins, which bind the histone H3 N-terminal tail in Rpd3–H3–HDAC complexes40; RbBP4 and RbBP7, which are required for the human Hat1 acetyltransferase complex activity and bind the complex's histone H4–tail substrate39; and the HIRA histone H3 chaperone, which associates with HDAC-1 in several complexes41. From an evolutionary standpoint, there is some logic and efficiency to the notion of delegating specific substrate recognition and catalysis to separate components: recognition modules could evolve without the constraint of maintaining catalytic power and combinatorially associate with alternative catalytic subunits. Although it is intriguing to speculate that these WD40-repeat proteins may serve similar histone peptide–recognition and substrate-presentation roles in their complexes, attribution of these functions awaits further study.
METHODS
WDR5 protein and histone peptide preparation
WDR5 was PCR-amplified from MGC clone 1025 and inserted into pMCGS7 by ligation-independent cloning to furnish tobacco etch virus protease–cleavable N-terminal His6 fusions corresponding to amino acid residues 2–335 (full length) and 23–335 (ΔN-WDR5). For the surface plasmon resonance experiments, a sequence-specific BirA biotinylation sequence (SGLNDIFEAQKIEWH) was fused N-terminally to the full protein (b-WDR5) for endogenous biotinylation at the lysine indicated (bold). Mutagenesis to make S91K, D107A, Y131A, F133A, F149A, Y191F, F219A, E322A and combinations thereof was performed with the QuikChange kit (Stratagene). All proteins were expressed in the Rosetta 2(DE3)PlysS expression host (Novagen) and purified to homogeneity on a series of nickel–nitrilotriacetic acid (Qiagen), Heparin Hitrap, then Superdex 75 chromatography columns, with intervening tobacco etch virus protease cleavage of the His6 tag. An additional monomeric avidin (Pierce) chromatographic step was used to isolate biotinylated b-WDR5 before gel-filtration chromatography in GF buffer (50 mM Tris-HCl, 500 mM NaCl and 10 mM β-mercaptoethanol (βME) (pH 7.4)). Histone H3 peptides (1–9, 1–14 and 1–20-Y-biotin) were prepared by the conventional Fmoc-SPSS method (with protected amino acids purchased from EMD and Bachem), purified to >95% homogeneity by reverse-phase HPLC, then characterized and quantified by LC-ESI mass spectrometry and amino acid analysis. The protein and peptide preparation for H3K4me2 complex II and the unmodified peptide complex is detailed in Supplementary Methods online.
Crystallization and data collection
ΔN-WDR5 crystals (250 μm × 200 μm × 50 μm) were grown by hanging drop vapor equilibration in Nextal plates as follows: 1 μl of 10–15 mg μl−1 protein solution (10 mM Tris-HCl (pH 7.4), 50 mM NaCl and 10 mM bME) were mixed with 1 μl of well solution composed of 50 mM HEPES (pH 7.5), 100 mM potassium formate and 10%–20% (w/v) polyethylene glycol 3,350 and equilibrated at room temperature overnight against 1 ml of well solution. Upon equilibration, cat whisker streak seeding from smaller crystals grown in higher PEG concentrations consistently provided diffraction-quality crystals. The complexes of WDR5 with various H3 peptide methylation forms were grown similarly, by premixing 3 molar equivalents of peptide in distilled water with 1 ΔN-WDR5 equivalent just before hanging drop setup. Crystals of the H3K4me2 complex II and the unmodified peptide complex were grown under different conditions, detailed in Supplementary Methods. Truncation of the first 23 amino acids of WDR5 in the recombinant protein was requisite for effective crystallization; this region of the protein is highly divergent among higher eukaryotes and is predicted to be unstructured19, and it therefore is anticipated to be dispensable for function. All data was collected at 100 K at the locations described in Table 1.
Table 1.
Data collection and refinement statistics
| Apo-WDR5a | Unmodified H31–15–WDR5 complexb |
H31–9K4me1–WDR5 complexc |
H31–9K4me2–WDR5 complex Ic |
H31–9K4me2–WDR5 complex IId |
H31–9K4me3–WDR5 complexd |
|
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | C2 | P2 1 | C2 | C2 | C2 | C2 |
| Cell dimensions | ||||||
| a, b, c (Å) | 134.6, 46.6, 112.4 | 64.9, 47.0, 101.8 | 133.5, 46.5, 112.1 | 134.7, 46.1, 112.7 | 100.3, 46.8, 64.8 | 134.3, 46.4, 111.4 |
| α, β, γ (°) | 90.0, 117.2, 90.0 | 90.0, 107.0, 90.0 | 90.0, 116.9, 90.0 | 90.0, 117.1, 90.0 | 90.0, 107.1, 90.0 | 90.0, 116.7, 90.0 |
| Resolution (Å) | 50.0–1.79 (1.86–1.79)e,f | 20.0–2.25 (2.33–2.25)e,f | 50.0–1.89 (1.97–1.89)e,f | 50.00–1.50 (1.55–1.50)e,f | 20.0–2.10 (2.17–2.10)e,f | 50.00–1.88 (1.95–1.88)e,f |
| R merge | 11.9 (16.9) | 15.3 (56.5) | 7.1 (25.5) | 14.9 (46.4) | 7.5 (23.6) | 9.1 (27.1) |
| I / σI | 10.9 (7.5) | 15.8 (3.07) | 16.2 (3.8) | 9.2 (2.9) | 27.2 (5.1) | 19.8 (6.6) |
| Completeness (%) | 99.6 (99.8) | 100.0 (100.0) | 98.9 (92.9) | 99.9 (99.6) | 98.5 (97.0) | 99.4 (95.1) |
| Redundancy | 3.9 (3.5) | 6.7 (6.5) | 3.2 (2.5) | 4.3 (3.5) | 6.8 (6.0) | 6.9 (5.4) |
| Refinement | ||||||
| Resolution (Å) | 1.78 | 2.25 | 1.89 | 1.50 | 2.10 | 1.88 |
| No. unique reflections | 57,091 | 28,238 | 48,816 | 98,522 | 16,940 | 50,232 |
| Rwork / Rfree | 17.0 / 19.2 | 19.0 / 22.6 | 14.6 / 19.0 | 17.9 / 19.8 | 19.2 / 22.3 | 15.2 / 18.5 |
| No. atoms | ||||||
| Protein | 4,723 | 4,711 | 4,735 | 4,721 | 2,369 | 4,705 |
| Peptide | – | 84 | 141 | 134 | 44 | 114 |
| Water | 581 | 297 | 737 | 632 | 192 | 498 |
| B-factors | ||||||
| Protein | 15.3 | 23.0 | 16.1 | 15.8 | 29.6 | 16.2 |
| Ligand/ion | – | 25.4 | 24.1 | 17.6 | 27.2 | 21.7 |
| Water | 30.7 | 32.7 | 32.8 | 32.4 | 39.9 | 33.9 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.012 | 0.006 | 0.011 | 0.009 | 0.006 | 0.013 |
| Bond angles (°) | 1.38 | 1.53 | 1.26 | 1.29 | 1.59 | 1.37 |
Data collected at NSLS X-29.
Data collected at Memorial Sloan-Kettering Cancer Center on a RIGAKU R-AXIS IV++.
Data collected at Advanced Light Source 5.0.2.
Data collected at APS 24ID-C.
Highest-resolution shell is shown in parentheses.
Single-crystal data set.
Structure solution and refinement
Structures were solved using MOLREP42 and BEAST43 from the CCP4 suite44 and a loop-truncated polyalanine sequence with backbone coordinates derived from the C-terminal tup1 structure as a probe. Initial building and refinement was done in ARP/wARP45. Further rounds of simulated annealing, conjugate gradient energy minimization and B-factor refinement, carried out in CNS46 and interspersed with manual rebuilding in Coot47, furnished nearly complete models. The final stages of refinement were done in Refmac48 using TLS refinement49, and PRODRG50 generated parameter files for the methylated lysine species. Ramachandran statistics are as follows (residues in most favored regions, residues in additional allowed regions, residues in generously allowed regions, residues in disallowed regions): apo-WDR5, 89.8%, 10.1%, 0.0%, 0.2%; unmodified H31–15–WDR5 complex, 86.2%, 13.4%, 0.4%, 0.0%; H3K4me11–9–WDR5 complex, 88.9%, 11.1%, 0.0%, 0.0%; H3K4me21–9–WDR5 complex I, 90.0%, 10.0%, 0.0%, 0.0%; H3K4me21–9–WDR5 complex II, 89.8%, 10.1%, 0.0%, 0.4%; H3K4me31–9–WDR5 complex, 89.0%, 11.0%, 0.0%, 0.0%51. All structure figures were generated in PyMOL (http://pymol.sourceforge.net) with the APBS plugin52 for surface electrostatics. Superpositions were done using LSQKAB in the CCP4 suite44.
Binding studies
All experiments were performed using a Biacore 3000 sensor (Biacore AB). The change in response level is proportional to the change in mass at the surface, as the interacting partner (the analyte) binds during its injection over the surface with bound ligand.
Either b-WDR5 or biotinylated peptide were immobilized on a Sensor Chip SA (Biacore AB) after the surface had been conditioned with three injections of 50 mM NaOH and 1 M NaCl for 1 min at 10 μl min−1. Peptides were immobilized to relatively high levels to see the peptide response over the high nonspecific background: H3K4me1, 497 resonance/response units (RU); H3K4me2, 701 RU; H3K4me3, 675 RU. Site-specifically biotinylated WDR5 was immobilized to 4,000-5,000 RU in flow cells 2, 3 and 4, giving three traces per injection of analyte, with flow cell 1 used as the blank for subtraction. Any free avidin sites were blocked with an injection of 1 mM biotin (in 50 mM phosphate buffer (pH 8.0)) for 1 min at 10 μl min−1. HBS-EP (10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% (v/v) surfactant P20) plus 5 mM βME was used as the instrument running buffer. Peptides and proteins were diluted directly into this buffer from a stock in water or GF buffer by at least 500 and 85 times, respectively. The runs were started with six start-up cycles, where HBS-EP plus βME was injected instead of sample, followed by sample-injection cycles, interspersed every three injections with another buffer injection. The peptide analyte injection cycles consisted of a 250-μl sample injection (75 μl min−1), 180 s of buffer flow (dissociation phase), a 40-μl regeneration injection (1 or 1.5 M NaCl in 70%–80% (v/v) HBS-EP plus βME) and a 30-s buffer injection. The unbiotinylated protein analyte injection cycles consisted of a 200-μl sample injection (75 μl min−1), 180 s of buffer flow (dissociation phase), a 75-μl regeneration injection (50 mM NaOH and 1 M NaCl) and a 10-μl regeneration injection (0.25% (w/v) SDS, 25 mM NaOH, 0.5 M NaCl) needed to remove the nonspecifically but tightly bound protein from the SA-dextran surface. The flow cell temperature was 25 °C for all experiments.
Scrubber 2.0 (Center for Biomolecular Interaction Analysis, University of Utah) was used for subtraction of reference and blank data, to align traces and to calculate saturation fractions for use in the equilibrium Kd analysis. KaleidaGraph 3.0 (Synergy Software) was used to fit the equilibrium Kds, using a modified Langmuir binding isotherm: y = m1 + (1 – m1)(x/(x + Kd)), where y is the fraction of saturation, x is the concentration and m1 allows for curve fitting when the plots do not trend toward zero. Biacore 3000 BiaEvaluation software (Biacore AB) was used for kinetic fits.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the staff of National Synchrotron Light Source (NSLS) beamline X-29, in particular W. Shi and H. Robinson, as well as the staff of Advanced Light Source beamline 5.02, especially C. Trame, and the staff of Advanced Photon Source (APS) beamline 24-ID, importantly I. Kourinov and K. Rajashankar, for assistance in X-ray data collection. Financial support for NSLS px group beamlines comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy and from the National Center for Research Resources of the US National Institutes of Health (NIH). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the APS, which is supported by the National Center for Research Resources at the NIH. We thank the Dana Farber Cancer Research Center, F. Bernal and L. Walensky for amino acid analysis and peptide synthesis advice; S.S. Yi (Memorial Sloan-Kettering Cancer Center) for synthesis of H3K4me2 complex II and the unmodified peptides, the Dana Farber Cancer Research Center and the Harvard Center for Genomics Research for use of their Biacore instruments, and F. Bernal, Y. Dou, K. Herlihy, Y. Inuzuka, D. Pakotiprapha, T.A. Milne, R.G. Roeder, S.D. Taverna and J. Wysocka for valuable discussions and scientific input. This work was supported by an NIH grant to G.L.V.; D.J.P. is supported by funds from the Abby Rockefeller Mauze Trust and the Dewitt Wallace and Maloris Foundations; and C.D.A. is supported by an NIH MERIT award and funds from Rockefeller University.
Footnotes
AUTHOR CONTRIBUTIONS
A.J.R. is responsible for the X-ray studies of the apo structure and the H3K4me1, H3K4me2 and H3K4me3 structures, W.-K.W. and H.L. are responsible for the unmodified peptide structure and an additional H3K4me2 structure (complex II), and D.M.G. performed the binding studies with some assistance from A.J.R. and W.-K.W. G.L.V., D.J.P. and C.D.A. supervised the structural and biochemical aspects of the project and take overall responsibility for their joint research. All authors discussed the results and commented on the manuscript.
Accession codes. Protein Data Bank: coordinates have been deposited with the accession codes 2H68, 2CO0, 2H6K, 2H6N, 2CNX and 2H6Q, representing the apo structure, unmodified H3 complex, H3K4me1 complex, H3K4me2 complex crystal form I, H3K4me2 complex crystal form II and H3K4me3 complex, respectively.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Wang Y, et al. Linking covalent histone modifications to epigenetics: the rigidity and plasticity of the marks. Cold Spring Harb. Symp. Quant. Biol. 2004;69:161–169. doi: 10.1101/sqb.2004.69.161. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 6.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 7.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 8.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs SD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller T, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 13.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura T, et al. ALL-1 is a histone methyltransferase that assembles a super-complex of proteins involved in transcriptional regulation. Mol. Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 15.Roguev A, et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem. Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 17.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol. Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Hughes CM, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 19.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 20.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 21.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. USA. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Z, et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Sprague ER, Redd MJ, Johnson AD, Wolberger C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 2000;19:3016–3027. doi: 10.1093/emboj/19.12.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson R. Structure of crystalline alpha-chymotrypsin. IV. The structure of indoleacryloyl-alpha-chyotrypsin and its relevance to the hydrolytic mechanism of the enzyme. J. Mol. Biol. 1970;54:341–354. doi: 10.1016/0022-2836(70)90434-1. [DOI] [PubMed] [Google Scholar]
- 25.Li, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature advance online publication 21 May. 2006 doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 27.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao B, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 31.Scheiner S, Kar T, Gu Y. Strength of the Calpha H.O hydrogen bond of amino acid residues. J. Biol. Chem. 2001;276:9832–9837. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 32.Case DA, et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. doi: 10.1038/nsmb1128. in the press. [DOI] [PubMed] [Google Scholar]
- 34.Chin HG, Patnaik D, Esteve PO, Jacobsen SE, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–3284. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- 35.Hu P, Zhang Y. Catalytic mechanism and product specificity of the histone lysine methyltransferase SET7/9: an ab initio QM/MM-FE study with multiple initial structures. J. Am. Chem. Soc. 2006;128:1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 36.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 37.Milne TA, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc. Natl. Acad. Sci. USA. 2005;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 40.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad A, Takami Y, Nakayama T. WD dipeptide motifs and LXXLL motif of chicken HIRA are essential for interactions with the p48 subunit of chromatin assembly factor-1 and histone deacetylase-2 in vitro and in vivo. Gene. 2004;342:125–136. doi: 10.1016/j.gene.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 43.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project Number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Morris RJ, et al. Breaking good resolutions with ARP/wARP. J. Synchrotron Radiat. 2004;11:56–59. doi: 10.1107/s090904950302394x. [DOI] [PubMed] [Google Scholar]
- 46.Brunger AT, et al. Crystallography & NMR system: a new software suite for macro-molecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 50.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystal-lography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 51.Laskowski RJ, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–290. [Google Scholar]
- 52.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.