Abstract
Genetic and genomic approaches have implicated hundreds of genetic loci in neurodevelopmental disorders and neurodegeneration, but mechanistic understanding continues to lag behind the pace of gene discovery. Understanding the role of specific genetic variants in the brain involves dissecting a functional hierarchy that encompasses molecular pathways, diverse cell types, neural circuits and, ultimately, cognition and behaviour. With a focus on transcriptomics, this Review discusses how high-throughput molecular, integrative and network approaches inform disease biology by placing human genetics in a molecular systems and neurobiological context. We provide a framework for interpreting network biology studies and leveraging big genomics data sets in neurobiology.
Large-scale genetic association studies have begun to unravel the genetic architecture of neurodevelopmental and neurodegenerative disorders and have found that hundreds to thousands of genetic loci are involved in disease risk1. To understand how genetic variants contribute to disease, neuroscientists are faced with the task of measuring and understanding phenotypes in the central nervous system (CNS), a hierarchically organized complex system (FIG. 1a). This leads to a reliance on models that only account for a few features of the CNS at a time, as is done in most laboratory experiments. Although this has been fruitful for some highly penetrant variants that yield clear phenotypes, it has been less successful for genetically complex diseases.
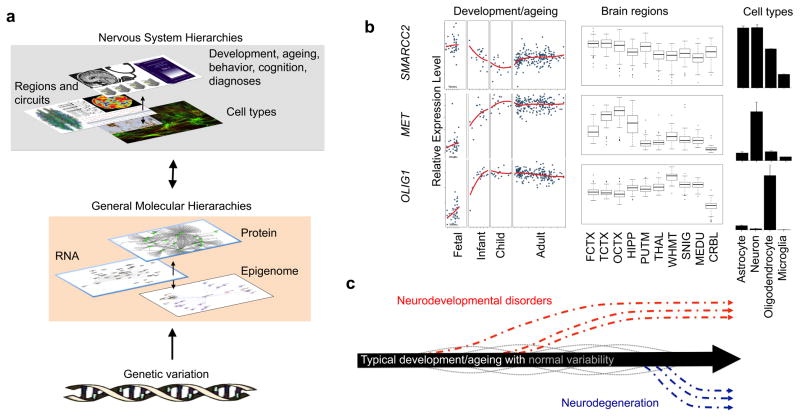
Figure 1. Molecular systems and the neurobiological hierarchy.
a | Genetic variants exert their effects on cognitive and behavioural phenotypes associated with neurodevelopmental or neurodegenerative disease through a neurobiological hierarchy that includes multiple molecular levels (transcriptomic, proteomic and epigenomic) that can be modelled as networks on the basis of physical interactions and correlations within and across multiple molecular levels (BOX 2). These molecular levels of organization can vary at multiple neurobiological phenotypic levels (cells, circuits, and cognition and behaviour) across the lifespan. b | Gene expression levels vary dramatically across development and ageing, brain regions and cell types, as illustrated by three genes: SMARCC2, which is a pan-regional neurodevelopmental gene; MET, a regionally patterned adult neuronal gene; and OLIG1, a gene most highly expressed in white matter and oligodendrocytes. Development and ageing data are from BrainCloud17, regional data are from Braineac16 and cell type expression data are from fluorescent-activated cell sorted transcriptomes from mouse cortex162 (http://web.stanford.edu/group/barreslab/brainrnaseq.html). c | Both the molecular and phenotypic levels exhibit a typical trajectory with normal variation during development and ageing that can be altered in disease, resulting in abnormal temporal trajectories. The x axis on this plot reflects the progression of time, and the y axis reflects theoretical deviation from the normal trajectory for any molecular or phenotypic measurement. CPi, inner cortical plate; CPo, outer cortical plate; CRBL, cerebellum; FCTX, frontal cortex; HIPP, hippocampus; ISVZ, inner subventricular zone; IZ, intermediate zone; lncRNA, long noncoding RNA; MEDU, brainstem medulla; miRNA, microRNA; OCTX, occipital cortex; OSVZ, outer subventricular zone; PUTM, putamen; SNIG, substantia nigra; SP, subplate; TCTX, temporal cortex; THAL, thalamus; VZ, ventricular zone; WHMT, subcortical white matter.
To understand how genes contribute to CNS phenotypes, it is necessary to adopt rigorous data-driven frameworks that operate at a systems or a network level2–4. Methods have recently become available that permit the measurement of large-scale molecular4,5, cellular6 and circuit-level3 phenotypes, and additional methods are currently in development7. One goal of these approaches is to connect genetic risk and mechanism by combining a molecular systems or integrative network approach with systems neuroscience to understand the molecular regulatory networks and pathways that underlie circuit function, behaviour and cognition in health and disease. Collaborative and consortium-level efforts have made substantial progress by mapping transcriptomic, epigenomic and proteomic landscapes in the brain8–10. Recent important advances include the evaluation of spatial and temporal transcriptomes by the Allen Brain Institute and BrainSpan8,11–13, the quantification of the epigenetic landscape in CNS tissue and cell types by the Roadmap Epigenomics Mapping Consortium14, and the integration of genetic variation with gene expression in the brain by the Genotype-Tissue Expression (GTEx) project15, as well as others16,17. These efforts have provided the first systematic view of the immensely complex molecular landscape across brain development, between brain regions and among major cell types (FIG. 1b). However, the molecular signatures of specific cell types, finer-grained temporal dynamics and causal or reactive alterations in CNS diseases remain mostly uncharacterized (FIG. 1c). Nevertheless, these new resources serve as an important foundation and proof of the value of such tissue- and stage-specific profiling data.
Molecular profiling and network approaches in disease-relevant neuroscience research face several major challenges when applied to the CNS: the complexity of molecular phenotypes owing to cell type, spatial and temporal heterogeneity throughout nervous system development and maturation (BOX 1); a dearth of human tissue and model systems with definitive human relevance (the ‘translational’ and ‘evolutionary’ problems4,18,19); and poor knowledge of appropriate intermediate phenotypes to measure. Although these challenges are not unique to studying the CNS, neuroscience has historically struggled with each of them owing to the extent that they affect the ability to link molecular function to behavior and cognition. Foundational aspects of each point have not been agreed: the definition of a cell type in the brain remains controversial20,21; the relationships of human disease phenotypes to developmental trajectories are relatively unknown; model systems in many neurobiological studies are often chosen on the basis of convenience and history; and most phenotypes are based on clinical and behavioural symptomatology rather than on biological mechanism or aetiology22–24.
Box 1. The unique cytoarchitecture and development of the brain.
Most neurodevelopmental and neurodegenerative disorders are defined by perturbations in specific cognitive and/or behavioural domains, pointing to a selective vulnerability of specific cells. Regional and cellular heterogeneity pose obstacles for transcriptomic studies in the central nervous system (CNS)100,200, but whole-tissue investigations in post-mortem human brain tissue are essential for identifying human-relevant global changes. These changes can be compared across regions to identify the most vulnerable regions and time points for further investigation. In general, the value of whole-tissue profiling in post-mortem brain tissue depends on the disease. In neurodevelopmental disorders, the specific brain regions, cell types or time points that are most affected remain poorly defined and whole-tissue profiling still holds great value. By contrast, for many neurodegenerative diseases, the selective death of certain cellular populations and the infiltration of inflammatory cells is well characterized, so transcriptomic studies focusing on sorted cellular populations are now necessary to identify new associations with disease.
To maximize neurobiological understanding from whole-tissue profiles, global changes can be related to cell type-specific gene expression profiles30,32,149,163,201, and targeted experiments can be carried out to identify novel insights, as highlighted by several recent studies202–205. However, it will be impossible to study disease-affected cell types without a complete knowledge of cell identities in normal brain development and ageing. A priority is to develop a complete knowledge of the cellular identity and cytoarchitectural changes that occur over time. This will necessitate surveying the diversity of cellular types and deciphering their molecular identities using single-cell approaches206–208.
Additionally, neuronal gene expression and epigenetic programmes also undergo changes at finer spatial and temporal scales, including changes induced by activity-dependent transcription in the nucleus and translation209 at the synapse. Locally regulated translation of these subcellular transcriptomes210 has a crucial role in synaptic function and plasticity211. Deeper characterization of these events at a high spatiotemporal resolution in normal brains followed by integration with coarser profiles from specific diseases will identify cellular compartments and mechanisms for more targeted study that are currently missed. Network approaches are particularly useful for relating whole-tissue-level changes to data from these high-resolution experiments11–13,26 (FIG. 2a).
In this Review, we provide an overview of integrative genomics approaches that have been applied to understand the basis of CNS disorders, and we anchor this discussion around transcriptomics (BOX 1). However, the themes discussed can be generalized to genomic, proteomic and epigenomic methods. We describe how large-scale molecular data sets and gene network approaches provide organizing principles that permit the development of testable hypotheses on a genome-wide scale. We discuss new insights into neurodevelopmental disorders and neurodegenerative diseases from these studies, highlight emerging themes and provide recommendations for designing and executing future molecular profiling studies.
Network biology and transcriptomics in the brain
Despite challenges in studying the CNS, dozens of informative transcriptional analyses of neurodevelopmental and neurodegenerative disorders have been carried out in the human brain. A major challenge, which has mostly been surmounted at the theoretical level and which now requires reduction to practice, has been measuring and identifying which genes are altered in disease in specific cells, circuits and regions. Differential gene expression analysis (DGE analysis) addresses this issue, albeit one gene at a time, but does not take into account the relationships between genes. This leads to additional challenges, including the interpretation of long lists of differentially expressed genes and integration of DGE sets with other data. Network methods (BOX 2) relate genes to each other using the measured or predicted relationships between them4 and provide an essential organizing framework that places each gene in the context of its molecular system. Gene network methods are now being applied to integrate genetics with transcriptomics, epigenomics and proteomics to identify causal molecular drivers of cellular, circuit-level and brain-wide pathology in disease. We review the principles of network analysis below and also delve into applications of molecular systems and integrative network approaches in neuropsychiatric and neurodegenerative disease.
Box 2. A framework for interpreting gene network analysis.
Molecular profiling data can be modelled as a network in which molecules or gene products are nodes and their functional relationships with each other are edges. Gene network analysis can be summarized in five basic steps.
Node specification
Seeded (prior-based) networks have nodes that are selected using prior knowledge, such as genetic variants that are associated with a disorder, and unseeded (genome-wide) networks use all available measurements from the genome.
Edge specification
In order to define edges, studies need to include one or more of the following: experimentally observed pairwise statistical relationships25,212,213 evaluating shared patterns of molecular levels across experiments, such as co-expression; experimentally observed or literature-curated physical interactions, such as protein interactions from immunoprecipitation and yeast two-hybrid (Y2H) experiments; or computationally predicted relationships, such as transcription factor binding based on DNA motifs. Notably, edges are susceptible to ascertainment biases52,214,215 and confounding factors that can induce spurious relationships178 (FIG. 2b).
Module identification
Modules are identified from an adjacency matrix to simplify biological relationships at a higher-order level, identifying interacting or highly correlated gene products (FIG. 2c). Assessing node connectivity or position within the module can identify hubs and enables the comparison of changes between health and disease at the module level.
Annotation of modules or gene connectivity
There are several common approaches to annotate modules. External measures of gene importance (such as cell type specificity or genome-wide association study (GWAS) signals) can be related to module membership, intra-modular connectivity or network-wide gene connectivity. Module summary or hub gene measurements, such as module eigengenes or average expression levels, can be associated with biological traits. Any differential gene expression (DGE) test that can be applied at a single-gene level can be applied to module-level summaries, such as eigengenes. Module-level association reduces the problem of multiple comparisons, as there are far fewer modules than genes in a network. The preservation or changes in network connectivity for specific genes or modules can be assessed between health and disease. Data can be integrated at the edge level or the module level across biological levels, such as different cell types or brain regions, or different regulatory levels, such as gene expression and ChIP–seq signals.
Validation
The crucial issue of reproducibility is addressed by validating network observations in independent data or experiments (BOX 3; TABLE 1). Biological validation may involve experimental testing of mechanistic predictions.
Networks organize biology
For gene expression studies, co-expression network analysis leverages the fact that gene expression reflects the state of the cellular or tissue system that is being analysed25. A major advantage of network analysis over DGE analysis is that it can identify multiple levels of molecular organization within the hierarchy of brain region, cell type, organelle and molecular pathways using only transcriptional data, and can thus enable integration with other information, such as known pathway annotations, protein interactions and other molecular profiling data11,12,26,27 (BOX 1; FIG. 2a). Furthermore, when thousands of genes might be differential between conditions, network analysis can subdivide changes into smaller, more biologically coherent sets of modules for further experimental analyses.
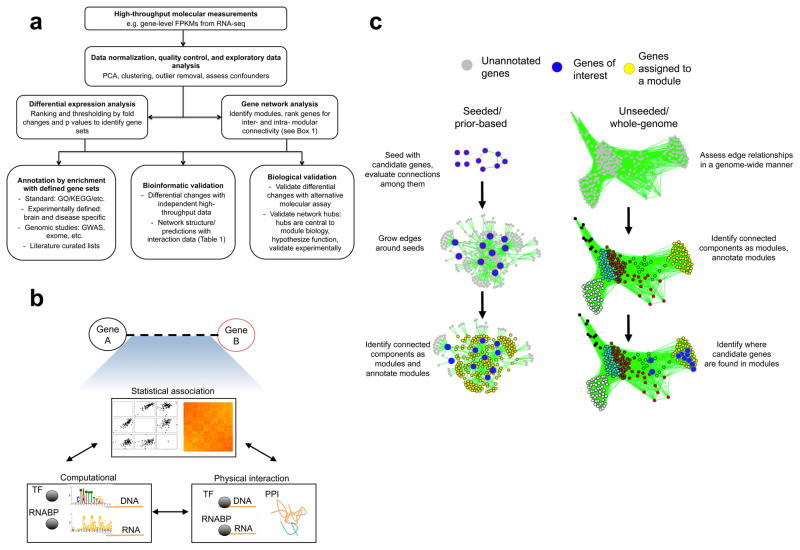
Figure 2. Flowchart of transcriptomic analysis and illustration of seeded and genome-wide approaches to network analysis.
A flowchart demonstrating the general approach to a transcriptomic study that uses differential gene expression (DGE) and network analysis (part a). Network-level features, such as connectivity ranking and module-level enrichment, allow the integration of many external data sources and experiments. Network analysis involves first (part b) connecting genetic or molecular nodes with information about pairwise relationships, which may be one or more of the following: statistical associations relating molecular patterns measured across experiments, such as variation in gene expression levels across brain regions; physical interaction data from experiments or curated from the literature such as transcription factor (TF) or RNA-binding protein (RNABP) binding or protein–protein interactions (PPIs); or computational predictions about TF or RNABP binding using motif enrichment analysis (here, U on the RNA motif is depicted as T). Next, the structure of the network is used to (part c) define modules using a seed-based or genome-wide approach, which groups together the genes that share similar edge-level properties. The seeded (prior-based) approach is shown on the left-hand side, and the unseeded (genome-wide) approach on the right-hand side. The seeded approach involves starting with genes of interest, expanding edges to bring in additional (unannotated) genes and identifying highly connected components as modules. The unseeded approach (right-hand side) involves starting with unannotated genes, using edges to identify interconnected components as modules and then evaluating where genes of interest fall in the resultant network structure. Modules from either approach can be further annotated with external information such as genetic associations and known pathways, integrated with additional data or used to prioritize targets for experimental validation (see BOX 2 and TABLE 1 for more details). Alternative depictions of the network analysis process are also available elsewhere28,41,169. GO, Gene Ontology; KEGG, Kyoto Encyclopaedia of Genes and Genome Elements.
Networks organize genome-wide molecular data by modelling molecules as nodes (typically genes or gene products) and the relationships between nodes as edges. Edges are not necessarily physical interactions — they may also reflect statistical similarity (for example, correlation or mutual information), computational inference or combinations of these edge types (FIG. 2b). Edges define the connectivity of nodes to each other in a network, and this connectivity can be used to organize and analyse the nodes. Many biological networks have a hierarchical structure such that their nodes can be organized into a relatively small collection of highly interconnected modules4,28,29 (FIG. 2c). Inter-modular connectivity reflects a higher-order structure of biological relationships in a gene network, and intra-modular connectivity can identify which genes are biological hubs within modules. In co-expression networks, hubs are highly connected genes; being a hub is an indication of the importance of a gene in the process of interest. Hubs can be key molecular drivers, such as transcriptional regulators that drive co-expression30,31, or they may annotate a module by reflecting the predominant biological role of the module. For example, when evaluating co-expression across brain regions, hubs in modules that are associated with specific regions, such as the cerebellum, are usually markers for predominant cell types, such as granule cells11,12,26,32.
Modularity is very useful, and although it provides a general organizing principle in biology, it need not be present in all constructed networks, and network biology provides many module-free analytical approaches; for example, nodes can be organized in relation to each other by ranking direct and indirect connectivity. If two gene products share an edge, they are said to be neighbours in the network; the more highly interconnected, the closer the neighbours. Thus, gene products that are involved in an unknown cell type or biological process can be annotated on the basis of their proximity to marker genes of known function (‘guilt by association’)26,33,34. Additionally, both modularity and connectivity rankings can be compared between studies to assess whether they are preserved35, and how a module or the position of specific genes within a module change in health and disease can be evaluated to prioritize those that show the most significant changes for further evaluation35–37.
Different approaches to gene co-expression
The most common workflow in gene co-expression network analysis in neuroscience involves the construction of co-expression relationships from microarray or RNA sequencing (RNA-seq) data, identifying modules and then annotating modules on the basis of the known function of module hubs, enrichment for gene sets and module-level association with biological factors such as disease (FIG. 2a). Discussion of the various options and the technical merits of specific network approaches is beyond the scope of this Review38–41. Comparisons among methods have indicated several important points: weighted networks are more reproducible and powerful than binary networks42; signed networks are more predictive of protein interactions and shared pathway relationships than unsigned networks38,42,43; weighted networks constructed with the topological overlap of correlation (for example, by weighted gene co-expression network analysis (WGCNA)42,44) have similar sensitivity and specificity for detecting true network structure for experiments involving monotonic relationships as do networks constructed with nonlinear association measures such as mutual information (for example, by the Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNE)45)38,39; and edge relationships using mutual information or other association measures might be necessary to accurately detect modules in time-series data, which can be non-monotonic46–48. Differential co-expression or connectivity methods36,37 are additional means for determining gene connectivity changes between conditions and can identify disruption or gain of function in pathways.
We provide guidelines in BOX 3 to aid co-expression network reproducibility regardless of the method used. Importantly, the replication of major conclusions in independent data and experimental validation lend the greatest confidence to a network analysis. There is a need for studies that rigorously compare network analysis in human CNS transcriptome data using experimental validation as a gold standard, similar to what has been done in the Dialogue on Reverse Engineering Assessment and Methods (DREAM) regulatory network inference challenge49. The DREAM challenge identified that the integration of multiple network methods yields the most robust regulatory relationship predictions49. This leveraged the availability of hundreds of gene expression profiles in single-cell organisms (bacteria and yeast) and compared regulatory predictions between methods with gold standard experimental validations. Building such regulatory networks in complex tissues such as the CNS is a step beyond current co-expression networks in the brain. Large amounts of data, ideally from homogeneous cellular populations, are necessary to systematically and accurately predict gene regulatory relationships in network studies.
Box 3. Recommendations and general guidelines for transcriptomic studies.
Experimental design
Randomize or balance sample preparation and data collection over all known factors to reduce confounding variation from batch effects, which can introduce spurious correlations. For RNA-seq, we recommend barcoding and multiplexing samples (over eight per lane) to reduce batch effects216.
Evaluate the contribution of both biological and technical factors via unsupervised methods such as principal component analysis178 and apply appropriate methods to remove unwanted variation from the data181,217.
RNA-seq studies with degraded RNA (RNA integrity number <9; essentially all post-mortem studies) should use ribosomal RNA depletion library preparation218. Sequencing samples with a read length of 50 bp with 10 million unique reads (20 million paired-end reads) will detect most highly expressed genes. Deeper sequencing and longer read lengths may be required to accurately and systematically detect noncoding RNAs, splicing or novel features, and pilot experiments are recommended for these scenarios.
DGE analysis
In most experiments, biological variability is greater than technical variability, so biological replicates are of greater value than technical replicates174,175,219.
For well-controlled experiments with expected changes of >twofold in many genes, three or more independent samples per condition are recommended175,219. For post-mortem samples, in which the detection of lower-fold changes may be important and variation may be greater owing to clinical heterogeneity and technical factors, at least 15 case and 15 control samples are recommended in an initial cohort.
Appropriately transformed and normalized sequencing data can be treated similarly to microarray data as far as statistical modelling and multiple corrections are concerned175,220. For differential gene expression (DGE), RNA-seq studies should observe existing analytical and statistical guidelines for microarrays221 and, if possible, should carry out pilot experiments to estimate power222.
Co-expression network analysis
The power of network analysis is dependent on similar factors to DGE but is also dependent on the network features of interest. At currently available sample sizes, networks are most reproducible at a module level35,38,39, then at the hub gene level41,223 and, last, at the level of precise gene connectivity rankings or precise module memberships of genes40,49.
To obtain module-level reproducibility, 20 independent samples are usually sufficient40, but systematic and accurate reconstruction of specific edges, particularly for systematic regulatory relationship discovery, may require hundreds of samples49. For studies comparing conditions, we recommend a minimum of 20 samples per condition. More samples may be necessary if many additional factors vary; for example, age, sex and different brain regions.
Given the large number of parameters in network analysis, there is no ‘one-size fits all’ solution. The most rigorous approach is to apply the empirical reproducibility criteria discussed below.
Reproducibility and biological value
Apply permutation analyses to ensure that gene network modules are significantly co-expressed (interconnected).
To reduce over-fitting and to improve reproducibility, select the outcome of interest (fold change for each gene and gene membership in a module) and apply cross-validation or the bootstrap method224.
Demonstrating reproducibility of major findings (for example, module definitions, top DGE genes and changes in gene network position between conditions) is the most convincing form of validation of a particular analysis. Replication involves identifying the outcome of interest, applying the same analysis as in the original study but to independent data, and demonstrating statistical replication of the same finding.
Generate hypotheses from the DGE and/or network analyses and test them bioinformatically or with wet-laboratory experiments to demonstrate predictive biological value.
To allow other researchers to examine the data sets, raw data should be deposited in a public database (such as GEO, SRA or dbGaP).
To allow for a comparison of analysis methods, always publish clear and usable code along with the publication reporting this analysis.
Literature-curated data
There are many databases that aggregate experiments to construct genome-wide data sets that can be utilized for network construction (TABLE 1). Gene networks that are built on data that contain even a small fraction of literature-curated components can contain substantial bias. Furthermore, when data are from non-neuronal tissue, the database may contain relationships not found in neural tissues (TABLE 1). Although reliant on data from non-neuronal tissue, pathway databases such as the Gene Ontology (GO50), the Kyoto Encyclopaedia of Genes and Genome Elements (KEGG51), Ingenuity Pathway Analyses and MetaCore are valuable for evaluating specific genes and pathways. However, networks with edges that are derived from shared pathway membership can reflect cellular states that might not be found in the CNS, and they will certainly lack many important CNS-specific relationships. In a worst-case scenario, hubs in these networks may be the most studied genes in other areas of biology, and therefore may not reflect neurobiological relevance. It is therefore important to distinguish between networks that are constructed using edges from pathway databases and those using edges derived from tissue-specific primary molecular profiling experiments.
Table 1.
Different edge types in gene networks: practical and theoretical considerations.
| Gene co-expression | Protein-protein interaction | Motif enrichment for transcription factors | |
|---|---|---|---|
| Edge relationships | Statistical association (correlation or mutual information) | Physical binding (interacting or not interacting) | Computational inference (motif binding scores) |
| Main advantages | Indirectly predicts co-regulation, physical interactions and cell type specificity; easiest to measure from tissue of interest | Based on direct physical interactions; predicts protein complexes and signalling pathways | Identifies putative co-regulatory relationships without needing to carry out new experiments |
| Completeness of data across the genome | Most genes are similarly covered genome-wide | Incomplete assessment for most interactions; biased towards most well-studied molecules | Predictions restricted to availability and accuracy of available motif information |
| Tissue specificity | Primary data are often tissue specific | Primary data are rarely tissue specific | Primary data not usually tissue specific |
| Module-level interpretation | Reflects cell types and transcriptionally co-regulated biological processes | Protein complexes; signalling cascades; subcellular structures | Groups of transcriptionally co-regulated genes |
| Interpretation of hubs | Cell type-specific markers; molecular regulators such as transcription factors or RNA-binding proteins | Key proteins in complexes; converging points of signalling cascades | Gene to which many transcription factors bind, perhaps under more complex regulation |
| Sources of bias | Technical artefacts (RNA quality and batch effects); biological confounders (age and sex); post-mortem artefacts (cause of death) | Literature-curated data contain biases towards more well-studied interactions, which tend to be non-neuronal | Unlikely to reflect tissue-specific interactions or regulation without additional data |
| Examples of bioinformatic validation | Preservation of co-expression in independent data; enrichment of physical interactions in modules | Enrichment of co-expression from independent data | Enrichment of predicted binding sites from independent ChIP–seq data |
| Examples of experimental validation | Showing cell type specificity of hubs by in situ hybridization; demonstrating regulatory potential of hubs by hub gene knockdown | Co-immunoprecipitation of proteins of interest; disruption of protein complexes when hubs are targeted | Showing changes in transcription of targets on perturbation of regulators |
Protein–protein interaction (PPI) databases, which compile known physical interactions between proteins, are another example of literature-curated data. PPI experiments may focus on a few proteins and evaluate interactions in a tissue-specific manner using co-immunoprecipitation followed by proteomics. Alternatively, most genome-wide PPI experiments use methods such as yeast two-hybrid (Y2H) screens or tandem affinity purification and are cell type agnostic. The genome-wide approaches yield many more interactions, so most databases typically combine both target-focused and genome-wide experiments52. Similar to pathway databases, these PPI data sets are biased to highly studied gene categories (for example, those implicated in cancer biology) and are still generally incomplete2,53 (TABLE 1). A particularly salient example of the utility of defining tissue-relevant networks is the power obtained by using PPIs derived from cardiac tissue to identify new human loci for long QT syndrome54. To reduce bias and improve tissue specificity for genome-wide networks in the absence of tissue-specific PPIs, one approach is to intersect tissue-specific RNA expression or co-expression with literature-curated PPI data55,56.
These considerations also apply to other physical interaction data, including CLIP–seq, ChIP–seq and miRNA binding data, unless they come from experiments using relevant tissues57. Computational approaches to predict physical interactions can partly circumvent bias (TABLE 1), but they do not address tissue specificity, and there may be relatively low reproducibility across different methods 58,59. There is compelling evidence that using DNase hypersensitivity or ATAC-seq data to infer open chromatin, followed by combining transcription factor binding with open chromatin footprinting, can provide a powerful and comprehensive way to identify tissue-specific transcription factor regulation60,61. The increasing availability of large amounts of relevant data sets within the public domain10,14 now permits the evaluation of network modules for complex regulatory relationships by combining network edges from statistical associations, time-series data, physical binding and computational predictions (FIG. 2b).
When combining multiple molecular levels in networks, it is important to recognize that transcriptomics, epigenomics and proteomics all query unique levels of cellular or tissue organization. For example, most proteins found only in mitochondria do not physically interact with most proteins found only in ribosomes or proteasomes, and these proteins would normally form distinct (but possibly connected) modules in PPI networks. However, in circumstances such as cellular stress or neurodegeneration, the genes encoding these organelle-specific proteins might be transcriptionally co-regulated and hence highly connected at a co-expression level. In this case, transcriptomics can provide a novel view of cellular mechanisms. In general, tissue-, time- or disease-specific data sets aid in conferring specificity to otherwise non-neuronal data. Until such data are available, we suggest beginning with genome-wide tissue-specific data such as transcriptomics, followed by combining literature-curated or non-tissue-specific evidence with gene co-expression modules.
Neurodevelopmental disorders
Neurodevelopmental disorders are characterized by abnormal behavioural or cognitive phenotypes originating either in utero or during early postnatal life, and can be accompanied by clinical features outside the CNS. Various genetic approaches have been successful in identifying the causes of more than 1,000 Mendelian, and fewer non-Mendelian, forms of neurodevelopmental disorders: prototypical examples are intellectual disability62–68, autism spectrum disorder (ASD)69–77, epilepsy78,79 and schizophrenia80–82.
As more genetic risk variants for these disorders have been discovered, studies have found remarkable pleiotropy1,1,83,84. Several rare, highly penetrant mutations in evolutionarily constrained fetal brain-expressed genes are associated with ASD, schizophrenia and intellectual disability, as well as epilepsy83,85–87. We frame this issue using the concept of developmental canalization88, whereby natural selection on developmental programmes in humans has led to robustness in a range of genetic or environmental perturbations89,90: typical development occurs along a ‘track’ (FIG. 1c). Under this framework, the observed pleiotropy is consistent with the notion that disrupting highly evolutionarily constrained genes leads to the ‘derailment’ of typical development off this track, rather than setting the brain on a path to a specific clinically defined disorder (FIG. 1c). Thus, many severe mutations do not converge on one specific phenotype but instead seem to cause a range of clinical disorders74,76,80,81,84,87,91. This formulation leads to several important questions that can be informed by integrative genomic studies, including whether diverse genetic lesions affect similar pathways and where disease specificity emerges. We provide examples below of gene network studies that use co-expression, PPIs and integrated networks to understand ASD and schizophrenia.
Dysregulated networks in the brains of individuals with ASD or schizophrenia
ASD is a phenotypically and aetiologically heterogeneous neurodevelopmental disorder that is defined by deficits in social communication and mental flexibility, with an onset in the first few years of life75. More transcriptional studies of ASD post-mortem brains have been limited by the paucity of available tissue, which has made them underpowered to identify reproducible pathways with statistical rigour92–95. Nevertheless, some themes have emerged across studies, including the increased expression of immune-microglial genes and the decreased expression of synaptic genes in the cerebral cortex. The first ASD study to identify reproducible, genome-wide findings used WGCNA42 to identify two modules, one containing upregulated genes and another containing downregulated genes that defined coherent biological processes in ASD brains30. This study used co-expression module eigengenes (the first principal component of the gene expression levels of each module) to identify modules associated with ASD and to ensure that they were unrelated to potential confounders such as RNA integrity, age or seizure history. This module-level association approach reduces the problem of multiple comparisons and highlights the advantages of using networks as an organizing framework96. The integration of genetic data with co-expression modules showed that the downregulated neuronal signalling module has a potential causal role in ASD, and that the upregulated ASD module was probably a response, which is consistent with its enrichment in microglia and astrocyte genes30. These results supported the findings of several previous smaller studies92,93. Synaptic and microglial modules have been replicated in ASD cortex using RNA-seq in larger independent cohorts97.
Schizophrenia is defined by prolonged or recurrent episodes of psychosis (characterized by hallucinations and delusions) as well as negative symptoms and deficits in cognitive function98. Although diagnosis is usually made in late adolescence or early adulthood, extensive evidence indicates a neurodevelopmental origin99. Transcriptional studies of schizophrenia have benefited from considerably larger sample sizes than those of ASD. However, patients with schizophrenia have greater comorbidity of confounders such as smoking, alcohol and substance abuse than those with ASD. Overcoming potential confounders requires careful matching of patient and control individuals and must take into account potential covariate effects when possible, as has been done in many studies100,101. Despite variable results, consistent findings across studies can be identified, including dysregulation of GABAergic signalling102; downregulation of oligodendrocyte- and myelination-related genes103, mitochondrial function or energy metabolism104, and synaptic genes105; and upregulation of immune and inflammatory genes106.
One of the first studies to put schizophrenia transcriptomics into a genome-wide co-expression network used mutual information and WGCNA107. This study showed that, as in ASD, the overall transcriptomic structure that is observed in control brains is intact but that a neural differentiation module that is associated with schizophrenia does not follow the normal trajectory of downregulation with age. Another study confirmed that a dysregulated neuronal differentiation module was consistently observed in schizophrenia post-mortem brains and suggested that the same pathways were involved in bipolar disorder108. Moreover, genome-wide association study (GWAS) signal enrichment analysis30 found that common variants associated with schizophrenia and bipolar disorder were enriched in the neuronal differentiation module, suggesting that disorders sharing a genetic architecture84 may also share functional transcriptional alterations: a hypothesis that warrants rigorous testing.
Mapping risk genes onto developmental networks
A shortcoming of studies using post-mortem brain tissue is that the tissue is usually obtained long after the disease-causing changes have occurred. Given that the human brain transcriptome has a reproducible structure12,26, one useful way to explore how mutations in risk genes perturb typical brain development is to map risk genes onto transcriptional networks that represent normal brain structure or development (FIG. 1c). The first study to do this identified co-expression modules that had cell type-specific and region-specific expression patterns using nearly 1,000 adult brain regions12,109, and identified neuronal gene-enriched modules containing ASD candidate genes and ASD GWAS signals. Moreover, this study found that genes in these modules have dynamic developmental trajectories, demonstrating a role for ASD risk genes in neural development.
The identification of genetic risk factors by whole-exome sequencing70–73 and the availability of transcriptome data spanning multiple brain regions and developmental stages13,17 created new opportunities to map disease risk genes onto developmental transcriptional networks. One network study defined robust co-expression modules that were reproducible in independent data and identified five developmentally regulated co-expression modules that were enriched for PPIs and ASD risk genes27. By comparing these genes with genes that cause intellectual disability, this study identified molecular processes that are preferentially disrupted by ASD risk genes, including transcriptional regulation, chromatin regulation and synaptic development, and it identified disruption of specific pathways, such as BAF (SWI/ SNF) complex-mediated neuronal development 30,110,111. A complementary study identified developmental co-expression networks enriched for ASD risk genes seeded around nine genes with the highest ASD association signal from whole-exome sequencing112. These investigators asked if, when and where ASD genetic risk converges during brain development by evaluating seeded co-expression networks. They started with the nine ‘high-confidence’ risk genes and expanded the network using combinations of spatial and temporal expression data from post-mortem brain tissue. They identified three spatiotemporal combinations that passed stringent correction for multiple testing: frontal cortical regions during the fetal period, and thalamic and cerebellar regions from birth to 6 years of postnatal age. Interestingly, there was no pathway or PPI enrichment in these modules, probably owing to the inclusion of both positive and negative correlations when computing co-expression relationships (unsigned networks), which is a method that is less sensitive to pathway and protein interaction detection38,43.
Importantly, both of these studies found convergence for rare de novo ASD-associated mutations during early fetal and mid-fetal development, with the greatest enrichment for risk in genes found in cortical glutamatergic neurons. Thus, despite the fact that the same gene is rarely hit recurrently by rare de novo variants in ASD, this class of variation preferentially disrupts projection neurons. Notably, the genome-wide approach27 assessed both ASD and intellectual disability genes, and further suggested that the disruption of the upper neocortical layers (layers 2–4) is related to ASD-like phenotypes and not intellectual disability. Other studies have also found that fetal cortical development and glutamatergic neurons are affected by mutations in ASD, suggesting that it is a robust finding11,113,114.
A seeded co-expression approach has also been used to identify risk convergence in schizophrenia, identifying fetal development of the prefrontal cortex as a point of convergence for de novo mutations115. This study did not extend the network to genes beyond the seed set, and it did not investigate cellular, laminar or regulatory relationships among these genes. As larger sets of risk genes are becoming available72,77,80,81, a more refined view will emerge of how mutations in ASD, schizophrenia, intellectual disability and other psychiatric disorders overlap and diverge to affect cells and circuits.
Regulatory hubs in neurodevelopment and disease
Another promising approach to identify disease-associated networks is to experimentally construct a seed-based network for a candidate regulatory molecule. Using CLIP–seq, investigators identified the RNA binding targets of the translational regulator fragile X mental retardation protein (FMRP)116, and a subsequent analysis found that these targets are highly enriched for de novo mutations in ASD72. Both genome-wide27 and seeded114 co-expression network analysis further connected FMRP targets with multiple forms of ASD genetic risk, including copy number variations (CNVs)114. Additionally, whole-exome sequencing studies of other neurodevelopmental disorders have found enrichment for FMRP targets in rare mutations in schizophrenia81, intellectual disability68 and epilepsy78. As many FMRP targets are highly conserved and are under purifying selection72,87,117, FMRP-related activity-dependent regulation during fetal brain development might be particularly vulnerable to genetic perturbations, with severe mutations resulting in disruption of developmental canalization.
At the transcriptional regulation level, ChIP–seq in induced pluripotent stem cell-derived neurons has been used to define the network of genes regulated by chromodomain helicase DNA-binding protein 8 (CHD8)118, which is the gene most frequently affected by ASD-associated rare de novo variation72,119–121. Integration of ChIP–seq, CHD8 knockdown and gene co-expression suggested that CHD8 directly regulates co-expression modules that are enriched for rare de novo mutations and genes found in the proliferating layers of the fetal cortex27. Another study applied a similar approach but evaluated CHD8 targets in the fetal brain in vivo57. This study identified stronger enrichment for ASD mutations, suggesting that ChIP–seq in the human brain at the right time point identifies interactions that are more disease relevant57. Given the emerging role of fetal brain-expressed transcriptional and chromatin regulators in ASD27,77,122, integrating ChIP–seq of other transcriptional regulators with developmental co-expression networks may help to elucidate a shared, evolutionarily constrained regulatory network that is susceptible to disruption in brain development.
PPI networks define new interactions
Genetic investigations in ASD have constructed seed-based networks with literature-curated PPIs to identify the convergence of ASD risk genes71,73. This approach was applied to identify a highly interconnected PPI subnetwork among rare de novo variants in ASD71. Genes in this subnetwork were evaluated in a larger cohort in a targeted sequencing study120, which identified more risk variants compared with chance and demonstrated that PPI connectivity can be a predictor of ASD risk mutations. However, the biases inherent to literature-curated data and the lack of tissue specificity in these PPI networks limit the identification of novel pathways or circuits with this approach (TABLE 1).
Recently, one study used global literature-curated PPI interactions in a genome-wide network analysis to identify modules that are enriched for ASD-associated genes123. This identified a PPI module that is enriched for genes related to synaptic function and weakly enriched for mutations from individuals with ASD. Integration with transcriptomics annotated the module as highly expressed in oligodendrocytes and the corpus callosum, demonstrating that tissue-specific data are essential for a neurobiological interpretation of PPI modules123. Given the biases inherent to global PPIs discussed above and in TABLE 1, these findings warrant replication with new PPI data. Understanding why these relationships are detected at the PPI level but not at the co-expression level will be valuable.
To evaluate whether ASD risk genes interact at the protein level in an unbiased manner, Sakai and colleagues124 carried out a Y2H screen of 35 syndromic or candidate ASD genes and identified many novel PPIs. This study was the first of its kind in neurodevelopmental disorders and showed that the PPI network seeded around these 35 genes was indeed highly interconnected. Another Y2H study assessed a larger seed set of ASD candidate genes that corresponded to spliced isoforms identified by whole-brain RNA-seq125, hypothesizing that isoform-level PPIs would allow for the discovery of tissue-specific PPI networks126. The genes in the most interconnected component of the PPI network formed a module that was modestly enriched for gene co-expression, gene co-regulation and known ASD genes. These results further demonstrated convergence among known disease-relevant genes at the PPI level and also demonstrated that evaluating tissue-specific isoforms can be used to identify novel interactions. Both of these PPI studies used state-of-the-art quality control and validation, and identified many novel interactions. However, even with knowledge of isoform-specific interactions, the tissue environment for interaction cannot be efficiently recapitulated with current PPI approaches at a genome-wide scale (TABLE 1). This, and other recent work studying cardiac tissue54, highlights how tissue-specific molecular data improve PPI analyses to identify or prioritize genetic variants that specifically function in that tissue, in this case causing cardiac arrhythmia.
Integrating multiple molecular levels
The idea that multiple lines of evidence may increase the power to detect disease-relevant interactions has motivated the integration of literature-curated, molecular and genetic evidence to support specific genes or pathways. The Network-Based Analysis of Genes (NETBAG)127 approach combines multiple forms of literature-curated data using an integrated network approach that has been demonstrated to be effective for predicting gene essentiality in yeast128. The goal of NETBAG is to construct a network in which highly interconnected genes are likely to participate in a similar phenotype. Edges in NETBAG are predominantly derived from multiple PPI databases, GO50 and KEGG51, which are all literature-curated databases, and thus NETBAG is susceptible to the biases discussed above. The first study with NETBAG evaluated CNV-hit genes implicated in ASD and found a highly interconnected module related to synaptic function129. Furthermore, genes in CNVs from females contributed more to the module connectivity than those from males, suggesting that females are affected by more severe genetic hits in ASD, an observation that has been replicated in exome-sequencing studies76,117. Another approach130 has evaluated CNV duplications in addition to CNV deletions and also found an interconnected PPI network that was enriched for proteins involved in synaptic transmission, validating the observation that pathogenic CNVs affect similar gene networks127. An extension of the NETBAG approach (dubbed NETBAG+) has also been applied to simultaneously evaluate large sets of single-nucleotide variants (SNVs) and CNVs in schizophrenia131 and ASD132, confirming the convergence of disease genes onto shared pathways.
An exciting approach is to simultaneously integrate PPIs, co-expression and mutational burden in neurodevelopmental disorders, as has been done by Merging Affected Genes into Integrated networks (MAGI133). This approach begins with mutation-affected genes in their known pathways and then adds genes to these ‘seed pathways’ on the basis of high co-expression or PPI connectivity. The extent to which genes are added to make a module is determined by an objective function that maximizes pathogenic mutations from cases compared with controls in the module. MAGI identified modules containing functionally related genes enriched for deleterious mutations in ASD, many of which are under strong purifying selection, and are also found in epilepsy, schizophrenia and intellectual disability133.
Neurodegenerative disease
Neurodegenerative diseases are characterized by a progressive loss of neural tissue that results in a decline in cognitive and behavioural function. Many of these diseases have known causes that involve mutations in ubiquitously expressed proteins134, but they follow stereotyped patterns of degeneration that selectively affect certain cell subsets more severely, resulting in disease-specific spatial and temporal patterns of degeneration135–137 (BOX 1). Neuropathological investigations have identified protein-centric mechanisms that might be involved in disease pathogenesis, but causal mechanisms are difficult to pinpoint, as post-mortem samples reflect the consequence of years of ageing and disease progression. Important disease-associated molecular changes can be confounded by environmental and clinical factors. Additionally, although positional cloning has identified genes and pathways that are involved in many neurodegenerative diseases, pathological mechanisms, modulators of pathogenesis and disease biomarkers have remained elusive, suggesting that genome-wide approaches are needed. Transcriptional and PPI network studies have recently identified many new insights into these diseases. Below, we focus on representative transcriptomic studies of two genetically complex diseases (Alzheimer disease and frontotemporal dementia (FTD)) and PPI studies of two diseases for which causal genes are well defined (Huntington disease and inherited ataxia), but for which disease mechanisms are still poorly understood.
Post-mortem transcriptomic analysis in dementia
The major challenge in Alzheimer disease and FTD transcriptomics has been the identification of changes that are independent of alterations in cell type proportions, which accompany cell death and inflammation. Three major study design principles have been used to overcome this issue: transcriptomes in differentially vulnerable brain regions or cellular populations can be compared to identify vulnerable or protected pathways138,139 (BOX 1); preclinical changes in at-risk individuals with a milder disease presentation can identify genes and pathways that might lead to disease140; and cell type-specific markers can be used in combination with bioin-formatic analyses to account for the effect of changes in cell proportion on the overall transcriptome141,142.
Multiple transcriptomic studies of Alzheimer disease have been carried out in the human brain at varying spatial resolutions143. Large studies using quantitative metrics of severity140 and differentially vulnerable regions144 have identified pathway-level changes in transcriptional regulation, apoptosis, cell proliferation, energy metabolism and synaptic transmission. One particularly powerful approach involved the use of the pattern of regional vulnerability to guide a microarray study that identified a defect in the retromer complex, which is responsible for endosome-mediated recycling of membrane proteins145. The involvement of this pathway in Alzheimer disease was experimentally validated146. The first large transcriptomic study (involving 188 controls and 176 individuals with Alzheimer disease)147 connected genetic variation to expression changes by using expression quantitative trait locus analysis (eQTL analysis) in controls and Alzheimer disease, and further supported the pathway-level findings related to transcriptional regulation and energy metabolism in Alzheimer disease140,148. Integration of eQTLs can identify causality in transcriptomic studies in the context of Alzheimer disease risk, adding a crucial mechanistic element to studies of post-mortem gene expression.
In FTD, transcriptional signatures related to differential regional vulnerability have helped to identify modulators of neurodegeneration. The first of two well-powered studies that applied this approach carried out transcriptomic analysis in a mouse model of FTD, identifying the gene Npepps138. Cross-species analyses in flies and humans confirmed the expression pattern and neuroprotective effect of NPEPPS138. The second study139 compared post-mortem tissue from patients with FTD harbouring dominant mutations in the pro-granulin (GRN) gene, patients who had FTD but who did not have a known family history or mutations, and control individuals. This study also leveraged regional vulnerability by comparing transcriptome profiles in the frontal cortex, hippocampus and cerebellum, identifying a diminishing hierarchy of susceptibility to FTD. The findings demonstrated that GRN-positive individuals were a transcriptomically distinct group from those with sporadic FTD139. Both of these studies in FTD demonstrate the value of using selective vulnerability and differential genetic risk in study design.
From individual genes to networks and mechanism
Most early post-mortem studies from individuals with Alzheimer disease or FTD generated long gene lists and were followed by analysis of GO or KEGG pathway enrichment139,140,147. In an early network study, Miller and colleagues149 applied network analysis to compare the transcriptome in normal ageing and Alzheimer disease, finding many shared features that were downregulated in Alzheimer disease and normal human ageing149. They subsequently150 incorporated more than 1,000 microarrays from mouse models of Alzheimer disease and human patients with Alzheimer disease from public databases to reproduce and extend these results, identifying additional co-expression modules that are related to mitochondrial dysfunction and synaptic plasticity. This work also found major differences in dementia susceptibility genes between humans and mice, potentially identifying why some mouse models might not recapitulate human neuropathology. Another study used similar methods to identify overlap in transcriptional networks between vascular disease (a major risk factor for dementia) and Alzheimer disease, identifying potential molecular mechanisms that might underlie their co-occurrence151. Forabosco and colleagues152 used network analysis to explore the function of TREM2 (triggering receptor expressed on myeloid cells 2), an Alzheimer disease risk gene, suggesting a role for microglial function and further implicating neuroinflammation in Alzheimer disease. In FTD, two studies re-analysed published transcriptome data139 to discover a role for WNT signalling in GRN-mediated FTD153,154. Both involved extensive bioinformatic analyses of expression data from in vitro neural progenitor models and identified transcriptomic changes shared across the post-mortem human brain, human neural cell lines and the mouse brain. Experimental validation of predictions from these networks showed that this cross-species approach can identify consistent, high-confidence perturbations in neurodegenerative disease46,153. Additionally, the use of previously published human data in many of these studies highlights the value of policies supporting data sharing, especially from patient cohorts. Finally, studies of the regulatory networks and targets of specific miRNAs such as miR-339-5p in dementia are in their early stages155–157 but promise to reveal novel regulators of neurodegeneration.
Although transcriptomic studies have furthered our understanding of disease mechanisms beyond neuropathology and single genes, the effects of cell type loss have not been completely accounted for in most studies. Purifying cell populations or carrying out transcriptional analyses on single cells158–163 can identify important changes that are not apparent in whole-tissue transcriptomes141,142. Combining bioinformatics approaches with single-cell sequencing will increase the resolution at which regional vulnerability can be assessed and will enhance the ability of gene co-expression networks to identify key changes associated with dementia.
Protein interaction networks with known disease genes
The causal mutations for Huntington disease and many inherited ataxias have been known for more than a decade, and thus the focus of molecular investigations has been on understanding disease mechanisms and modifiers. Lim and colleagues164 used a seed-based approach based on a Y2H screen to identify interactors of the protein products of multiple causal and candidate genes in inherited ataxias. Analysis of the resultant PPI network identified an interconnected network of proteins related to inherited ataxias. Importantly, interactors in the network were potential modifiers of disease progression, and, in subsequent work, gain of function mediated by a newly identified protein complex was found to mediate disease pathogenesis165. This Y2H approach has also been used to identify potential modulators of Huntington disease166, in which it is thought that inter-actors of huntingtin (the causally mutated pathological protein) might modulate disease severity. Interestingly, in vivo PPI screening by large-scale co-immunoprecipitation and mass spectrometry provided tissue- and time-specific information that was not found by Y2H studies167. WGCNA identified spatially and temporally specific modules associated with mutant Htt (which encodes huntingtin); and proteins with high intra-modular connectivity (hub proteins) modulated neurodegeneration in flies. This work further emphasizes the importance of considering tissue context in the studies examining disease-relevant protein associations.
Integrating genetic variation and transcriptome networks
The most ambitious and exciting goal in systems biology is to elucidate the functional genetic architecture of diseases by systematically identifying causal effects using genome-wide variation to disambiguate primary and secondary changes that occur in disease168,169. A recent study shows that this goal is possible in the CNS by using genetic variation as a causal anchor to define genetically driven network-level changes in Alzheimer disease and to provide experimental validation for network predictions170. Zhang and colleagues170 applied WGCNA to hundreds of post-mortem brain samples from individuals with Alzheimer disease, other neurodegenerative diseases and controls. They showed that multiple transcriptional modules were remodelled in Alzheimer disease: gain of connectivity was observed in immune and neurogenesis pathways, and loss of connectivity was predominant in pathways related to GABA signalling and myelination. An eQTL analysis followed by module-level genetic signal enrichment identified several modules in which genetic association signals were enriched. Given that gene expression changes are caused by genetic variation, this suggested these modules were causally involved171. The researchers then applied Bayesian network analysis to evaluate causal relationships in an Alzheimer disease-related microglial module, implicating TYROBP (TYRO protein tyrosine kinase-binding protein) as a regulatory hub. The role of Tyrobp was experimentally validated in mice170, showing that network structure is predictive, as had previously been demonstrated with co-expression networks32. Overall, integrating genetics with co-expression networks using large sample sizes (with a minimum of 100 cases and controls) and establishing causality by evaluating genotype–phenotype relationships and eQTL is very promising.
Specificity and convergence across CNS disorders
Many of the most influential studies using gene networks to probe neuropsychiatric disease mechanisms integrate multiple data types (for example, RNA expression, GWAS signals and PPI) or data sets (for example, human post-mortem, mouse and in vitro), emphasizing the value of publicly available data sets. The further availability of raw molecular profiling data with necessary metadata amplifies the value of individual studies. In addition to generating new hypotheses, molecular systems approaches integrating data from diverse studies can reveal unexpected and distinct relationships that are common to different CNS disorders. FIGURE 3 describes an example of a network-based meta-analysis of brain transcriptional profiles from publicly available data in ASD, schizophrenia and Alzheimer disease, which identifies shared and distinct biological processes across disorders. Several modules are shared by two of the three disorders, including the red module (ASD and schizophrenia), which contains voltage-gated calcium channels, and the green module (ASD and Alzheimer disease), which contains microglial markers (FIG. 3b–d). This demonstrates how cross-disorder analyses can systematically reveal shared and distinct biological processes among disorders, even when the data are from different studies (see Supplementary information S1 (box)). It will be fruitful to combine more CNS disorders and diseases and to integrate GWASs and rare mutations to identify which variants affect gene expression across diagnostic boundaries and which are more specific. Prioritizing the disease-specific genes for further investigation may also aid in clarifying the molecular processes that lead to behavioural and cognitive alterations that are specific to a particular disease.
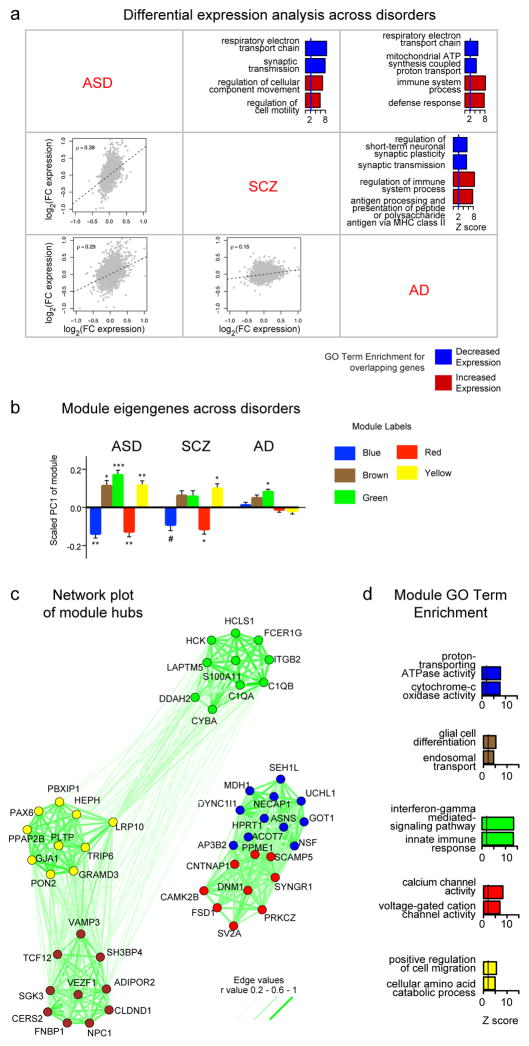
Figure 3. Transcriptomic convergence and divergence across central nervous system disorders.
Transcriptomics can systematically compare genes and pathways across neurobiological disorders. To provide a simple example, we compare genome-wide expression patterns in the cerebral cortex across published microarray studies of autism spectrum disorder (ASD)30, schizophrenia (SCZ)225 and Alzheimer disease (AD)226 (part a). We applied differential gene expression (DGE) analysis across these disorders in a pairwise manner and performed a meta-analysis with weighted gene co-expression network analysis (WGCNA). Please see Supplementary information S1 (box) for details. The bottom-left half of the comparisons shows pairwise comparison of DGE across conditions. ASD–SCZ and ASD–AD are significantly correlated in DGE changes, as demonstrated by Spearman correlations (ρ values) between genome-wide DGE effect sizes in each disorder. On the upper-right half of the comparisons, Gene Ontology (GO) term enrichment of pairwise shared upregulated and downregulated changes demonstrates that upregulated inflammation and downregulated synaptic function and oxidative phosphorylation are common to all three disorders. Results are shown as enrichment Z scores for pathway enrichment, Z > 2 suggests enrichment227. WGCNA across these three conditions identified five modules (labelled with different colours) that are perturbed across at least one condition, as demonstrated by differences in eigengene expression (*p < 0.05,**p < 0.01,***p < 0.001, two-tailed t-test) (part b). The top ten interconnected (hub) genes in each module with edges reflecting the strength of correlation between genes reveals (part c) and GO term enrichment for each module (part d). MHC, major histocompatibility complex.
Guidelines for transcriptomic and network studies
Given the promise of molecular systems and integrative network approaches, it is perhaps surprising that there are few universally agreed on metrics, power analysis tools or methodological comparisons to guide experimental design, execution and analysis, such as there are for genetic association studies172. For DGE and network analysis, there are many studies with guidelines that are based on theoretical models and empirical assessments40,173–176, but most studies use data from experiments that do not have the spatial, temporal or disease-relevant complexities that occur in studies of the CNS or post-mortem tissue. There is no experimental design that suits all aims, but we suggest criteria for initial experimental design, ensuring reproducibility and improving biological interpretability for transcriptomic analyses in BOX 3.
In general, it is helpful to think of all variation in gene expression or other molecular profiling data as a consequence of technical, biological and unmeasured factors177, rather than assuming that differences are due to experimental interventions or disease status41. Optimal methodological choices and study designs ensure that the biological signals from the main factors are not confounded by variation from unwanted factors178. Notably, molecular profiles in post-mortem gene expression studies are affected by RNA degradation and post-mortem intervals179, but other technical factors including library preparation and sequencing depth in RNA-seq analysis should also be carefully evaluated180,181.
Additionally, we note two important points about studies that construct predictive models and studies that make causal claims. For studies that develop predictive models, such as disease classifiers, experimental design should include the estimation of a model on initial data followed by evaluation of accuracy in held out or, preferably, independent data182. As far as causality is concerned, most molecular profiling studies, especially those using post-mortem tissue, cannot show causality without follow-up controlled experiments or genetic evidence169–171. We also strongly suggest the experimental validation of key network predictions, as this provides avenues for refinement and biological grounding of the network30,32,153,170.
Gene set enrichment with networks
As shown by multiple studies, gene network analyses can aid in understanding genetic association studies. Grouping genetic findings using disease- and tissue-relevant modules can increase the power to detect genetic associations with disease by combining signals that reflect similar underlying biology while simultaneously informing biological mechanism by functionally annotating genetic findings30,109,122,183. Enrichment for genetic variants in a module can be evaluated using gene set enrichment methods, which rely on comparing enrichment in a module relative to a control gene set. However, studies have demonstrated biases in gene length, gene mutability and other factors that can drive gene set enrichment instead of a true biological signal. For example, longer genes are more likely to be implicated by CNVs and SNVs87,184,185, and genes highly expressed in the brain, particularly those involved in synaptic function, are longer on average than other genes186. These biases inflate enrichment results and can result in false positives, so it is important to identify appropriate control sets or to apply the correct statistical methods (permutation tests or covariate modelling187). We note that each of the points discussed here and in BOX 3 is applicable to many other types of high-throughput data and that there are many valid variations to FIG. 2a.
Future directions
In this Review, we have discussed how transcriptomic and integrative network approaches have been applied to provide a systems-level understanding of CNS disorders in an unbiased and reproducible manner. Mapping genetic variants to gene expression and PPI networks has been fruitful, but most disease-associated variation in complex diseases lies in the noncoding regulatory regions of the genome188. The next crucial step for high-throughput molecular studies in the brain will be to understand regulatory alterations and interactions during development with histone mark profiling and chromosome conformation capture approaches189,190. Additionally, understanding transcriptomic and epigenetic changes in more homogeneous cellular populations, or at a single-cell resolution, will greatly improve our mechanistic understanding of normal human brain development. Initial maps of these neurobiologically relevant epigenetic landscapes and cell type differences, mostly at the tissue level, are under construction by the PsychENCODE consortium.
As noted throughout this Review, studies of proteomic data are highly complementary to co-expression data and have revealed a crucial level of organization and regulation at the translational and post-translational levels. A particularly salient example is the synaptic signalling apparatus, more specifically the postsynaptic density (PSD), which has been extensively characterized at the protein level in humans and mice, showing key areas of overlap and divergence191,192. However, developmental and cell subtype differences in the PSD are not well understood, so obtaining PSD co-expression and PPI networks in relevant neural tissue and time points, similar to what BrainSpan has done for gene expression, will be invaluable. Currently, high-throughput, high dynamic range spatial and temporal data from minute sample quantities with proteomics are not available, so creative integration of cell type-specific transcriptional data with more generic PPI data may provide an approximation of the regional or cellular differences in synaptic structure in the near future. The development of methods, including benchmarking and refining methods for the integration of different forms of data (for example, PPI and gene expression data), developing tools for exploring network structure at a more fine-grained level, and empirically defining the most sensitive and robust network approaches, will also be crucial.
One of the greatest challenges is to systematically infer causality in molecular networks with a systems genetics approach168,169. This will necessitate more comprehensive eQTL studies, particularly in early brain development and disease170. Core molecular pathways that are confirmed to be perturbed in disease can then be interrogated with drug or environment perturbation data193,194 to identify interventions that will perturb networks from a disease state into a healthy state. This area of in silico drug screening based on DGE or network modules has barely been explored in the CNS, but it has considerable promise193. Additionally, with the advent of mandatory electronic medical records, population-level studies, including longitudinal data for many simple phenotypes, coupled with biobanking, can provide the scale needed to more fully understand genetic contributions to disease risk as well as disease relationships across the lifespan195–197.
Even once we have the information from thousands of genomes, biological insights into the CNS require the assessment of relevant behavioural and cognitive phenotypes, which are not well defined for most neuropsychiatric diseases198,199 and are rarely collected in large populations. Genome-wide transcriptomic approaches provide a quantitative endophenotype, or a biomarker, that genetic association studies can use to further refine the measurement of disease states. Transcriptomic and other molecular systems measurements can also be correlated with systems neuroscience phenotypes, such as MRI and functional MRI measurements, or behavioural phenotypes to identify non-invasive indicators of disease state4 (FIG. 1a).
Conclusions
Currently, much basic and translational neuroscience research is still focused on candidate genes and candidate hypotheses, so sceptics may question the value of measuring entire systems. However, biological complexity cannot be ignored; genome-wide measurements, in conjunction with studying individual genes and pathways, are essential to address the true underlying mechanisms of neurodevelopmental and neurodegenerative disorders. Well-designed, reproducible molecular profiling studies allow biologists to simultaneously evaluate hypotheses in an unbiased manner and to generate new hypotheses. Although certainly vast and seemingly complex, gene networks provide an organizational framework that simplifies the process of hypothesis generation and testing. The general paradigm of using correlational and physical interaction molecular networks in neurobiology to understand molecular systems changes can be applied across methodologies and enables the investigation of relationships that span multiple levels of analysis. The results of high-quality genome-wide studies will be essential to develop and test hypotheses that look beyond where our current knowledge ends to develop a more encompassing view of the problems posed by neurodevelopmental and neurodegenerative disorders, and their potential solutions.
Supplementary Material
Acknowledgments
The authors thank L. de la Torre-Ubieta and H. Won for assistance with Figure 1, as well as members of the Geschwind laboratory and K. Lage for critical reading of the manuscript. This work is supported by the US National Institute of Mental Health grants (5R37MH060233 and 5R01MH094714, D.H.G.), an Autism Center for Excellence network grant (9R01MH100027), the Simons Foundation (SFARI 206744, D.H.G.), NIMH Training and NRSA Fellowships (T32MH073526 and F30MH099886, N.N.P.), and the Medical Scientist Training Program at University of California, Los Angeles (UCLA).
Glossary
- Genetic architecture
For genetic variants, the relationship among allele frequency, effect size, number of contributing variants and how they quantitatively influence a given trait
- Molecular systems or integrative network approach
Systems biology methods that use high-throughput quantification, analysis and interpretation of the molecular relationships within and across molecular levels, including the genome, transcriptome, epigenome, proteome and other ‘omes’
- Systems neuroscience
An area of neuroscience that focuses on short- and long-range circuits that are usually related to specific behavioral or cognitive functions (vision, motor function, attention and so on)
- Gene network
A graph consisting of genes as nodes connected by edges that represent relationships between genes
- Differential gene expression analysis (DGE analysis)
An approach commonly used in transcriptomic studies that serially compares thousands of genes between groups (for example, disease and controls) to evaluate the mean difference and its significance for each gene independently
- Modules
Also known as clusters, cliques and communities. Highly interconnected subsets of genes in a gene network; for example, genes in a transcriptomic network sharing highly similar patterns of gene expression
- Nodes
Molecular entities that constitute a network; for example, genes in a gene network or proteins in a protein interaction network
- Edges
The relationships between nodes in a network delineating some measure of shared function; for example, correlations or physical interactions
- Mutual information
A measure of dependence between two variables that can capture complex relationships, including nonlinear and nonmonotonic patterns, that could be missed by linear correlation measures
- Hubs
Genes in a network or module that are highly connected; that is, they have a relatively high number of edges compared with other genes
- RNA sequencing (RNA-seq)
An assay for measuring RNA transcript levels in a genome-wide manner that involves the extraction of RNA followed by construction of cDNA libraries that undergo high-throughput sequencing
- Weighted networks
Networks in which the edges have continuous values, with higher values reflecting an increased strength or probability of connectivity
- Binary networks
Networks in which the edges are all or nothing, either because this is inherent to the edge measurement (for example, physically interacting or not) or because a cut-off or threshold has been applied to a continuous measurement (for example, by applying a rule that all correlation values≥0.7 are 1, all others are 0)
- Signed networks
Networks in which the direction of association is taken into consideration in addition to the magnitude of the correlation; for example, in a signed correlation network, high positive correlations are assigned high edge values, but high negative correlations are assigned low edge values
- Unsigned networks
Networks in which any high magnitude association is assigned a high edge value regardless of the direction of the association
- Topological overlap
A computation on direct edge relationships in a network that transforms them into indirect edge values that reflect the sharing of neighbourhoods between genes
- Seeded (prior-based) networks
Network analysis approaches in which edges are ‘grown’ around ‘seed’ genes that are selected on the basis of previous experiments or prior hypotheses, and the network structure is dependent on these seed genes
- Unseeded (genome-wide) networks
Network analysis approaches in which edges are evaluated in a genome-wide manner, and network structure is not dependent on prior knowledge of a particular set of genes
- Adjacency matrix
A matrix of pairwise node–node relationships that quantifies all possible edges in a network. Edge relationships may be determined from one data type or by weighting the contribution from multiple types of data
- CLIP-seq
An assay for measuring the binding sites of a protein on RNA transcripts in a genome-wide manner that involves crosslinking immunoprecipitation followed by high-throughput sequencing
- ChIP-seq
An assay for measuring the binding sites of a protein on DNA across the genome that involves chromatin immunoprecipitation followed by high-throughput sequencing
- DNase hypersensitivity or ATAC-seq
Sequencing methods that infer regions of the genome in a particular cell or tissue with open chromatin by exploiting the fact that these regions are preferentially accessible to the DNase I enzyme or a transposase
- Eigengenes
Module-level summaries of expression utilized in co-expression networks calculated by taking the first principal component of the expression levels of genes in a module
- Psychosis
A mental state defined by a loss of contact with reality and characterized by exaggerations or distortions of normal perception
- Negative symptoms
Symptoms involving a loss of normal emotional responses, including a lack of motivation, an inability to experience pleasure and reduced expression through speech
- Unsupervised methods
Analysis approaches that use the intrinsic variation in data to define shared patterns without explicit prior knowledge of how the data should be grouped (for example, hierarchical clustering). This can identify novel clusters or groupings of data points
- Expression quantitative trait locus analysis (eQTL analysis)
A specific case of genotype-to-phenotype association that uses RNA transcript levels as the phenotype in order to identify genetic loci that regulate RNA levels
- Selective vulnerability
The relative susceptibility of specific brain regions, cell populations or time points to genetic or environmental insults that result in disease
- Causal anchor
A causal factor, such as genetic variation, that can be used to orient edges to transform an undirected correlational network to a directed causal network
- Gene set enrichment
An analysis approach that assesses the statistical significance of the overlap between two gene sets, one set is usually an annotated reference set, and the other is an unannotated set of interest
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
Allen Brain Institute: http://www.brain-map.org/
BioGRID: http://thebiogrid.org/
BrainCloud: http://braincloud.jhmi.edu/
Braineac: www.braineac.org/
BRAIN Initiative: http://braininitiative.nih.gov/
BrainSpan: http://www.brainspan.org/
DAPPLE: http://www.broadinstitute.org/mpg/dapple/dapple.php
dbGaP: www.ncbi.nlm.nih.gov/gap
DREAM challenges: http://dreamchallenges.org/
GEO: www.ncbi.nlm.nih.gov/geo/
GTEx: http://www.gtexportal.org/
Ingenuity Pathway Analyses: www.ingenuity.com
KEGG: www.genome.jp/kegg
MetaCore: www.portal.genego.com
PsychENCODE consortium: http://www.psychencode.org
Roadmap Epigenomics Mapping Consortium: http://www.roadmapepigenomics.org/
SRA: http://www.ncbi.nlm.nih.gov/sra
VISTA Enhancer Browser: http://enhancer.lbl.gov/
References
- 1.Gratten J, Wray NR, Keller MC, Visscher PM. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 2014;17:782–790. doi: 10.1038/nn.3708. A comprehensive review of GWASs and exome studies across major neuropsychiatric disorders. It discusses the role of common variants and rare variants across disorders, and the concept of explaining heritability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat Rev Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 4.Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant S. Systems biology in neuroscience: bridging genes to cognition. Curr Opin Neurobiol. 2003;13:577–582. doi: 10.1016/j.conb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kandel ER, Markram H, Matthews PM, Yuste R, Koch C. Neuroscience thinks big (and collaboratively) Nat Rev Neurosci. 2013;14:659–664. doi: 10.1038/nrn3578. [DOI] [PubMed] [Google Scholar]
- 8.Sunkin SM, et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramasamy A, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. A large eQTL study across multiple brain regions that identified region-specific eQTLs and demonstrated the value and promise of eQTL analysis in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolmetsch R, Geschwind DH, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jucker M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med. 2010;16:1210–1214. doi: 10.1038/nm.2224. [DOI] [PubMed] [Google Scholar]
- 20.Nelson SB, Sugino K, Hempel CM. The problem of neuronal cell types: a physiological genomics approach. Trends Neurosci. 2006;29:339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 21.DeFelipe J, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casey BJ, et al. DSM-5 and RDoC: progress in psychiatry research? Nat Rev Neurosci. 2013;14:810–814. doi: 10.1038/nrn3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Insel T, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 25.Carter SL, Brechbühler CM, Griffin M, Bond AT. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics. 2004;20:2242–2250. doi: 10.1093/bioinformatics/bth234. [DOI] [PubMed] [Google Scholar]
- 26.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. A study that constructs genome-wide co-expression networks to identify modules spanning prenatal human brain development and demonstrates how module structure can be validated with multiple data sources and how tissue-specific and temporally specific co-expression modules can provide new biological insights into genetic variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra K, Carvunis AR, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nat Rev Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter H, Hofree M, Ideker T. Genotype to phenotype via network analysis. Curr Opin Genet Dev. 2013;23:611–621. doi: 10.1016/j.gde.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. This study identifies common transcriptomic changes in a heterogeneous neuropsychiatric disorder and illustrates how gene network modules can be validated experimentally and how network modules can be related to GWAS findings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winden KD, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of ‘guilt-by-association’ within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dougherty JD, et al. PBK/TOPK, a proliferating neural progenitor-specific mitogen-activated protein kinase kinase. J Neurosci. 2005;25:10773–10785. doi: 10.1523/JNEUROSCI.3207-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. A description and comparison of multiple metrics for measuring modular structure in networks, which provides a statistical framework for demonstrating module preservation that is included in the R package WGCNA.[Au:OK?] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson NJ, Reverter A, Dalrymple BP. A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation. PLoS Comput Biol. 2009;5:e1000382. doi: 10.1371/journal.pcbi.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi JK, Yu U, Yoo OJ, Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- 38.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics. 2012;13:328. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen JD, Xie Y, Chen M, Girard L, Xiao G. Comparing statistical methods for constructing large scale gene networks. PLoS ONE. 2012;7:e29348. doi: 10.1371/journal.pone.0029348. This study compares multiple approaches for constructing large-scale gene networks, including methods based on correlation and mutual information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballouz S, Verleyen W, Gillis J. Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv118. http://dx.doi.org/10.1093/bioinformatics/btv118. An evaluation of sample size and power for constructing co-expression networks with RNA-seq. [DOI] [PubMed]
- 41.Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2013;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:17. doi: 10.2202/1544-6115.1128. This paper describes the value of weighted co-expression networks over binary co-expression networks and discusses the theory behind the widely used WGCNA package. [DOI] [PubMed] [Google Scholar]
- 43.Ramani AK, et al. A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol Syst Biol. 2008;4:180. doi: 10.1038/msb.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong J, Horvath S. Understanding network concepts in modules. BMC Syst Biol. 2007;1:24. doi: 10.1186/1752-0509-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wexler EM, et al. Genome-wide analysis of a Wnt1-regulated transcriptional network implicates neurodegenerative pathways. Sci Signal. 2011;4:ra65. doi: 10.1126/scisignal.2002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoppoli P, Morganella S, Ceccarelli M. TimeDelay-ARACNE: reverse engineering of gene networks from time-course data by an information theoretic approach. BMC Bioinformatics. 2010;11:154. doi: 10.1186/1471-2105-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. This paper, from the DREAM challenge on regulatory network reconstruction, describes the results of applying multiple regulatory network inference algorithms to three large data sets from bacteria and yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hakes L, Pinney JW, Robertson DL, Lovell SC. Protein–protein interaction networks and biology — what’s the connection? Nat Biotechnol. 2008;26:69–72. doi: 10.1038/nbt0108-69. [DOI] [PubMed] [Google Scholar]
- 53.Hart GT, Ramani AK, Marcotte EM. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundby A, et al. Annotation of loci from genome-wide association studies using tissue-specific quantitative interaction proteomics. Nat Methods. 2014;11:868–874. doi: 10.1038/nmeth.2997. This study experimentally defines PPIs specific to cardiac tissue for four genes known to cause long QT syndrome and demonstrates how tissue-relevant PPI networks can be used to prioritize genetic association signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 56.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cotney J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tompa M, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 60.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pique-Regi R, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ropers HH. Genetics of intellectual disability. Curr Opin Genet Dev. 2008;18:241–250. doi: 10.1016/j.gde.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 63.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- 64.Matson JL, Shoemaker M. Intellectual disability and its relationship to autism spectrum disorders. Res Dev Disabil. 2009;30:1107–1114. doi: 10.1016/j.ridd.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 65.Lubs HA, Stevenson RE, Schwartz CE. Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet. 2012;90:579–590. doi: 10.1016/j.ajhg.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 67.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 68.Gilissen C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 69.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Epi4K Consortium & Epilepsy Phenome/Genome Project. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poduri A, Lowenstein D. Epilepsy genetics — past, present, and future. Curr Opin Genet Dev. 2011;21:325–332. doi: 10.1016/j.gde.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu B, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu X, Need AC, Petrovski S, Goldstein DB. One gene, many neuropsychiatric disorders: lessons from Mendelian diseases. Nat Neurosci. 2014;17:773–781. doi: 10.1038/nn.3713. [DOI] [PubMed] [Google Scholar]
- 84.Cross-Disorder, Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doherty JL, Owen MJ. Genomic insights into the overlap between psychiatric disorders: implications for research and clinical practice. Genome Med. 2014;6:29. doi: 10.1186/gm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17:764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 89.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. This paper discusses the concept of canalization and its implications for molecular biology and evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suliman R, Ben-David E, Shifman S. Chromatin regulators, phenotypic robustness, and autism risk. Front Genet. 2014;5:81. doi: 10.3389/fgene.2014.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 92.Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- 93.Garbett K, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ginsberg MR, Rubin RA, Falcone T, Ting AH, Natowicz MR. Brain transcriptional and epigenetic associations with autism. PLoS ONE. 2012;7:e44736. doi: 10.1371/journal.pone.0044736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chow ML, et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet. 2012;8:e1002592. doi: 10.1371/journal.pgen.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gupta S, et al. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748. doi: 10.1038/ncomms6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 99.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 100.Mirnics K, Pevsner J. Progress in the use of microarray technology to study the neurobiology of disease. Nat Neurosci. 2004;7:434–439. doi: 10.1038/nn1230. [DOI] [PubMed] [Google Scholar]
- 101.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 102.Hashimoto T, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2007;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hakak Y, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Altar CA, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 105.Faludi G, Mirnics K. Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci. 2011;29:305–309. doi: 10.1016/j.ijdevneu.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arion D, Unger T, Lewis DA, Levitt P, Mirnics K. Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia. Biol Psychiatry. 2007;62:711–721. doi: 10.1016/j.biopsych.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Torkamani A, Dean B, Schork NJ, Thomas EA. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res. 2010;20:403–412. doi: 10.1101/gr.101956.109. This work applies mutual information-based co-expression network analysis to transcriptomic data from post-mortem brains of individuals with schizophrenia to identify several schizophrenia-associated modules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen C, et al. Two gene co-expression modules differentiate psychotics and controls. Mol Psychiatry. 2012;18:1308–1314. doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ben-David E, Shifman S. Networks of neuronal genes affected by common and rare variants in autism spectrum disorders. PLoS Genet. 2012;8:e1002556. doi: 10.1371/journal.pgen.1002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stein JL, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steinberg J, Webber C. The roles of FMRP-regulated genes in autism spectrum disorder: single- and multiple-hit genetic etiologies. Am J Hum Genet. 2013;93:825–839. doi: 10.1016/j.ajhg.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gulsuner S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet. 2014;15:133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 118.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol Psychiatry. 2012;18:1054–1056. doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- 123.Li J, et al. Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol. 2014;10:774–774. doi: 10.15252/msb.20145487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sakai Y, et al. Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med. 2011;3:86ra49. doi: 10.1126/scitranslmed.3002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Corominas R, et al. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat Commun. 2014;5:3650. doi: 10.1038/ncomms4650. This study rigorously identifies the interactors of proteins encoded by autism candidate genes using brain-relevant isoforms and identifies interactions among CNV-affected genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellis JD, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 127.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. This study applies a rigorous framework to integrate multiple levels of molecular data and evaluates whether genes affected by CNV in autism were functionally interconnected. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee I, et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 129.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 130.Noh HJ, et al. Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism. PLoS Genet. 2013;9:e1003523. doi: 10.1371/journal.pgen.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gilman SR, et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hormozdiari F, Penn O, Borenstein E, Eichler EE. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2015;25:142–154. doi: 10.1101/gr.178855.114. This study uses a network analysis method that combines gene co-expression and PPIs to identify modules that are highly interconnected in the network but that are also more likely to be mutated in individuals with neurodevelopmental disorders compared with controls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Taylor JP. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 135.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Forman MS, Trojanowski JQ, Lee VMY. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- 138.Karsten SL, et al. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron. 2006;51:549–560. doi: 10.1016/j.neuron.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 139.Chen-Plotkin AS, et al. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum Mol Genet. 2008;17:1349–1362. doi: 10.1093/hmg/ddn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Blalock EM, et al. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuhn A, Thu D, Waldvogel HJ, Faull RLM, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods. 2011;8:945–947. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- 143.Miller JA, Geschwind DH. In: Systems Biology for Signaling Networks. Choi S, editor. Ch 25. Springer; 2010. pp. 611–643. [Google Scholar]
- 144.Liang WS, et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Small SA, et al. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 146.Muhammad A, et al. Retromer deficiency observed in Alzheimer’s disease causes hippocampal dysfunction, neurodegeneration, and Aβ accumulation. Proc Natl Acad Sci USA. 2008;105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Webster JA, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liang WS, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miller JA, Oldham MC, Geschwind DHA. Systems level analysis of transcriptional changes in Alzheimer’s disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ray M, Ruan J, Zhang W. Variations in the transcriptome of Alzheimer’s disease reveal molecular networks involved in cardiovascular diseases. Genome Biol. 2008;9:R148. doi: 10.1186/gb-2008-9-10-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Forabosco P, et al. Insights into TREM2 biology by network analysis of human brain gene expression data. Neurobiol Aging. 2013;34:2699–2714. doi: 10.1016/j.neurobiolaging.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rosen EY, et al. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wexler EM, Paucer A, Kornblum HI, Palmer TD, Geschwind DH. Endogenous Wnt signaling maintains neural progenitor cell potency. Stem Cells. 2009;27:1130–1141. doi: 10.1002/stem.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Long JM, Ray B, Lahiri DK. MicroRNA-339-5p down-regulates protein expression of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer disease subjects. J Biol Chem. 2014;289:5184–5198. doi: 10.1074/jbc.M113.518241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lau P, et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol Med. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schonrock N, Matamales M, Ittner LM, Götz J. MicroRNA networks surrounding APP and amyloid-β metabolism — implications for Alzheimer’s disease. Exp Neurol. 2012;235:447–454. doi: 10.1016/j.expneurol.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 158.Ginsberg SD, et al. Single-cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem Res. 2004;29:1053–1064. doi: 10.1023/b:nere.0000023593.77052.f7. [DOI] [PubMed] [Google Scholar]
- 159.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 160.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 162.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. An RNA-seq database of gene expression and splicing differences between major cell types in the mouse CNS that provides cell type-specific profiles that can be used to query cell type specificity in other studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lim J, et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 165.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Goehler H, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 167.Shirasaki DI, et al. Network organization of the Huntingtin proteomic interactome in mammalian brain. Neuron. 2012;75:41–57. doi: 10.1016/j.neuron.2012.05.024. Illustrates the power of network analysis for defining protein interaction modules across brain regions and time points to understand the huntingtin interactome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen JC, et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014;159:402–414. doi: 10.1016/j.cell.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2013;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. This study combined network analysis in post-mortem tissue, eQTL mapping and Bayesian causal inference to identify a causal role for the gene TYROBP in Alzheimer disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Aten JE, Fuller TF, Lusis AJ, Horvath S. Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst Biol. 2008;2:34. doi: 10.1186/1752-0509-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- 173.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 2013;20:970–978. doi: 10.1089/cmb.2012.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Robles JA, et al. Efficient experimental design and analysis strategies for the detection of differential expression using RNA-sequencing. BMC Genomics. 2012;13:484. doi: 10.1186/1471-2164-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rapaport F, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14:R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ching T, Huang S, Garmire LX. Power analysis and sample size estimation for RNA-seq differential expression. RNA. 2014;20:1684–1696. doi: 10.1261/rna.046011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hansen KD, Wu Z, Irizarry RA, Leek JT. Sequencing technology does not eliminate biological variability. Nat Biotechnol. 2011;29:572–573. doi: 10.1038/nbt.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. A must-read paper prior to pursuing the design or analysis of a high-throughput experiment, it contains advice and analyses for evaluating the contribution of technical and biological variation in data sets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Trabzuni D, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;120:473–473. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hoen PA, Friedländer MR, Almlöf J. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31:1015–1022. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 181.Mostafavi S, et al. Normalizing RNA-sequencing data by modeling hidden covariates with prior knowledge. PLoS ONE. 2013;8:e68141. doi: 10.1371/journal.pone.0068141. This study presents a comprehensive framework for thinking about signal and noise in gene expression data and unifies most known methods into one framework. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning. Springer Science & Business Media; 2013. [Google Scholar]
- 183.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shohat S, Shifman S. Bias towards large genes in autism. Nature. 2014;512:E1–E2. doi: 10.1038/nature13583. [DOI] [PubMed] [Google Scholar]
- 185.Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Raychaudhuri S, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25:211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gusev A, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nord AS, Pattabiraman K, Visel A, Rubenstein JLR. Genomic perspectives of transcriptional regulation in forebrain development. Neuron. 2015;85:27–47. doi: 10.1016/j.neuron.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Maze I, et al. Analytical tools and current challenges in the modern era of neuroepigenomics. Nat Neurosci. 2014;17:1476–1490. doi: 10.1038/nn.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bayés À, et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci. 2010;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bayés À, et al. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS ONE. 2012;7:e46683. doi: 10.1371/journal.pone.0046683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Qu XA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discov Today. 2012;17:1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 194.Lamb J. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 195.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol. 2006;24:55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Blair DR, et al. A nondegenerate code of deleterious variants in mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci USA. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Freimer N, Sabatti C. The Human Phenome Project. Nat Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- 199.Congdon E, Poldrack RA, Freimer NB. Neurocognitive phenotypes and genetic dissection of disorders of brain and behavior. Neuron. 2010;68:218–230. doi: 10.1016/j.neuron.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Coppola G, Geschwind DH. Technology Insight: querying the genome with microarrays — progress and hope for neurological disease. Nat Clin Pract Neurol. 2006;2:147–158. doi: 10.1038/ncpneuro0133. [DOI] [PubMed] [Google Scholar]
- 201.Jaffe AE, et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2015;18:154–161. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dougherty JD, et al. The disruption of Celf6, a gene identified by translational profiling of serotonergic neurons, results in autism-related behaviors. J Neurosci. 2013;33:2732–2753. doi: 10.1523/JNEUROSCI.4762-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34:1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Heiman M, et al. Molecular adaptations of striatal spiny projection neurons during levodopa-induced dyskinesia. Proc Natl Acad Sci USA. 2014;111:4578–4583. doi: 10.1073/pnas.1401819111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dalal J, et al. Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 2013;27:565–578. doi: 10.1101/gad.207654.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. The first single-cell RNA-seq study of the adult mouse cortex and hippocampus that uses unsupervised clustering to identify dozens of cell types, including many distinct interneuron subtypes. [DOI] [PubMed] [Google Scholar]
- 207.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lovatt D, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493:327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 211.Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010;33:173–182. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Butte AJ, Kohane IS. In: Pacific Symposium on Biocomputing 2000. Altman RB, et al., editors. World Scientific; 2000. pp. 418–429. [DOI] [PubMed] [Google Scholar]
- 213.Horvath S. Weighted Network Analysis: Applications in Genomics and Systems Biology. Springer; 2011. [Google Scholar]
- 214.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lee I, Marcotte EM. Effects of functional bias on supervised learning of a gene network model. Methods Mol Biol. 2009;541:463–475. doi: 10.1007/978-1-59745-243-4_20. [DOI] [PubMed] [Google Scholar]
- 216.Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185:405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Li S, et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol. 2014;32:915–925. doi: 10.1038/nbt.2972. A comprehensive evaluation of different sequencing platforms and methodologies that identifies optimal parameters for RNA-seq, including for degraded RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liu Y, Zhou J, White KP. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics. 2014;30:301–304. doi: 10.1093/bioinformatics/btt688. A comparison of multiple RNA-seq differential expression methodologies that demonstrated biological replicates are more important than technical replicates and provided guidelines on sequencing depth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics. 2013;14:91. doi: 10.1186/1471-2105-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 222.Tibshirani R. A simple method for assessing sample sizes in microarray experiments. BMC Bioinformatics. 2006;7:106. doi: 10.1186/1471-2105-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Langfelder P, Mischel PS, Horvath S. When is hub gene selection better than standard meta-analysis? PLoS ONE. 2013;8:e61505. doi: 10.1371/journal.pone.0061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Good PI. Permutation, Parametric, and Bootstrap Tests of Hypotheses. Springer; 2010. [Google Scholar]
- 225.Narayan S, et al. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Berchtold NC, et al. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol Aging. 2013;34:1653–1661. doi: 10.1016/j.neurobiolaging.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zambon AC, et al. GO-Elite: a flexible solution for pathway and ontology over-representation. Bioinformatics. 2012;28:2209–2210. doi: 10.1093/bioinformatics/bts366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.