To the Editor
Asthma is a chronic inflammatory disorder mainly characterized by reversible airflow obstruction, bronchial hyperresponsiveness and dyspnea. The functional alterations in the airway smooth muscle usually observed in asthmatics have been associated with increased expression of the smooth muscle myosin light chain kinase isoform (smMLCK).1 This constitutes a key cytoskeleton effector of the smooth muscle contractile machinery and is encoded by the myosin light chain kinase (MYLK) gene. Two MYLK single nucleotide polymorphisms (SNPs), which are common in Africans (>30%) but rare in Europeans (≤1%), have been associated with asthma in African Americans and Afro-Caribbeans.2,3 However, none of the MYLK variants have attained the strict Bonferroni-corrected level of genome-wide significance in any genome-wide association study (GWAS) of asthma. This discrepancy may arise from the low coverage of this region on commercial arrays and/or the genetic specificities of the populations studied, since risk factors in one population might not generalize to another.4 Here we aimed to fine map the association of SNPs from MYLK gene with asthma in case-control studies from Spanish (n=3,219) and Latino (n=4,650) populations.
In the discovery stage, DNA samples from 606 physician-diagnosed asthmatics from the Genetics of Asthma (GOA) study in the Spanish population (GOA I) were compared with 1,258 non-asthmatic subjects. Top associated SNPs were then replicated in two independent Spanish studies: GOA II and The Genetics of Asthma study in the Spanish population from Malaga (GOAM). GOA II included 248 asthma cases and 537 controls genotyped with the Axiom® Genome-Wide CEU 1 Array (Affymetrix, Santa Clara, CA), whereas GOAM comprised 320 asthma cases and 250 controls genotyped using TaqMan allelic discrimination assays (Life Technologies, Carlsbad, CA). Further replication was assessed in two Latino studies using existing genome-wide genotyping data: The Genetics of Asthma in Latino Americans (GALA I),4 consisting of 529 cases and 347 controls, and Genes-Environments & Admixture in Latino Americans (GALA II) study,5 including 1,893 cases and 1,881 controls. Detailed descriptions of the study design and sample characteristics can be found in Figure E1, Tables E1 and E2, and Text E1 in the Online Repository at www.jacionline.org.
In the discovery stage, a total of 29 tagging SNPs (tSNPs) were selected from HapMap II to analyze variants with minor allele frequency (MAF)≥5% from the MYLK gene in European individuals. Genotyping was performed using the iPLEX® Gold assay on MassARRAY® system (Sequenom Inc., San Diego, CA). A total of 26 SNPs passed quality control (after removing monomorphic SNPs or with p≤1.7×10−3 for Hardy-Weinberg equilibrium in controls) (Table E3). After imputation, 272 SNPs with MAF≥5%, and a squared correlation between imputed and true genotypes (Rsq)≥0.3, were kept for association testing using logistic regression models (88% of the total variants with MAF≥5% in Europeans). Principal components were used as covariates to adjust for population stratification, as previously described.6
Three significantly associated SNPs were observed after Bonferroni correction (p-value ≤1.8×10−4). Two intronic SNPs in perfect linkage disequilibrium (LD, r2=1) and with MAF=9% showed the most significant associations: rs77820417 and rs78442149 (for both SNPs: OR=2.71 [95%CI=1.79–4.11] for the minor allele, p=2.78×10−6) (Figure 1, Table E4). We followed up the SNP rs77820417 for replication in four independent studies. Association was significant in GOAM (OR=2.32, 95%CI=1.23–4.36, p=4.17×10−3), in GALA I (OR=1.73, 95%CI=1.03–2.93, p=.040), and in GALA II (OR=1.31, 95%CI=1.05–1.64, p=.019), but not in GOA II (OR=0.88, 95%CI=0.48–1.61, p=.669). Due to the heterogeneity of effects among studies (Cochran’s Q test, p=.007), a random effects meta-analysis of the 7,869 individuals was performed, confirming the strong association of rs77820417 with asthma susceptibility (OR=1.66, 95% CI=1.14–2.42, p=1.57×10−7; Table 1). We further explored the association of rs77820417 with asthma exacerbations in GALA II, defined by the presence of ≥1 asthma-related events (hospitalizations, emergency department visits, and oral steroid use) over the 12 months prior to recruitment and adjusting for the use of medication during the same period. The A allele, associated with asthma risk, was also associated with increased risk of asthma exacerbations (OR=1.80, 95% CI=1.08–2.99, p=.023). The associated SNP is located within the MYLK gene region encoding the smMLCK isoform, which participates in smooth muscle cell contractility. smMLCK activation is a critical step in the cytoskeletal rearrangements, providing dynamic regulation of cell shape, cell motility and adhesion, which are involved in the remodeling processes underlying asthma.7
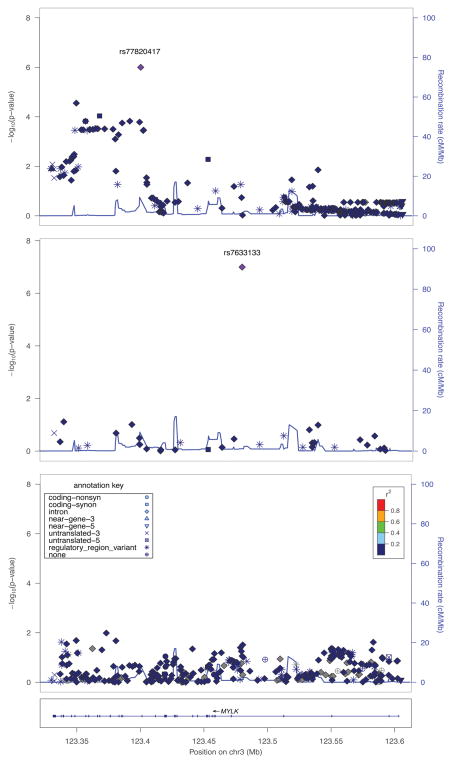
Figure 1. Regional plots of association results in the discovery sample (upper panel), GABRIEL (middle panel), and EVE (lower panel).
The -log10 transformed p-values for association tests are plotted as symbols according to their functional annotation. The SNP rs number indicated on the plot denotes the result for the most significantly associated SNP with asthma risk. The results for the remaining SNPs are color coded to reflect their degree of linkage disequilibrium with the most significant SNP based on pairwise r2 values from the European population of the 1KGP. Estimated recombination rates (light blue line) are plotted on the right y-axis.
Table 1.
Summary of association testing of rs77820417 with asthma susceptibility.
| Study | Sample size (Cases:Controls) | MAF | OR (95% CI) | p-value |
|---|---|---|---|---|
| GOA I | 1,864 (606:1,258) | 0.090 | 2.71 (1.79–4.11) | 2.78×10−6 |
| GOA II | 785 (248:537) | 0.084 | 0.88 (0.48–1.61) | .669 |
| GOAM | 570 (320:250) | 0.052 | 2.32 (1.23–4.36) | 4.17×10−3 |
| GALA I | 876 (529:347) | 0.073 | 1.73 (1.03–2.93) | .040 |
| GALA II | 3,774 (1,893:1,881) | 0.044 | 1.31 (1.05–1.64) | .019 |
| Meta-analysis | 7,869 (3,596:4,273) | - | 1.66 (1.14–2. 42) | 1.57×10−7 |
p-values≤.05 are in boldface.
To date, GWAS have firmly identified susceptibility genes underlying asthma risk, although most of the studies were performed using HapMap-based inferences, where the coverage for genetic variation is limited compared to the information provided by the 1000 Genomes Project (1KGP). In fact, neither the top hit observed in the current study nor its proxy (rs78442149) were tested for association in the GABRIEL (http://www.cng.fr/gabriel/results.html) or the EVE consortia, the largest GWAS meta-analyses in asthma performed in Europeans8 and multi-ethnic groups.9 No other variants from the 1KGP are in moderate LD with those two SNPs (highest r2=0.25). However, one MYLK variant was associated with asthma in the GABRIEL consortium (rs7633133, p=1.01×10−7) (Figure 1). This SNP shows a MAF=1% in 1KGP Europeans and, therefore, was not tested in our study. In the EVE consortium, although no MYLK SNP showed an outstanding significance (minimum p=.01) (Figure 1), a 4-fold enrichment of significant associations was observed in Latinos (Fisher exact test p=7.03×10−5), but not in European or African Americans (p=.720 and p=1.0, respectively) (Table E5 and Text E1).
One striking aspect of our study is the large effect sizes found for the association of rs77820417 with asthma susceptibility and exacerbations, which is only comparable to the effect reported for a SNP in GSDMB with early childhood asthma with severe exacerbations.10 However, this effect size may be confounded by the large gender and age differences among cases and controls in the discovery sample. Despite this, the validation across multiple studies of both children and adults with different gender balance suggests that the result from the discovery study is not a false positive. The catalog of asthma susceptibility genes could be more comprehensive if imputation based on 1KGP data would be exploited to meta-analyze existing GWAS data. In addition, analysis of diverse populations also contributes with new susceptibility loci, as many GWAS hits are not transferable to all populations.4 In fact, higher North African ancestry is detected in southwestern Europe and is decreased in northern latitudes.11 Therefore, novel susceptibility loci for asthma could be revealed in Spanish-descent populations.
In summary, we identified a MYLK SNP association with asthma in Spanish descent individuals, showing suggestive genome-wide significance. Future studies will be needed to confirm its importance in other populations.
Supplementary Material
Acknowledgments
Sources of funding:
This work was supported by Instituto de Salud Carlos III (FIS PI11/00623) and co-financed by the European Regional Development Funds, “A way of making Europe” from the European Union, by the University of Chicago Core Subsidy Mini Award (ITM/CTSA UL1 RR024999) and by a grant from the 7th Framework Programme (FP7-REGPOT-2012-CT2012-31637-IMBRAIN). This work was also supported by grants from National Institutes of Health to E.G.B.: the National Heart, Lung and Blood Institute (RC2 HL101651, HL088133, HL078885, HL004464, HL104608 and HL117004); the National Institute of Environmental Health Sciences (ES015794); the National Institute on Minority Health and Health Disparities (MD006902); the National Institute of General Medical Sciences (GM007546). E.G.B was also funded by the American Asthma Foundation, the RWJF Amos Medical Faculty Development Award, the Sandler Foundation and the Flight Attendant Medical Research Institute. MPY was supported by a postdoctoral fellowship from Fundación Ramón Areces (http://www.fundacionareces.es). MAH and ABL were supported by fellowships from the Instituto de Salud Carlos III (FI11/00074 and FI12/00493, respectively).
The authors thank Servicio de Apoyo Informático a la Investigación, ULL (SAII) for the HPC support; the GALA investigators, recruiters, participants and study coordinators; and the EVE consortium from granting access to summary data.
Abbreviations
- 1KGP
1000 Genomes Project
- CI
confidence interval
- LD
linkage disequilibrium
- MLCK
myosin light chain kinase
- MYLK
myosin light chain kinase gene
- GWAS
genome-wide association study
- GALA I
The Genetics of Asthma in Latino Americans
- GALA II
Genes-Environments & Admixture in Latino Americans
- GOAM
Genetics of Asthma study in the Spanish population from Malaga
- GOA I
Genetics of Asthma study in the Spanish population
- GOA II
Genetics of Asthma study in the Spanish population II
- OR
odds ratio
- SNP
Single nucleotide polymorphism
- tSNPs
tagging SNPs
References
- 1.Benayoun L, Druilhe A, Dombret MC, Aubier M, Pretolani M. Airway structural alterations selectively associated with severe asthma. Am J Respir Crit Care Med. 2003;167:1360–8. doi: 10.1164/rccm.200209-1030OC. [DOI] [PubMed] [Google Scholar]
- 2.Flores C, Ma SF, Maresso K, Ober C, Garcia JG. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol. 2007;31:296–305. doi: 10.1002/gepi.20210. [DOI] [PubMed] [Google Scholar]
- 3.Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, et al. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol. 2007;119:1111–8. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Galanter JM, Gignoux CR, Torgerson DG, Roth LA, Eng C, Oh SS, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: The Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol. 2014;134:295–305. doi: 10.1016/j.jaci.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012;130:76–82. e12. doi: 10.1016/j.jaci.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pino-Yanes M, Corrales A, Acosta-Herrera M, Perez-Rodriguez E, Cumplido J, Campo P, et al. HLA-DRB1*15:01 allele protects from asthma susceptibility. J Allergy Clin Immunol. 2014;134:1201–3. doi: 10.1016/j.jaci.2014.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Garcia JG, Lazar V, Gilbert-McClain LI, Gallagher PJ, Verin AD. Myosin light chain kinase in endothelium: molecular cloning and regulation. Am J Respir Cell Mol Biol. 1997;16:489–94. doi: 10.1165/ajrcmb.16.5.9160829. [DOI] [PubMed] [Google Scholar]
- 8.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–5. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 11.Botigue LR, Henn BM, Gravel S, Maples BK, Gignoux CR, Corona E, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci U S A. 2013;110:11791–6. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.