Abstract
Dynamic clusters of lipid-anchored Ras proteins are important for high-fidelity signal transduction in cells. The average size of Ras nanoclusters was reported to be independent of protein expression levels, and cholesterol depletion is commonly used to test the raft-preference of nanoclusters. However, whether protein concentration and membrane domain stability affect Ras clustering in a reversible manner is not well understood. We used coarse-grained molecular dynamics simulations to examine the reversibility of the effects of peptide and cholesterol concentrations as well as a lipid domain-perturbing nanoparticle (C60) on the dynamics and stability of H-Ras lipid-anchor nanoclusters. By comparing results from these simulations with previous observations from the literature, we show that effects of peptide/cholesterol concentrations on the dynamics and stability of H-Ras peptide nanoclusters are reversible. Our results also suggest a correlation between the stabilities of lipid domains and Ras nanoclusters, which is supported by our finding that C60 penetrates into the liquid-disordered domain of the bilayer, destabilizing lipid domains and thereby the stability of the nanoclusters.
Main Text
Previous experiments have shown that Ras proteins assemble into dynamic nanoclusters on the plasma membrane (PM) (1), as well as in synthetic model membranes (2, 3). Nanoclustering is essential for the biological function of Ras proteins, which mediates signal transduction pathways involved in cell growth and development (4). Somatic mutations that lead to unregulated Ras function are found in 15–25% of all human cancers (4). There are three common Ras proteins in humans (namely, H-, N-, and K-Ras A/B) that share a highly conserved catalytic domain but differ at the C-terminus, where they undergo posttranslational lipid modification(s) before binding to the PM (1). The minimal PM anchor of H-Ras, a farnesylated and doubly palmitoylated heptapeptide (tH), forms nanoclusters similar to the full-length protein (5). Therefore, tH is an excellent model system to study Ras clustering in atomic detail.
Recently, we used coarse-grained (CG) molecular dynamics (MD) simulations to show that (6, 7, 8): 1) tH nanoclusters in model bilayers predominantly localize at the interface between liquid-ordered (Lo) and liquid-disordered (Ld) domains and modulate bilayer curvature; 2) there is a critical concentration below which tH does not form clusters; and 3) cholesterol stabilizes lipid domains and thereby tH nanoclusters, but it is not required for cluster formation. However, it was not clear whether these are reversible processes. Reversibility has important implications on the apparent independence of the average size of Ras nanoclusters on protein expression levels (1, 5) and the relevance of cholesterol depletion (e.g., by methyl-β-cyclodextrin (9) treatment) to test raft-preference of nanoclusters. Reversibility is also crucial for the regulation of Ras function through nanocluster dynamics (10). Here we studied, with the same CG MD approach we have used previously, possible declustering or changes in the stability/dynamics of preformed tH nanoclusters upon a systematic depletion of tH and cholesterol from a system containing clustered tH. Our goal is to test whether effects of peptide and cholesterol concentration on nanocluster size, stability, and lateral distribution are reversible.
We used the MARTINI CG model (Ver. 2.1) (11) and the GROMACS program (Ver. 4.5.4) (12) for all of these simulations (see Table S1 in the Supporting Material), which were started from a snapshot at 32 μs of a previous simulation (7) on a bilayer composed of 960 DPPC (dipalmitoylphosphatidylcholine), 576 DLiPC (dilinoleoylphosphatidylcholine), 576 cholesterol, 64 tH, and 17,789 water molecules plus 128 Na+ and 192 Cl− ions. The 64 tH molecules were all on one leaflet to mimic Ras binding to the inner leaflet of the PM. Of these, we deleted 0, 16, 32, and 48 tH molecules from the nonclustered fraction (monomer, dimers, or trimers; see Janosi et al. (6) and Li et al. (7)) to examine the effect of decreasing tH concentration on clustering; nanoclusters of size 4 or higher were left intact to allow for potential spontaneous declustering. Similarly, we studied the impact of cholesterol depletion by deleting 0, 192, 384, and 576 cholesterol molecules (half from each leaflet), resulting in 27, 20, 11, and 0% cholesterol systems while keeping the number of tH fixed at 64. Subsequent steps of system equilibration, production, and data analysis were identical to previous work (6, 7); a brief description is provided in the Supporting Material.
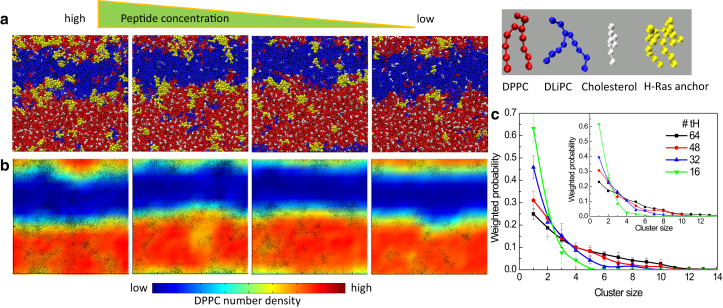
Results from the simulations with decreasing tH concentration are summarized in Figs. 1 and S5 a. They show that tH depletion did not lead to changes in lipid domain organization; both the DPPC/cholesterol-enriched Lo and DLiPC-enriched Ld domains remained intact (Fig. 1, a and b). However, there is significant variation between the smallest tH concentration (16 tH) and the rest in terms of tH cluster size distribution (Fig. 1 c), especially in the fraction of aggregates of size 4 or higher that represent nanoclusters based on our previous definition (6, 7). Specifically, the cluster size distributions before tH depletion (i.e., with 64 tH) and after removal of 25 and 50% of the peptides are similar, but further reduction to 16 peptides led to negligible nanoclustering. This is consistent with our previous estimate that a peptide/lipid of ∼1:100 might be required for cluster formation (7). Moreover, projection of the center-of-mass positions of tH molecules on a two-dimensional heat map of the mean DPPC positions shows that the peptides frequently sample the interface between the Lo and Ld domains. The same observation has been made in our previous studies (6, 7). We conclude that the effect of peptide concentration on tH nanoclustering is reversible. Although not the main focus of this study, we note that asymmetric tH insertion could cause curvature to the bilayer (6, 7, 8); however, this does not have adverse effect on clustering (Fig. S1).
Figure 1.
Effect of decreasing peptide concentration on tH clustering. (a, left to right) The last snapshot from a 12 μs trajectory of systems with 64, 48, 32, and 16 tH molecules (color scheme is the same as the molecular models, right top). (b, left to right) Two-dimensional number-density map of DPPC colored from low (blue) through high (red) and the instantaneous location of tH molecules (black points) derived from analysis of each of the last 4 μs trajectories. (c) Probability distribution of tH cluster sizes upon reduction of tH molecules (error bars represent SD from averaging over eight 500 ns blocks of the last 4 μs of the 12 μs data; see Section S8 in the Supporting Material for details). (Inset) Results from one of the two additional simulations with different initial velocity assignments. To see this figure in color, go online.
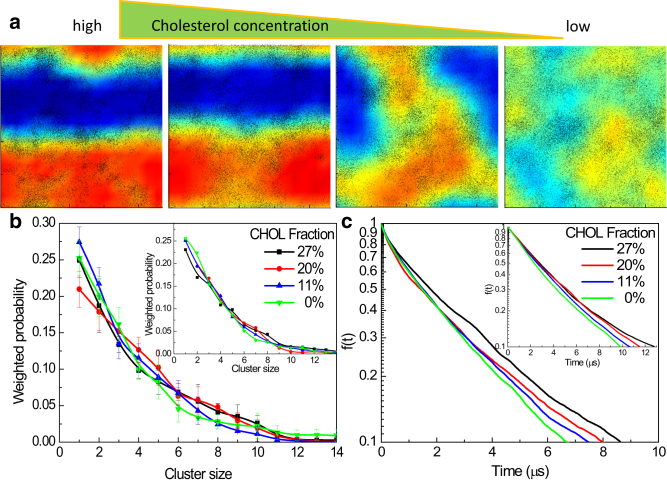
We previously showed that increasing cholesterol concentration enhances the stability of tH nanoclusters but does not significantly affect their formation or size distribution (7). This suggested that cholesterol is not required for tH clustering, but is important for stability (7). We hypothesized that is a reversible process; namely, decreasing cholesterol concentration in a system of preformed tH nanoclusters would have a similar effect as increasing cholesterol before clustering. To test this hypothesis, we ran four simulations with progressively lower cholesterol content (27, 20, 11, and 0%), all started from a previously simulated bilayer system containing a fixed number of 64 tH molecules. The reduction in cholesterol fraction led to progressively increasing lipid mixing (Fig. 2 a), but the size distribution of the tH clusters remained almost unaffected (Figs. 2 b and S5 b). This result not only confirms our previous observation that cholesterol is not required for tH clustering, but also indicates that cholesterol depletion does not decluster tH. On the other hand, analysis of the rate of decay of selected clusters (Fig. 2 c) in terms of a molecular expulsion autocorrelation function f(t) described in the Supporting Material suggest that cholesterol depletion increases cluster dynamics and, thereby, the rate of exchange between the pools of clustered and nonclustered fractions. Combined with our previous observations (7), these results strongly suggest that cholesterol depletion, despite being a common technique to test raft-preference of nanoclusters (9), does not lead to declustering. Instead, it reversibly affects cluster dynamics.
Figure 2.
Cholesterol depletion reduces lipid domain stability and enhances tH cluster dynamics. (a, left to right) Two-dimensional number-density map of DPPC colored from low (blue) through high (red) along with the instantaneous location of tH molecules (black points) based on analysis over the last 4 μs trajectories. (b) Probability distribution of tH cluster sizes upon cholesterol depletion; error from averaging over eight 500-ns blocks of the last 4 μs of data. (Inset) Data from another independent set of simulations. (c) Stability of tH nanoclusters upon cholesterol depletion. Considering that on average there are ∼6 Ras proteins per cluster (5), here we show f(t) for clusters of size 6, although f(t) values for other cluster sizes exhibit similar trends (as an example, the inset shows f(t) for cluster size 4). To see this figure in color, go online.
The results described above suggest a correlation between the stabilities of Ras nanoclusters and lipid domains (i.e., extent of lipid demixing). It follows that compounds that directly or indirectly affect lipid domain stability might also affect protein clustering. To test this hypothesis, we used a nanoparticle (C60) as a perturbant of domain integrity because it has been shown to partition to the Ld phase of multidomain bilayers (13). C60 belongs to a class of biomedically relevant (e.g., as drug-delivery vehicles) carbon-based nanoparticles whose interaction with biomembranes is the subject of intense investigations. We simulated the 64-tH/27% cholesterol system after adding an aggregate of 16 C60 molecules into the water box. Consistent with previous reports (13), we found that the C60 aggregate quickly adheres to the bilayer, enters into the hydrophobic core of the Ld domain, and dissolves into individual C60 molecules (Fig. S4, a and b). This led to significant lipid remixing and thus destabilization of the striped lipid domains (Fig. S4 c). Consequently, the stability/dynamics of tH clusters was altered in a manner that is similar to cholesterol depletion (Figs. S4 e and 2 c). Also, just as cholesterol depletion did not decluster tH, the size distribution of tH clusters was not significantly affected by the presence of C60 (see Fig. S4 d). These results support our conclusion that the stability (and lateral distribution) of tH clusters is a function of the stability of lipid domains.
In summary, previous experiments have shown that Ras nanoclusters are important for high-fidelity signal transduction (10) while MD studies provided detailed information about the driving forces for Ras clustering in model lipid bilayers (6, 7). Here we examined the effects of peptide and cholesterol depletion on the dynamics and stability of the H-Ras lipid anchor nanoclusters using a tens-of-microseconds timescale CG MD. As summarized in Fig. S5, we found that the effect of systematic depletion of the nonclustered tH fraction is equivalent to increasing tH concentration. Similarly, depletion of cholesterol destabilizes lipid domains and thereby the stability of tH clusters, but it has no effect on cluster formation per se. We conclude that effects of changes in both peptide and cholesterol concentration are reversible. Additional evidence for the correlation between the stabilities of lipid domains and tH clusters was provided by C60, which partitioned into the core of the Ld domain and destabilized lipid domains as well as tH nanoclusters.
Author Contributions
A.A.G. and X.L. designed research; X.L. and Z.L. performed research; X.L., A.A.G., and Z.L. analyzed data; and X.L. and A.A.G. wrote the article.
Acknowledgments
We thank the National Institutes of Health (grant No. RO1 GM100078) for financial support and the Texas Advanced Computing Center for computational resources.
Editor: Scott Feller.
Footnotes
Supporting Material, six figures, two tables, and simulation and analysis details are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01166-2.
Supporting Material
References
- 1.Abankwa D., Gorfe A.A., Hancock J.F. Ras nanoclusters: molecular structure and assembly. Semin. Cell Dev. Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weise K., Triola G., Winter R. Influence of the lipidation motif on the partitioning and association of N-Ras in model membrane subdomains. J. Am. Chem. Soc. 2009;131:1557–1564. doi: 10.1021/ja808691r. [DOI] [PubMed] [Google Scholar]
- 3.Weise K., Kapoor S., Winter R. Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 2011;133:880–887. doi: 10.1021/ja107532q. [DOI] [PubMed] [Google Scholar]
- 4.Cox A.D., Fesik S.W., Der C.J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y., Hancock J.F. Ras nanoclusters: versatile lipid-based signaling platforms. Biochim. Biophys. Acta Mol. Cell Res. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Janosi L., Li Z., Gorfe A.A. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. USA. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Janosi L., Gorfe A.A. Formation and domain partitioning of H-Ras peptide nanoclusters: effects of peptide concentration and lipid composition. J. Am. Chem. Soc. 2012;134:17278–17285. doi: 10.1021/ja307716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., Gorfe A.A. Deformation of a two-domain lipid bilayer due to asymmetric insertion of lipid-modified Ras peptides. Soft Matter. 2013;9:11249–11256. doi: 10.1039/C3SM51388B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niv H., Gutman O., Henis Y.I. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian T., Harding A., Hancock J.F. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 11.Monticelli L., Kandasamy S.K., Marrink S.J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 12.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 13.Sun D., Lin X., Gu N. Cholesterol affects C60 translocation across lipid bilayers. Soft Matter. 2014;10:2160–2168. doi: 10.1039/c3sm52211c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.