Abstract
Radiomics is being explored for potential applications in radiation therapy. How various imaging protocols affect quantitative image features is currently a highly active area of research. To assess the variability of image features derived from conventional [three-dimensional (3D)] and respiratory-gated (RG) positron emission tomography (PET)/computed tomography (CT) images of lung cancer patients, image features were computed from 23 lung cancer patients. Both protocols for each patient were acquired during the same imaging session. PET tumor volumes were segmented using an adaptive technique which accounted for background. CT tumor volumes were delineated with a commercial segmentation tool. Using RG PET images, the tumor center of mass motion, length, and rotation were calculated. Fifty-six image features were extracted from all images consisting of shape descriptors, first-order features, and second-order texture features. Overall, 26.6% and 26.2% of total features demonstrated less than 5% difference between 3D and RG protocols for CT and PET, respectively. Between 10 RG phases in PET, 53.4% of features demonstrated percent differences less than 5%. The features with least variability for PET were sphericity, spherical disproportion, entropy (first and second order), sum entropy, information measure of correlation 2, Short Run Emphasis (SRE), Long Run Emphasis (LRE), and Run Percentage (RPC); and those for CT were minimum intensity, mean intensity, Root Mean Square (RMS), Short Run Emphasis (SRE), and RPC. Quantitative analysis using a 3D acquisition versus RG acquisition (to reduce the effects of motion) provided notably different image feature values. This study suggests that the variability between 3D and RG features is mainly due to the impact of respiratory motion.
Introduction
Positron emission tomography (PET) is a beneficial technology in the process of cancer diagnosis and staging [1], [2], monitoring tumor response to treatment [3], detecting necrosis and tumor heterogeneity, identifying the primary tumor location [4], and delineating tumors from atelectasis [5], particularly in lung cancers. In fact, studies have shown that the use of PET/computed tomography (CT) improves confidence in diagnosis, increases the number of “definite” lesions by 41% in patients with non–small cell lung cancer (NSCLC) [6], and improves delineation accuracy for gross tumor volumes (GTVs) in radiation therapy [4], [5].
Standardized uptake value (SUV) is the standard PET metric in image analysis. However, it is clear that SUV is dependent on many technical as well as physiologic factors [7]. It is proportionately dependent upon fluorodeoxyglucose (FDG) uptake, which in turn is affected by dose calibration, clock (decay) synchronization, patient weight and blood sugar level, documentation of unused tracer remains, and other setup specifics [7]. Moreover, SUV indirectly depends on the method of obtaining raw data, radionuclide uptake time, hardware platform, and the applied reconstruction algorithm [7], [8].
Studies have shown SUV’s predictive ability of therapy response and survival [9], [10], although, particularly in NSCLC, discrepancy remains regarding whether maximum or minimum SUV is a predictor based on treatment modality [11]. The prognostic ability of SUV parameters has also been shown, but conflict emerges when defining the best cutoff value [9]. Nonetheless, SUV can be unreliable. A clinical study done on the test-retest reproducibility of SUV demonstrated greater than expected SUV variability within a single institution [7], and 10% to 25% SUV variability was detected in a multicenter consortium before biological effects or protocol influences [12]. Based on these results and the SUV’s reliance on various nonstandardized factors, it is clear that there is a need for additional indicators that are more robust than SUV or complementary to SUV-based findings. Quantifiable and robust (radiomic) image features, such as texture features, may be candidates for such indicators.
Texture is a global pattern resulting from repetition of local subpatterns [13]. Features associated with image texture describe the relationships between the gray-scale intensity of pixels [or voxels in three-dimensional (3D) image] on a local or global image scale. These features have been used for classification and segmentation purposes, identifying regions of interest in an image and estimating tumor heterogeneity [14].
Image features are quite vast in number and can be subdivided into shape features and first-, second-, and higher-order features. First-order features provide information about gray-scale intensities and are derived from intensity distributions and histograms, whereas second-order texture features are derived from gray-tone spatial dependency matrices which are constructed from the intensity values of an image as described below [14], [15]. In this paper, we refer to all of these as image features.
In medical imaging, CT image texture analysis has been studied extensively, dating back to the early 1980s [16]. More recent studies in CT image analysis have uncovered feature correspondence with lung tumor aggressiveness and tumor heterogeneity; demonstrated potential as a marker for survival in NSCLC; and revealed relationships between features, tumor stage, and metabolism [17], [18], [19]. The reproducibility and robustness of specific identifiers in NSCLC CT images have also been studied [20].
The application of image feature analysis to PET images has been explored more recently. Prior studies in PET/CT image texture analysis have demonstrated its potential as a predictor of tumor and normal tissue response to therapy, a quantifier of tumor heterogeneity and radiosensitivity, and an indicator for adaptive therapy schemes [9], [21], [22]. Conclusive and beneficial results regarding tumor response to treatment using multimodality imaging [23] have been drawn, and PET texture analysis is currently being explored for application to predictive/prognostic models of treatment outcome, partnered with genomics and proteomics patterns [21]. In addition, various features have been investigated using test-retest and interobserver stability in FDG-PET [24]. PET image analysis has also been shown to predict response to radiochemotherapy in esophageal cancer [25] and to quantify tumor heterogeneity as a response predictor [26]. Partnered with a multimodality modeling system, PET texture analysis could lead to more individualized treatment planning in lung radiotherapy [23].
Moreover, certain FDG-PET–based texture features have demonstrated association with nonresponse to chemoradiotherapy for NSCLC tumors [27]. These and many other studies are part of a more general systematic approach, namely radiomics, which is an emerging framework relating image features to molecular medicine where large amounts of quantitative features (400 +) are extracted for diagnostic, prognostic, and predictive information [21], [28], [29], [30]. In other words, PET/CT image feature analysis is an emerging and promising quantitative imaging field.
Although radiomics shows much promise, there have been several studies showing image features’ dependency on various factors in the production of images. For example, in a recent study, 45 of 50 texture features showed 10% to 200% variability across acquisition protocols and reconstruction algorithms [31]. Therefore, several investigators have pointed to the need for standardization in texture analysis [32], [33], [34]. The usefulness of radiomics depends on the reliability of feature values; thus, it is important to characterize feature behavior under many potential clinical conditions. Our goal in this study was to explore image feature value variability between respiratory-gated (RG) and conventional (3D) PET images acquired on the same patient during a single PET scan session.
Conventional (3D) PET images are influenced by motion because of their relatively long acquisition times. The acquired coincidence counts measured and used to form the images are spatiotemporally averaged over multiple breathing cycles [35]; consequently, for a point inside a mobile tumor, the signal is convoluted along its trajectory of motion. Respiratory-gated PET/CT aims to account for respiratory motion and thereby respiration-induced image blurring. One way to discern the effect of motion on feature values is by comparing image feature values between conventional and RG acquisition protocols, although other important factors stemming from the differences in the imaging protocols such as image noise may also be at play. Thus far, only one study accounting for motion in PET images has been reported by Yip et al. [36], which was limited to only five features. This report represents the first study that evaluates how 3D and RG acquisitions affect a large number of image features currently being used and tested in several medical applications. Robust features that emerge from this study may be suitable candidates for future quantitative imaging applications involving mobile tumors.
Materials and Methods
Preliminaries
Twenty-three lung cancer patients were retrospectively selected for a study of image feature variation between 3D and RG [RG is alternatively known as four-dimensional (4D)] PET/CT images and feature value variation among the 10 phases of an RG scan. The main selection criterion was for these patients to have both 3D and RG PET/CT scans performed during the same imaging session. The 3D images were acquired in free-breathing conditions for patients with regular breathing as required for radiotherapy planning. There were 13 female and 10 male patients ranging from age 47 to 83. All lung cancer patients were diagnosed with NSCLC. For each patient, 18F-FDG was the administered radiotracer, and the RG PET scan was acquired during the same session as the 3D PET scan in the same position per protocol for radiation treatment planning. A radiologist-approved 4D PET/CT protocol was applied for image acquisition [37]. The 3D PET/CT protocol was adapted for our institution from the Netherlands Protocol [38]. In routine clinical practice following these protocols, the average scan start times after the tracer administration were 118 ± 17.3 minutes (standard deviation) for 3D PET and 117 ± 36.0 minutes for RG PET, with average administered activity of 11.9 ± 2.0 mCi. A study on SUV variance in clinical FDG-PET/CT found that SUVmax and SUVmean were independent of variations in the uptake period [7].
PET/CT data were obtained using a GE Discovery STE PET/CT Scanner (for 21 cases) and a GE Discovery 600 PET/CT Scanner (for 2 cases). The 3D CT was a standard step and shoot CT (not helical), and the RG PET counts were binned into 10 phases with 3D CT attenuation correction applied to 4D PET data (standard protocol at our institution). The standard reconstruction protocol was the ordered subset expectation maximization algorithm with 20 or 28 subsets and 2 iterations. Full width at half maximum and field of view varied between patients: full width at half maximum of 4.29, 7, or 10 mm, and field of view of 50, 60, or 70 cm were standard PET settings. Standard of practice procedures at our institution were followed, and this study was approved with waived informed consent by the University of South Florida Institutional Review Board.
ROI Delineation and Tumor Segmentation
Following patient selection, the tumor region was identified using an advanced image viewing software (Mirada RTx, Mirada Medical, Oxford, UK), allowing identification of the primary tumor location and exportation of Digital Imaging and Communications in Medicine image data for 3D and RG PET/CT images. The image viewing software provided tumor visualization; easy access to X, Y, or Z slices of 3D PET data; and region-of-interest (ROI) delineation. A background-adapted thresholding method of segmentation defined by Dholakia et al., which accounted for background uptake, was applied to eliminate subjective errors and interobserver variability [39]. This method involved placing a 3-cm spherical contour inside the liver and extracting the mean SUV and standard deviation to calculate a threshold value for the lung tumor [Equation (1)]:
| (1) |
where SUVmean is the mean SUV value of the 3-cm sphere and SUVSD is the standard deviation of the 3-cm sphere's SUV values. We are aware of the many segmentation methods in the literature and that no one method is generally regarded optimal for general medical applications [40]. Because we were working with lung tumors, major structures and surrounding tissues were minimal. Consequently, there was little uptake outside the tumor volume. If, however, a tumor was close to the diaphragm or pleura, nearby metabolic structures were also segmented. Because of the adaptive segmentation method, six PET lesions were rendered too tiny and were not evaluated.
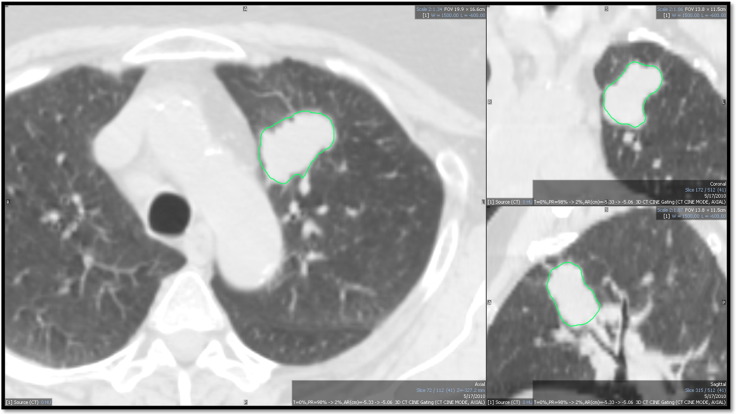
In CT images, tumors were contoured with CT threshold, a proprietary algorithm using Mirada RTx (RTx, Mirada Medical, Oxford, UK) (Figure 1). The 3D contours were drawn separately on the 3D CT image and on one phase (phase 1 or phase 10) of the corresponding RG CT image for each patient. In our clinic, CT contours are used for treatment planning purposes, whereas PET ensures that the entire metabolic tumor volume is included in the GTV.
Figure 1.
CT segmentation of one patient viewed in two dimensions (the ROI extends in 3D). This CT image is viewed in the window preset for the lung in Mirada DBx (RTx, Mirada Medical, Oxford, UK).
Feature Extraction
An internally developed application imported the ROI data file and extracted image intensity statistics, shape descriptors, co-occurrence matrices, run length matrices, and other second-order features from each ROI for a total of 56 image features (Tables 1 and 2). Although some authors have shown the instability of certain features from 3D images [25], [26], [31], we decided to include them here to analyze their stability for RG images. Moreover, some groups have used a large number of features (> 200 [21]). Nevertheless, we deemed 56 features sufficient to assess the variability between 3D and RG feature values.
Table 1.
Intensity and First-Order Image Features
| Feature | Description |
|---|---|
| Imin | Minimum intensity value in the 3D ROI |
| Imax | Maximum intensity value in the ROI |
| Imean | Mean intensity value in the ROI |
| SD | Variation from the average intensity in the ROI |
| Skewness | Measure of the symmetry of the intensity distribution |
| Kurtosis | Measure of the shape of the peak of the intensity distribution |
| Coeff Var | Normalized measure of the dispersion of the ROI (coefficient of variation) |
| TGV | Total summed intensity |
| I30⁎ | Intensity ranging from lowest to 30% highest intensity volume |
| I10-I90⁎ | Intensity ranging from lowest to 10% highest intensity volume minus intensity ranging from lowest to 90% highest intensity volume |
| V40⁎ | Percentage volume with at least 40% intensity |
| V70⁎ | Percentage volume with at least 70% intensity |
| V80⁎ | Percentage volume with at least 80% intensity |
| V10-V90⁎ | Percentage volume with at least 10% intensity minus percentage volume with at least 90% intensity |
| Sphericity | Measure of the spherical shape (roundness) of the ROI |
| Convexity | Measure of the spiculation of the ROI (ratio of true ROI volume to convex ROI volume) |
Denotes features that were derived from intensity volume histogram [10].
Table 2.
Selected Second-Order Features
| Name | Description | Mathematical Description |
|---|---|---|
| Energy⁎ | Also defined as Angular Second Moment. This feature describes the homogeneity of an image. 0 represents complete heterogeneity. 1 represents complete homogeneity [42]. | |
| Local homogeneity⁎ | Measures the relation of GLCM intensities to the diagonal GLCM matrix. A value of 1 represents total homogeneity. A value of 0 represents nonhomogeneity [42]. | |
| Entropy⁎ | Measures the pair contributions and information content. | |
| Correlation⁎ | Measures correlation between co-occurrence matrix values. | |
| SRE† | Measures short run distribution (short run emphasis). | |
| LRE† | Measures long run distribution (long run emphasis). | |
| RPC† | Ratio of total number of runs to total number of pixels in the image. Measures homogeneity and run distribution (run percentage). | |
| LGRE† | Measures low gray-level distribution (low gray-level run emphasis). | |
| SRLGE† | Measures short runs and low gray-level distribution (short run low gray-level emphasis). | |
| LRLGE† | Measures long runs and low gray-level distribution (long run low gray-level emphasis). | |
| RLNU† | Measures the nonuniformity of the run lengths (run length nonuniformity). | |
| GLNU† | Measures the nonuniformity of the gray levels (gray-level nonuniformity). |
Shape descriptors were calculated directly from the segmented ROI. First-order features (extracted from image intensity statistics) were calculated from volume intensity histograms. Second-order gray-level co-occurrence matrix (GLCM) features, originally described by Haralick et al. [14], [41], were implemented with feature descriptions provided by Liang [42]. The Haralick definition of second-order statistics (based on gray-level matrix metric), nearest neighbor spatial dependence matrices, provided texture information from the spatial relationship of image voxels [14].
The GLCM feature calculations were implemented as follows: the intensities of image voxels were binned into 256 gray-scale levels for PET (128 gray-scale levels for CT) with equal intervals. The resulting two-dimensional co-occurrence matrix was 256 × 256 (128 × 128) with unit (1) pixel distance. Co-occurrence matrices were calculated in 13 directions across a 3D image, and the resultant matrix was the average of the matrices in the 13 directions. Given a set of cubical voxels, the 13 directions were 3 axial directions, 2 diagonal directions per axial plane × 3 axial planes, and 4 diagonal directions cross cube [43]. These 13 directions were chosen so that the resulting matrix would represent the entire tumor texture without bias. The elements of the matrix were integers. Next, a probability matrix was calculated by dividing each element by the total sum of the matrix so that the sum of the probability matrix was 1. The features were then calculated using the probability matrix.
Galloway’s original run length features were also implemented [44]. Feature definitions were acquired from Galloway, Chu et al., and Dasarathy and Holder [44], [45], [46]. The run length matrix (RLM) had dimensions of L × R, where L was the number of gray-scale levels (256 for PET; 128 for CT) and R was the possible runs (determined case by case). The elements of the matrix were integers which represented runs. A run was defined as a set of pixels that possessed the same gray level in a specified direction [47]. The RLM was calculated in 13 directions across an image (similar to the co-occurrence matrix) [48]. The feature values were the summed values of all 13 directions normalized by the total runs in each direction. No probability matrix was involved for the run length features.
In PET, the image intensity was the number of registered counts per voxel. For CT, intensity represented the Hounsfield units in each voxel. All intensity levels were used. Normalization was applied only to the co-occurrence and run length matrices (in the form of binning; 128 bins). In addition, intensity values for PET images were not converted to SUV. Instead, stored image intensity values were analyzed directly. For each patient, image features were extracted from the 3D PET ROI, 3D CT ROI, all phases (bins) of the RG PET ROI, and one phase (bin) of the RG CT ROI.
Following feature extraction, 3D and RG PET/CT image feature differences were calculated using Equation (2):
| (2) |
where RGTVi is the RG image feature value for feature i and 3DTVi is the 3D image feature value for feature i. This method was chosen because it accounts for features that changed sign between 3D and RG cases. The maximum possible percent difference using Equation (2) is 200%, and differences greater than 100% were deemed large. The percent difference across cases was then averaged for each image feature and a paired, two-tailed t test was applied to 3D and RG feature data to compare the two data sets. We assumed normal distributions and that the t test was applicable. The concordance correlation coefficient (CCC) was calculated for all features across 3D and RG feature data to determine the correlation between the two data sets. The scale used for determining strength of agreement was as follows: high strength of agreement, CCC > 0.99; substantial strength of agreement, CCC: 0.95 to 0.99; moderate strength of agreement, CCC: 0.90 to 0.95; poor strength of agreement, CCC < 0.90 [49].
RG (4D) PET Phase Analysis
The previously described procedure of image feature extraction was applied to all RG PET bins. Mean percent difference was used to compare features between phases:
| (3) |
where i represents the bin, j represents the specific image feature, TVij represents the value for bin i and feature j, and μTV,j represents the mean value for image feature j. Image feature values were also normalized by average value across all bins:
| (4) |
The subscript definitions for Equation (3) also apply to Equation (4). In addition, a paired, two-tailed t test was applied to RG inhale (phase 1) feature data and RG exhale (phase 5) feature data to compare the two data sets. The CCC was calculated for phase 1 and phase 5 of the feature data to determine correlation between the two data sets.
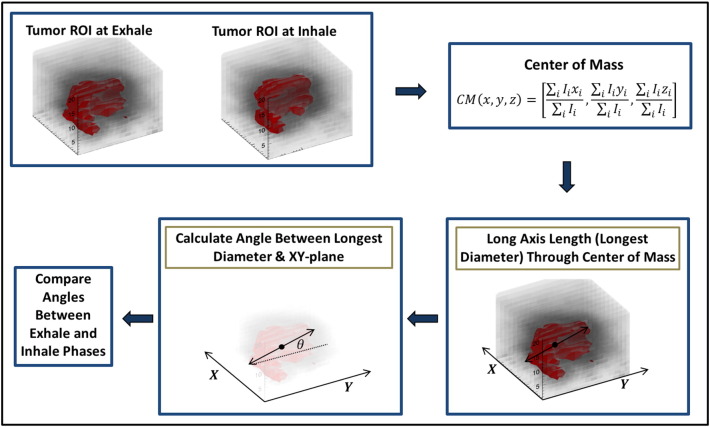
Long Axis Calculation and Rotation Analysis
The long axis length (through the center of mass) was calculated with an internally developed program for each bin of the RG cycle (PET only). The tumor’s center of mass location was calculated for the inhale and exhale phases of the RG PET image sets [Equation (5)].
| (5) |
where CM (x, y, z) is the center of mass for a tumor in a PET phase and Ii is the number of counts per voxel i. The center of mass motion (CMM) was calculated as the displacement between the center of mass for inhale phase and center of mass for exhale phase. The difference in long axis length and CMM were used to assess changes in internal tumor morphology. Tumor angle was defined as the angle between the long axis of the tumor and the XY plane (Figure 2). A Pearson’s correlation test was applied to identify correlation in tumor angle and long axis length between inhale and exhale images.
Figure 2.
Tumor rotation calculation method. First, the tumor volume is delineated at exhale (phase 1) and inhale (phase 5) on RG PET images. Second, the center of mass of each volume is calculated. The long axis length (longest diameter) through the center of mass of the tumor is calculated. Then, the angle between the long axis length and the XY plane is calculated. This angle is compared between the exhale (phase 1) and inhale (phase 2) to determine the pseudotumor rotation.
Results
3D and RG PET/CT Image Feature Analysis
Features from both PET and CT images demonstrated dependency on whether the acquisition was 3D, which is conventional (also called static), or RG (4D), where the coincidence counts are binned in multiple phases/bins composing the respiratory cycle. Large differences in some features were found between 3D PET/CT and one of the phases/bins of the corresponding RG data set. The percent differences between 3D and RG modalities were usually larger for CT than for PET. For PET, 10 of 56 features had a percentage difference (between 3D PET and RG PET for each patient) of less than 5% for more than half of the cases. In comparison, 11 of 56 CT features had a percentage difference (between 3D CT and RG CT for each patient) of less than 5% for more than half the cases. The percent differences between 3D PET and RG PET varied from 0% to 193%. The outlier of 193% was kurtosis. For 4 of 17 cases, kurtosis demonstrated the greatest percent difference between 3D and RG PET. Image feature average differences between 3D PET and RG PET are shown in Table 3.
Table 3.
Features Presenting Average Differences between 3D and RG PET Image Features
| < 2% Difference | < 5% Difference | < 10% Difference | < 15% Difference | < 20% Difference | > 50% Difference |
|---|---|---|---|---|---|
| SRE | Sphericity | Surface area/volume | Volume | V10-V90 | Minimum intensity |
| Spherical disproportion | Compactness | Surface area | Contrast (1st order) | Mean intensity | |
| Entropy (1st order) | Convexity | Long axis | Co-occurrence mean | Kurtosis | |
| Information measure of correlation 2 | Entropy (2nd order) | Short axis | Sum average | TGV | |
| RPC | Sum entropy | Local homogeneity (1st order) | Information measure of correlation 1 | RMS | |
| Difference entropy | Difference average | I30 | |||
| Difference variance | I10-I90 | ||||
| LRLGE |
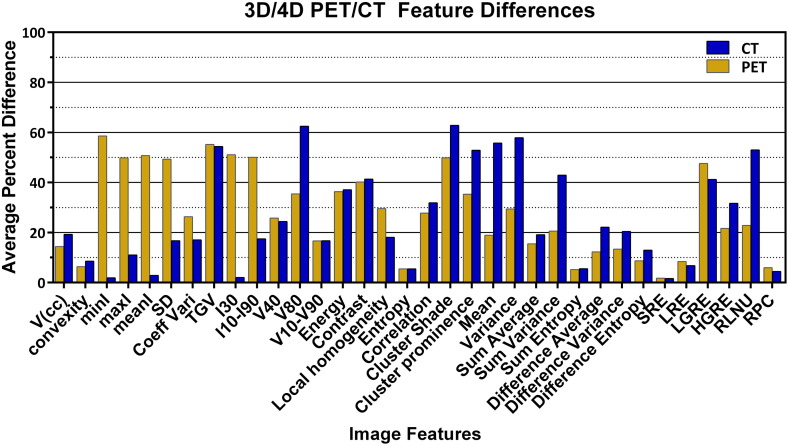
Percent differences between 3D CT and RG CT varied from 0% to 176%, kurtosis again being the outlier. Figure 3 shows selected feature percent differences, and Table 4 shows image feature average differences between 3D and RG CT. Forty-six percent of the CT features between 3D CT and RG CT presented average percent differences larger than 20%. In some cases, average percent differences were larger than 50%. Table 5 displays the number and percent of total features with specific percent differences for CT, PET, and PET RG phases.
Figure 3.
Average differences between 3D and RG image features. % Diffi3D/RG between selected image features from 3D PET/CT and RG PET/CT.
Table 4.
Features Presenting Average Differences between 3D and RG CT Image Features
| < 2% Difference | < 5% Difference | < 10% Difference | < 15% Difference | < 20% Difference | > 50% Difference |
|---|---|---|---|---|---|
| Minimum intensity | Mean intensity | Convexity | Surface area/volume | Volume | Kurtosis |
| SRE | RMS | LRE | Sphericity | SD | TGV |
| I30 | Compactness | Coefficient of variation | V70 | ||
| RPC | Spherical disproportion | I10-I90 | V80 | ||
| Difference entropy | Local homogeneity (2nd order) | Energy (1st order) | |||
| Sum average | Cluster shade | ||||
| Cluster prominence | |||||
| Co-occurrence mean | |||||
| Co-occurrence variance | |||||
| GLNU | |||||
| RLNU |
Table 5.
Percent Differences (% Diffi3D/RG) between Image Features of 3D and RG, PET and CT Images, and Conglomerate Image Features of RG PET Phases for All Cases (% DiffijMean)
|
CT |
PET |
PET RG Phases |
||||
|---|---|---|---|---|---|---|
| Percent Difference | No. of Features⁎(1288 Total) | % Total Features⁎ | No. of Features⁎(952 Total) | % Total Features⁎ | No. of Features⁎(9464 Total) | % of Total Features⁎ |
| < 5% | 342 | 26.6% | 249 | 26.2% | 5051 | 53.4% |
| < 10% | 498 | 38.7% | 405 | 42.5% | 7258 | 76.7% |
| < 15% | 617 | 47.9% | 515 | 54.1% | 8043 | 85.0% |
| < 20% | 697 | 54.1% | 585 | 61.4% | 8410 | 88.9% |
| > 20% | 591 | 45.9% | 367 | 38.6% | 998 | 10.5% |
Total number of features refers to 56 image features per tumor.
Overall, 249 of 952 (26.2%) of all PET features (56 features per patient) had a percent difference of less than 5% between 3D and RG protocols, whereas 342 of 1288 (26.6%) of all CT features (56 features per patient) had a percent difference of less than 5% between 3D and RG scans. Table 6 shows common features with percent differences between 3D and RG protocols for all cases and for both PET and CT modalities.
Table 6.
Image Features with Common Average Differences in 3D/RG PET and CT
| Percent Difference | Common Features |
|---|---|
| < 2% | SRE |
| < 5% | – |
| < 10% | Convexity, 1st and 2nd order entropy, sum entropy, LRE, RPC |
| < 15% | Surface area/volume, sphericity, compactness, spherical disproportion, difference entropy, information measure of correlation 2 |
| < 20% | Volume, long axis length,V10-V90, sum average |
| > 50% | Kurtosis, TGV |
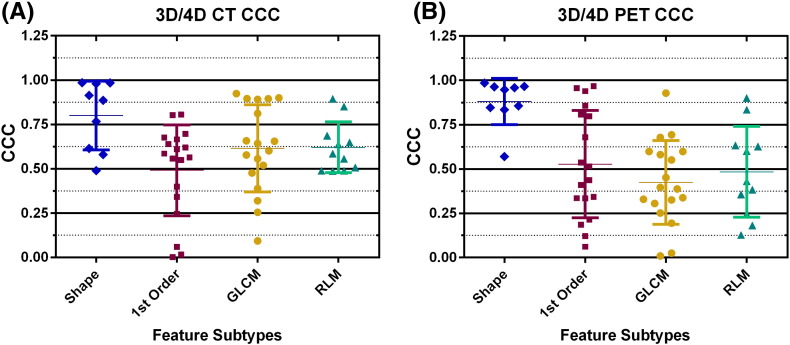
According to the CCC strength-of-agreement scale by Mcbride et al., PET and CT feature subtypes demonstrated poor correlation between 3D and RG images [49] (Figure 4). This was demonstrated by CCC strength-of-agreement values less than 0.90 for each feature subtype (shape, first order, GLCM, and RLM). However, there were specific features that demonstrated substantial strength of agreement. These were from the shape and first-order features in PET and shape features only in CT.
Figure 4.
Concordance correlation coefficients for each feature with mean and standard deviation for each feature subtype for (A) 3D/4D CT and (B) 3D/4D PET.
The paired, two-tailed, t test for 3D PET and RG PET features revealed 17 PET features with P values < .05 (indicating that these data sets are different). The t test for 3D CT and RG CT features revealed 12 CT features with P values < .05. Features with P values < .05 for both PET and CT were entropy (first order), compactness, and information measure of correlation 1 and 2.
RG (4D) PET Phase Analysis
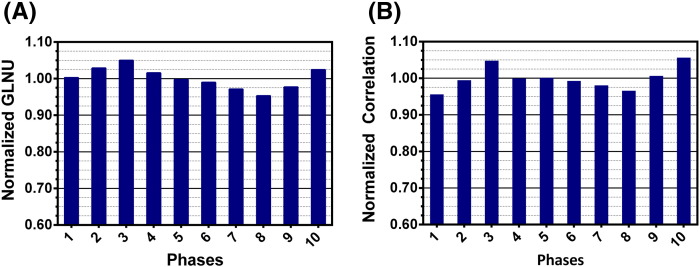
Results indicated a weak dependency (relative to the differences between 3D and RG presented above) of all PET features on respiration phase in RG scans of 10 phases (Figure 5). The most robust features (less than 5% difference among RG phases) belonged to select features from all categories (shape descriptors, and first- and second-order features). Sphericity, spherical disproportion, information measure of correlation 2, SRE, and LRE were within 10% difference of the average value for all cases across all phases. Normalized image features across 10 phases for RG PET demonstrated that, for all patients, 77% (7258:9464) of image features (56 features per phase per patient) varied less than 10% from the average values and 10.5% (998:9464) demonstrated more than 20% difference from average values (Table 3). Features with the largest difference (> 50%) were kurtosis, Low Gray-level Run Emphasis (LGRE), Short Run Low Gray-level Emphasis (SRLGE), and Long Run Low Gray-level Emphasis (LRLGE). The paired, two-tailed, t test for RG PET inhale and RG PET exhale feature data revealed one PET feature, namely, short axis length, with P value < .05. The CCC revealed that the shape features had the highest CCC strength of agreement between image data sets from phase 1 and phase 5 (mean CCC strength of agreement, 0.95; moderate). First-order features and GLCM had mean strength-of-agreement values of 0.93 (moderate), and RLM features exhibited mean CCC strength of agreement of 0.86 (poor).
Figure 5.
Feature dependency on respiration phase for selected features. (A) Normalized GLNU across 10 phases of RG PET image sets. (B) Normalized correlation across 10 phases of a RG PET image set.
Overall Feature Results
In comparisons of results among respiratory-phases and 3D-to-RG PET features, we concluded that the features with the least variability overall for PET images were sphericity, spherical disproportion, first-order entropy, information measure of correlation 2, and SRE. Features demonstrating the greatest variability were kurtosis and LRLGE. For CT images, features with the least variability were minimum intensity, mean intensity, RMS, SRE, and RPC, whereas features with the greatest variability were kurtosis, V70, V80, energy (first order), cluster shade, cluster prominence, co-occurrence mean, co-occurrence variance, Gray-level Nonuniformity (GLNU), and Run Length Nonuniformity (RLNU).
Long Axis Tumor Length, Rotation, and CMM
The long axis tumor length and rotation results demonstrated that tumors exhibited deformation over RG phases (Table 7). A Pearson’s correlation test demonstrated that there were a weak correlation between the tumor angle with respect to the XY plane at inhale and the same angle in the corresponding 3D image (R = 0.350) and a weak correlation between the tumor angle at the exhale phase and the corresponding 3D image (R = 0.319). There was a weak correlation between 3D image tumor volume and the 3D image tumor angle (R = − 0.399), and long axis length was not correlated to the breathing cycle. Table 7 shows that the long axis length of the tumor was inconsistent across inhalation phase (phase 1), 3D scan, and exhalation phase (phase 5). The long axis lengths of the tumor for 3D, phase 1, and phase 5 were highly correlated (3D and phase 1: R = 0.936; 3D and phase 5: R = 0.954; phase 1 and phase 5: R = 0.986); long axis lengths between phase 1 and phase 5 differed by more than 5 mm in some cases, indicating possible changes in tumor shape during the respiratory cycle. The largest difference was case 11 with long axis lengths of 124.5 mm and 139.9 mm for phase 1 and phase 5, respectively, whereas the long axis length for 3D was 147.4 mm. There was a weak to moderate correlation between tumor angle at the inhale phase and the exhale phase (R = 0.438), indicating tumor rotation during the respiratory cycle. Moreover, the long axis angle also changed in some cases from positive to negative, indicating tumor rotation. There was also a weak to moderate negative correlation between average percent difference in 3D and RG images (in PET) and center of mass motion (R = − 0.445).
Table 7.
Long Axis Lengths of Lung Tumors on 3D PET Images and RG PET Images at Exhale and Inhale
| Case |
Length (mm) |
Angle (Relative to XY Axis) |
Volume (cc) |
CMM (mm) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3D | Exhale | Inhale | 3D | Exhale | Inhale | 3D | Exhale | Inhale | ||
| 1 | 31.58 | 46.29 | 35.65 | 18.10 | − 25.08 | − 39.95 | 12.57 | 12.79 | 12.36 | 3.70 |
| 2 | 67.19 | 69.78 | 66.97 | − 25.98 | − 48.58 | − 22.99 | 45.77 | 40.45 | 40.02 | 4.21 |
| 3 | 62.73 | 76.31 | 71.78 | 38.72 | 0.00 | − 43.10 | 82.56 | 81.77 | 77.24 | 6.22 |
| 4 | 24.67 | 25.85 | 24.92 | 7.62 | − 14.65 | − 7.54 | 4.69 | 4.44 | 4.14 | 1.87 |
| 5 | 41.00 | 41.98 | 40.38 | − 13.84 | 22.92 | 34.53 | 23.47 | 24.94 | 21.81 | 13.30 |
| 6 | 49.21 | 44.18 | 43.71 | − 36.73 | 31.20 | − 8.61 | 30.41 | 30.18 | 29.75 | 2.99 |
| 7 | 125.95 | 133.22 | 126.72 | − 65.32 | − 68.87 | − 61.33 | 140.78 | 119.62 | 113.54 | 3.35 |
| 8 | 55.03 | 46.76 | 46.66 | − 40.82 | − 24.81 | − 57.25 | 24.84 | 20.84 | 19.90 | 1.29 |
| 9 | 28.11 | 21.67 | 21.96 | 13.45 | 26.91 | − 17.33 | 6.45 | 4.38 | 4.31 | 0.28 |
| 10 | 29.05 | 24.78 | 21.96 | 19.74 | − 15.30 | 17.33 | 6.85 | 4.67 | 4.74 | 1.71 |
| 11 | 147.42 | 124.53 | 139.92 | − 36.79 | 6.03 | − 37.42 | 571.04 | 419.89 | 427.80 | 0.16 |
| 12 | 63.53 | 68.11 | 63.27 | 27.60 | 22.59 | 14.98 | 64.35 | 58.92 | 53.38 | 4.08 |
| 13 | 46.96 | 48.25 | 55.16 | 16.17 | − 7.79 | − 62.77 | 26.21 | 24.21 | 28.17 | 2.05 |
| 14 | 54.12 | 54.02 | 54.02 | 17.58 | 14.01 | 14.01 | 33.25 | 33.34 | 32.91 | 0.49 |
| 15 | 10.94 | 47.98 | 47.98 | 0.00 | − 15.82 | − 15.82 | 35.40 | 27.38 | 30.46 | 0.29 |
| 16 | 24.59 | 20.40 | 19.86 | 23.51 | − 18.70 | − 19.23 | 6.75 | 3.81 | 2.80 | 4.60 |
| 17 | 41.55 | 47.61 | 39.99 | 18.35 | − 33.33 | 19.09 | 26.01 | 22.70 | 22.99 | 2.84 |
Discussion
RG PET scans can provide a “snapshot” of the tumor within a phase along the breathing cycle, thereby greatly reducing the effects of motion on a tumor’s shape, volume, and image feature values. In contrast, 3D PET scans convolute the absorbed activity distribution over the motion/deformation pattern a tumor and its surroundings experience during multiple respiration cycles [50]. Consequently, a 3D (static) PET may fail to provide accurate position, volume, and absorbed activity distribution for a mobile tumor. This is especially true in the thoracic region and regions with substantial internal motion. This agrees with Adams’ finding that respiratory motion affects the SUV with changes up to 30% and that any moving lesion would be inaccurately measured because of the effects of blurring [51]. In addition, patient motion/breathing is known to cause image artifacts due to a mismatch in registration between CT attenuation correction and emission scans [38]. Internal motion, as the results support, notably affected the image feature values of PET and CT images. The percent differences between 3D and RG CT were generally greater than those in PET. CT images have higher spatial resolution than PET images and therefore more voxels for texture formation and thus a greater sensitivity to motion. In addition, 3D CT may also be affected by motion depending on the acquisition protocol [52].
In addition to the affine tumor motion caused by respiration, we identified deformation of tumors (characterized by varying tumor axis lengths and angles with respect to the XY plane between 3D PET, RG PET at inhale, and RG PET at exhale). Conceivably, rotations and deformations also affect image feature values. Our results demonstrated a weak correlation between the long axis angles of RG images at inhalation and exhalation. There was also an inconsistency of long axis length between 3D images, RG images at inhale, and RG images at exhale, thus indicating that tumor shape and rotation varied between phases. The degree to which rotations and/or deformations affect image features, and in particular texture values, requires further investigation. Interestingly, there was no correlation between CMM, tumor volume, or long axis length with 3D/RG feature value differences based on Pearson’s correlation tests. There was, however, a weak to moderate correlation between CMM and average percent difference. Nonetheless, it is clear from our data that the feature value differences between RG phases are smaller than the differences between 3D images and RG images at a given phase. In other words, the rotational motion and/or deformation of the tumors in our patient cohort had a smaller effect on image feature values than the averaging effects of the static acquisition. Yip et al. also investigated variability of texture features between 3D and RG imaging [36]. In contrast to our study, they tested only five image features (contrast, busyness, coarseness, maximal correlation coefficient, and long run low gray). They found that differences between 3D and RG PET were significant [36] after having accounted for noise differences due to different acquisition times. This agrees with our findings that certain features (e.g., kurtosis and LRLGE) demonstrated large variability between 3D and RG protocols. There were, however, certain features in our study (e.g., SRE, first-order entropy, and RPC) that did not demonstrate large variability between protocols.
Figure 3 and Tables 3 and 5 show differences between feature values from 3D and RG protocols. The features with the smallest change across PET for all RG bins and for 3D PET were sphericity, spherical disproportion, entropy (first and second order), sum entropy, information measure of correlation 2, SRE, LRE, and RPC. Interestingly, a study by Galavis et al. on the variability of PET texture features caused by different acquisition modes and reconstruction parameters demonstrated that first-order entropy exhibited small variation (≤ 5%), whereas second-order entropy and sum entropy exhibited intermediate variability (10%-25%) [31]. Our results were comparable, showing that first-order entropy exhibited variation smaller than 5% and that second-order entropy and sum entropy exhibited less than 10% difference between 3D and RG PET protocols. Sum entropy, second-order entropy, and the information measure of correlation 2 are based on entropy calculations which measure randomness in a pattern. A portion of the randomness can be attributed to the noise intrinsic to the scanner, whereas the remaining can be attributed to statistical differences in counts (quantum noise). Hence, 3D images are less noisy than RG images because percentage image noise is given by , where N is the count density (counts/cm2). Thus, 3D/RG feature differences are a combination of both tumor motion and count statistics. This suggests that it would be informative to normalize for count density. Unfortunately, this study was retrospective and list-mode data were not accessible for normalization. Nevertheless, the number of counts and therefore the noise among RG images from the 10 phases can be assumed to be similar. Therefore, the differences in feature values from phase to phase may be attributed to the effect of motion and/or deformation.
The features LRE, SRE, and RPC, which demonstrated small change across PET for all RG bins and 3D PET, are features from the RLM. LRE measures the long run emphasis distribution. Correspondingly, SRE measures the short run emphasis distribution. Run percentage is the ratio of the number of runs to the number of pixels in an image (Table 2). We conclude that the cumulative number and length of short runs and cumulative number and length of long runs do not vary significantly between 3D and RG images and that the total number of runs does not vary significantly between 3D and RG images. These conclusions may depend on the algorithms used to calculate these features. For example, in this paper, we averaged runs from 13 directions; other definitions are possible.
Feature differences between 3D and RG in PET and CT images that showed large differences (> 50%) were typically features from intensity volume histograms such as kurtosis and TGV. Thus, the intensity histogram distributions between 3D and RG features were quite different in terms of symmetry about their means and the degrees of “peakness” of their distributions. Cluster shade and cluster prominence exhibited large differences in CT. These features measure the skewness of the GLCM [53]. According to Ion, a high cluster shade value reveals an asymmetric image [53].
Overall, it is clear that image feature values are different between 3D and RG images. As discussed above, this is due to both the smearing effects of tumor motion, both affine and nonaffine, and noise intrinsic to image acquisition, with the former apparently having a larger effect [36]. This is also supported by the relative variation in feature values from different phases of the RG scans even though the tumor VOIs varied from phase to phase because of motion and deformation. Thus, the motion convoluted into the 3D images seems to have a greater effect on feature values than noise given that the RG images are intrinsically noisier because of lower counts (acquisition times). This study suggests that it would be important to account for motion in quantitative image feature analysis, regardless of modality (PET or CT), as attempted by other investigators [23]. Alternatively, if the definition of any one feature includes details of the acquisition protocol, then 3D and RG features may be treated as “different” sets of features. Further studies are needed to elucidate the potential usefulness of this alternative definition.
Limitations
Though our results clearly demonstrated that image feature values were different between 3D and RG protocols, there were limitations to the study. First of all, we were unable to normalize for count density between 3D and RG protocols. Another limitation was the nonconformity of the uptake time with the protocol. This was mainly due to clinical logistics. Also, partial volume effects were not taken into account. Because 3D and RG data on same patient were acquired on the same scanner and hence partial volume effects were similar in both sets of images except for the effect of motion, we did not take these affects into account. In addition, binning artifacts and breathing irregularities were assumed negligible because only patients with regular breathing patterns are candidates for RG PET for radiotherapy in our institution [54]. Another limitation was that 4D PET received 3D CT attenuation correction. This is currently standard procedure at our institution. Lastly, our patient size was limited but comparable to other published studies [22], [23], [36]. We plan to address these limitations in future studies.
Conclusions
This study investigated the variation of image features between 3D and RG PET/CT images of lung tumors. To our knowledge, this is the first study that evaluates how 3D and RG acquisitions affect a large number of image features currently being used and tested in several medical applications. The data showed that image feature analysis using a static acquisition (3D) versus an RG acquisition (to account for motion of the ROI) revealed notably different feature values. The results support that these differences are mainly due to the effect that respiratory motion has on image features. We have also concluded that rotational motion and deformation of the tumor also affect the features of an image. However, the effect of rotational motion and deformation from phase to phase appears to be smaller than the averaging/smearing effects of static acquisition. In sum, this study calls attention to the differences in 3D and RG image feature values for mobile tumors. The predictive and/or prognostic power of RG versus 3D image feature values will be explored in future studies.
Acknowledgements
The late Caroline C. Kuykendall was the lead PET technologist during this study. She is acknowledged as a team member for time and expertise devoted to this study. Jongphil Kim is acknowledged for his contributions in the statistical analysis of this study.
References
- 1.Lardinois D, Weder W, Hany TF, Kamel EM, Korom S, Seifert B, von Schulthess GK, Steinert HC. Staging of non-small-cell lung cancer with integrated positron-emission tomography and computed tomography. N Engl J Med. 2003;348(25):2500–2507. doi: 10.1056/NEJMoa022136. [PubMed PMID: 12815135] [DOI] [PubMed] [Google Scholar]
- 2.Nestle U, Kremp S, Schaefer-Schuler A, Sebastian-Welsch C, Hellwig D, Rube C, Kirsch CM. Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-Small cell lung cancer. J Nucl Med. 2005;46(8):1342–1348. [PubMed PMID: 16085592] [PubMed] [Google Scholar]
- 3.Erdi YE, Macapinlac H, Rosenzweig KE, Humm JL, Larson SM, Erdi AK, Yorke ED. Use of PET to monitor the response of lung cancer to radiation treatment. Eur J Nucl Med. 2000;27(7):861–866. doi: 10.1007/s002590000258. [DOI] [PubMed] [Google Scholar]
- 4.Brunetti J. Clinical PET-CT in Radiology: Integrated Imaging in Oncology. Springer; New York: 2011. PET-CT in Radiation Treatment Planning; pp. 121–129. [Internet] [Google Scholar]
- 5.Bradley J, Thorstad WL, Mutic S, Miller TR, Dehdashti F, Siegel BA, Bosch W, Bertrand RJ. Impact of FDG-PET on radiation therapy volume delineation in non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2004;59(1):78–86. doi: 10.1016/j.ijrobp.2003.10.044. [PubMed PMID: 15093902] [DOI] [PubMed] [Google Scholar]
- 6.Osman MM, Wall A, Miller MD, Nguyen N, Oliver DA, Bucholz RD, Walz BJ. Robotic Radiosurgery Treating Tumors that Move with Respiration. Springer; 2007. 18FDG-PET/CT for Treatment Planning and Follow-Up; pp. 55–60. [Internet] [Google Scholar]
- 7.Kumar V, Nath K, Berman CG, Kim J, Tanvetyanon T, Chiappori AA, Gatenby RA, Gillies RJ, Eikman EA. Variance of SUVs for FDG-PET/CT is Greater in Clinical Practice Than Under Ideal Study Settings. Clin Nucl Med. 2013;38(3):175–182. doi: 10.1097/RLU.0b013e318279ffdf. [PubMed PMID: WOS:000315049200016. English] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nestle U, Kremp S, Grosu AL. Practical integration of [18F]-FDG-PET and PET-CT in the planning of radiotherapy for non-small cell lung cancer (NSCLC): the technical basis, ICRU-target volumes, problems, perspectives. Radiother Oncol. 2006;81(2):209–225. doi: 10.1016/j.radonc.2006.09.011. [PubMed PMID: 17064802] [DOI] [PubMed] [Google Scholar]
- 9.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40(1):133–140. doi: 10.1007/s00259-012-2247-0. [PubMed PMID: 23064544. Epub 2012/10/16. eng] [DOI] [PubMed] [Google Scholar]
- 10.El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, Chaudhari S, Yang D, Schmitt M, Laforest R. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recogn. 2009;42(6):1162–1171. doi: 10.1016/j.patcog.2008.08.011. [PubMed PMID: 20161266. Pubmed Central PMCID: PMC2701316. Epub 2010/02/18. Eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borst GR, Belderbos JS, Boellaard R, Comans EF, De Jaeger K, Lammertsma AA, Lebesque JV. Standardised FDG uptake: a prognostic factor for inoperable non-small cell lung cancer. Eur J Cancer. 2005;41(11):1533–1541. doi: 10.1016/j.ejca.2005.03.026. [PubMed PMID: 15953716. Epub 2005/06/15. eng] [DOI] [PubMed] [Google Scholar]
- 12.Fahey FH, Kinahan PE, Doot RK, Kocak M, Thurston H, Poussaint TY. Variability in PET quantitation within a multicenter consortium. Med Phys. 2010;37(7):3660–3666. doi: 10.1118/1.3455705. [PubMed PMID: WOS:000279845300019. English] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zucker SW, Terzopoulos D. Finding structure in Co-occurrence matrices for texture analysis. Comput Graph Image Process. 1980;12(3):286–308. [Google Scholar]
- 14.Haralick RM, Shanmugam K, Dinstein Ih. Textural Features for Image Classification. IEEE Trans Syst Man Cybern. 1973;SMC-3(6):610–621. [Google Scholar]
- 15.Aggarwal N. First and Second Order Statistics Features for Classification of Magnetic Resonance Brain Images. J Signal Inf Process. 2012;03(02):146–153. [Google Scholar]
- 16.Duncan JS, Ayache N. Medical image analysis: progress over two decades and the challenges ahead. IEEE Trans Pattern Anal Mach Intell. 2000;22(1):85–106. [Google Scholar]
- 17.Ganeshan B, Abaleke S, Young RC, Chatwin CR, Miles KA. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10:137–143. doi: 10.1102/1470-7330.2010.0021. [PubMed PMID: 20605762. Pubmed Central PMCID: PMC2904029. Epub 2010/07/08. eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol. 2012;22(4):796–802. doi: 10.1007/s00330-011-2319-8. [PubMed PMID: 22086561. Epub 2011/11/17. eng] [DOI] [PubMed] [Google Scholar]
- 19.Al-Kadi OS, Watson D. Texture analysis of aggressive and nonaggressive lung tumor CE CT images. IEEE Trans Biomed Eng. 2008;55(7):1822–1830. doi: 10.1109/TBME.2008.919735. [PubMed PMID: 18595800] [DOI] [PubMed] [Google Scholar]
- 20.Hunter LA, Krafft S, Stingo F, Choi H, Martel MK, Kry SF, Court LE. High quality machine-robust image features: identification in nonsmall cell lung cancer computed tomography images. Med Phys. 2013;40(12):121916. doi: 10.1118/1.4829514. [PubMed PMID: 24320527. Pubmed Central PMCID: PMC4108720] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [PubMed PMID: 22257792. Pubmed Central PMCID: PMC4533986. Epub 2012/01/20. eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willaime JM, Turkheimer FE, Kenny LM, Aboagye EO. Quantification of intra-tumour cell proliferation heterogeneity using imaging descriptors of 18F fluorothymidine-positron emission tomography. Phys Med Biol. 2013;58(2):187–203. doi: 10.1088/0031-9155/58/2/187. [PubMed PMID: 23257054. Epub 2012/12/22. eng] [DOI] [PubMed] [Google Scholar]
- 23.Vaidya M, Creach KM, Frye J, Dehdashti F, Bradley JD, El Naqa I. Combined PET/CT image characteristics for radiotherapy tumor response in lung cancer. Radiother Oncol. 2012;102(2):239–245. doi: 10.1016/j.radonc.2011.10.014. [PubMed PMID: 22098794. Epub 2011/11/22. eng] [DOI] [PubMed] [Google Scholar]
- 24.Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, Hoekstra CJ, Boellaard R, Dekker AL, Gillies RJ. Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52(7):1391–1397. doi: 10.3109/0284186X.2013.812798. [PubMed PMID: 24047337. Pubmed Central PMCID: PMC4533992] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tixier F, Le Rest CC, Hatt M, Albarghach N, Pradier O, Metges JP, Corcos L, Visvikis D. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52(3):369–378. doi: 10.2967/jnumed.110.082404. [PubMed PMID: 21321270. Pubmed Central PMCID: PMC3789272. Epub 2011/02/16. eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. 2012;53(5):693–700. doi: 10.2967/jnumed.111.099127. [PubMed PMID: 22454484. Pubmed Central PMCID: PMC3779464] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook GJ, Yip C, Siddique M, Goh V, Chicklore S, Roy A, Marsden P, Ahmad S, Landau D. Are pretreatment 18F-FDG PET tumor textural features in non-small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013;54(1):19–26. doi: 10.2967/jnumed.112.107375. [PubMed PMID: 23204495. Epub November 30, 2012] [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [PubMed PMID: 22898692. Pubmed Central PMCID: PMC3563280] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, Lambin P, Haibe-Kains B, Mak RH, Aerts HJ. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114(3):345–350. doi: 10.1016/j.radonc.2015.02.015. [PubMed PMID: 25746350. Pubmed Central PMCID: PMC4400248] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunliffe A, Armato SG, III, Castillo R, Pham N, Guerrero T, Al-Hallaq HA. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol Biol Phys. 2015;91(5):1048–1056. doi: 10.1016/j.ijrobp.2014.11.030. [PubMed PMID: 25670540. Pubmed Central PMCID: PMC4383676] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49(7):1012–1016. doi: 10.3109/0284186X.2010.498437. [PubMed PMID: 20831489. Pubmed Central PMCID: PMC4091820. Epub 2010/09/14. eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng NM, Fang YH, Yen TC. The promise and limits of PET texture analysis. Ann Nucl Med. 2013;27(9):867–869. doi: 10.1007/s12149-013-0759-8. [PubMed PMID: 23943197] [DOI] [PubMed] [Google Scholar]
- 33.Leijenaar RTH, Nalbantov G, Carvalho S, Elmpt WJCV, Troost EGC, Boellard R, Aerts HJWL, Gillies RJ, Lambin P. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5(11075) doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging. 2015;2(4) doi: 10.1117/1.JMI.2.4.041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicente AMG, Castrejon AS, Martin AAL, Garcia BG, Woll JPP, Munoz AP. Value of 4-Dimensional 18F-FDG PET/CT in the Classification of Pulmonary Lesions. J Nucl Med. 2011;39(2):91–99. doi: 10.2967/jnmt.110.082719. [DOI] [PubMed] [Google Scholar]
- 36.Yip S, McCall K, Aristophanous M, Chen AB, Aerts HJ, Berbeco R. Comparison of texture features derived from static and respiratory-gated PET images in non-small cell lung cancer. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0115510. [PubMed PMID: 25517987. Pubmed Central PMCID: PMC4269460] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuykendall CC, Budzevich MM, Latifi K, Moros EG, Hoffe SE, Dilling TJ, Zhang GG, Montilla-Soler JL, Eikman EA. 4D PET/CT: Radiology Imaging to Radiation Therapy. Pract Radiat Oncol. 2013;3(2 Suppl 1):S28. doi: 10.1016/j.prro.2013.01.096. [PubMed PMID: 24674534] [DOI] [PubMed] [Google Scholar]
- 38.Boellaard R, Oyen WJ, Hoekstra CJ, Hoekstra OS, Visser EP, Willemsen AT, Arends B, Verzijlbergen FJ, Zijlstra J, Paans AM. The Netherlands protocol for standardisation and quantification of FDG whole body PET studies in multi-centre trials. Eur J Nucl Med Mol Imaging. 2008;35(12):2320–2333. doi: 10.1007/s00259-008-0874-2. [PubMed PMID: 18704407] [DOI] [PubMed] [Google Scholar]
- 39.Dholakia AS, Chaudhry M, Leal JP, Chang DT, Raman SP, Hacker-Prietz A, Su Z, Pai J, Oteiza KE, Griffith ME. Baseline metabolic tumor volume and total lesion glycolysis are associated with survival outcomes in patients with locally advanced pancreatic cancer receiving stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89(3):539–546. doi: 10.1016/j.ijrobp.2014.02.031. [PubMed PMID: 24751410. Pubmed Central PMCID: PMC4372085] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster B, Bagci U, Mansoor A, Xu Z, Mollura DJ. A review on segmentation of positron emission tomography images. Comput Biol Med. 2014;50:76–96. doi: 10.1016/j.compbiomed.2014.04.014. [PubMed PMID: 24845019. Pubmed Central PMCID: PMC4060809] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haralick RM. Statistical and Structural Approaches to Texture. Proc IEEE. 1979;67(5):786–803. [Google Scholar]
- 42.Liang M. 2012. 3D co-occurrence matrix based texture analysis applied to cervical cancer screening: Uppsala Universitet. [Google Scholar]
- 43.Kurani AS, Xu D-H, Furst JD, Raicu DS, editors. Co-occurrence matrices for volumetric data. CGIM; Kauai, Hawaii, USA: 2004. (The 7th IASTED International Conference on Computer Graphics and Imaging). [Google Scholar]
- 44.Galloway MM. Texture Analysis Using Gray Level Run Lengths. Comput Graph Image Process. 1975;4:172–179. [Google Scholar]
- 45.Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recogn Lett. 1990;11(6):415–420. [Google Scholar]
- 46.Dasarathy BV, Holder EB. Image characterizations based on joint gray-level run-length distributions. Pattern Recogn Lett. 1991;12(8):497–502. [Google Scholar]
- 47.Tustison NJ, Gee JC. 2008. Run Length Matrices For Texture Analysis. [Google Scholar]
- 48.Xu D-H, Kurani AS, Furst JD, Raicu DS. The 4th IASTED International Conference on Visualization, Imaging and Image Processing. VIIP; Marbella, Spain: 2004. Run-length encoding for volumetric texture. [Google Scholar]
- 49.McBride GB. National Institute of Water & Atmospheric Research Ltd; New Zealand: 2007. Equivalence measures for comparing the performance of alternative methods for the analysis of water quality variables. [Google Scholar]
- 50.Park SJ, Ionascu D, Killoran J, Mamede M, Gerbaudo VH, Chin L, Berbeco R. Evaluation of the combined effects of target size, respiratory motion and background activity on 3D and 4D PET/CT images. Phys Med Biol. 2008;53(13):3661–3679. doi: 10.1088/0031-9155/53/13/018. [PubMed PMID: 18562782] [DOI] [PubMed] [Google Scholar]
- 51.Adams MC, Turkington TG, Wilson JM, Wong TZ. A systematic review of the factors affecting accuracy of SUV measurements. AJR Am J Roentgenol. 2010;195(2):310–320. doi: 10.2214/AJR.10.4923. [PubMed PMID: 20651185] [DOI] [PubMed] [Google Scholar]
- 52.Low DA, White BM, Lee PP, Thomas DH, Gaudio S, Jani SS, Wu X, Lamb JM. A novel CT acquisition and analysis technique for breathing motion modeling. Phys Med Biol. 2013;58(11):L31–L36. doi: 10.1088/0031-9155/58/11/L31. [PubMed PMID: 23640212. Pubmed Central PMCID: PMC3685480. Epub 2013 May 2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ion AL. Methods for Knowledge Discovery in Images. Inf Technol Control. 2009;38(1):43–50. [PubMed PMID: WOS:000264666200006. English] [Google Scholar]
- 54.Teo BK, Saboury B, Munbodh R, Scheuermann J, Torigian DA, Zaidi H, Alavi A. The effect of breathing irregularities on quantitative accuracy of respiratory gated PETCT. Med Phys. 2012;39(12):7390–7397. doi: 10.1118/1.4766876. [PubMed PMID: 23231288] [DOI] [PubMed] [Google Scholar]