Abstract
Examples of associations between human disease and defects in pre–messenger RNA splicing/alternative splicing are accumulating. Although many alterations are caused by mutations in splicing signals or regulatory sequence elements, recent studies have noted the disruptive impact of mutated generic spliceosome components and splicing regulatory proteins. This review highlights recent progress in our understanding of how the altered splicing function of RNA-binding proteins contributes to myelodysplastic syndromes, cancer, and neuropathologies.
Introduction
Nearly all protein-encoding human genes have multiple exons that are combined in alternative ways to produce distinct mRNAs, often in an organ-specific, tissue-specific, or cell type–specific manner. Although documenting the function of this vast collection of splice variants is a challenging endeavor, the regulated production of splice variants is required for important functions encompassing virtually all biological processes. The growing recognition of splicing and alternative splicing as critical contributors to gene expression was accompanied by many new examples of how splicing defects are associated with human disease. As several excellent reviews have reported on this expanding, and sometimes causal, relationship (Poulos et al., 2011; Singh and Cooper, 2012; Zhang and Manley, 2013; Cieply and Carstens, 2015; Nussbacher et al., 2015), the goal of this review is to highlight recent efforts in understanding how disease-associated mutations disrupt regulation of splicing. After an overview of basic concepts in splicing and splicing control, we discuss recently described defects in the control of splicing that suggest contributions to myelodysplastic syndromes (MDS), cancer, and neuropathologies.
Splicing and splicing control
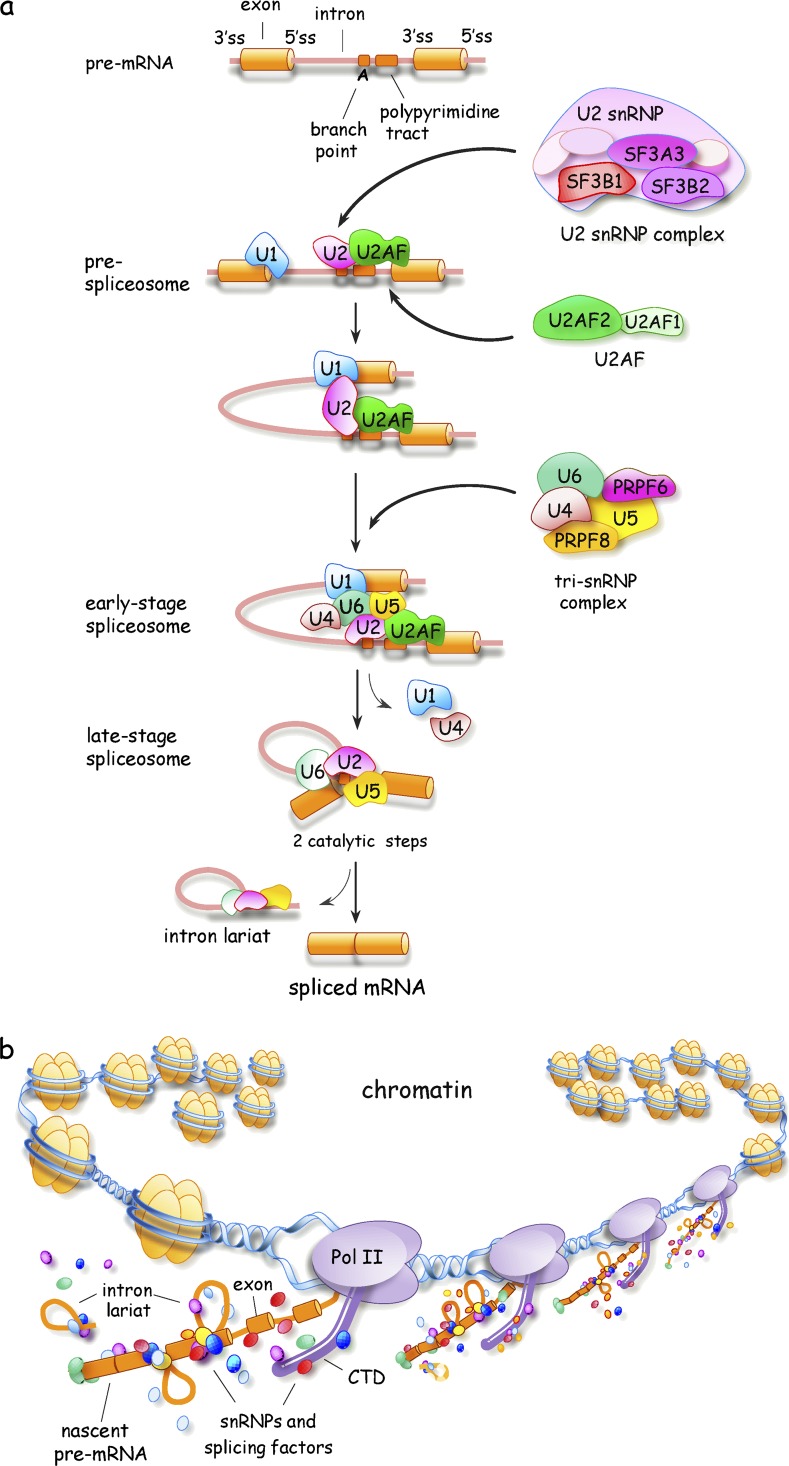
Intron removal is performed by the spliceosome (Fig. 1 A), whose assembly starts with the recognition of the 5′ splice site (5′ss), the 3′ splice site (3′ss), and the branch site by U1 small nuclear RNP (snRNP), U2AF, and U2 snRNP, respectively. Along with the U4/U6.U5 tri-snRNP, >100 proteins are recruited to reconfigure the interactions between small nuclear RNAs, between small nuclear RNAs and the pre-mRNA, and to position nucleotides for two successive nucleophilic attacks that produce the ligated exons and the excised intron (Wahl et al., 2009; Matera and Wang, 2014). Fewer than 1,000 introns (i.e., ∼0.3%) are removed by the minor spliceosome, which uses distinct snRNPs (U11, U12, U4atac, and U6atac) but shares U5 and most proteins with the major spliceosome (Turunen et al., 2013).
Figure 1.
Spliceosome assembly and transcription-coupled splicing. (A) Schematic representation of spliceosome assembly indicating the position of 5′ss, 3′ss, the branch point, and the polypyrimidine tract. Exons and introns are represented as solid cylinders and lines, respectively. Only a portion of spliceosome components are depicted, with some subunits of U2AF, U2 snRNP, and the tri-snRNP complex indicated. (B) Schematic representation of the chromatin-associated cotranscriptional assembly of splicing complexes on a nascent pre-mRNA. CTD, C-terminal domain of RNA polymerase II.
Definition of intron borders often requires the collaboration of RNA-binding proteins (RBPs), such as serine arginine (SR) and heterogeneous nuclear RNPs (hnRNPs), which interact with specific exonic or intronic sequence elements usually located in the vicinity of splice sites. As the combinatorial arrangement of these interactions helps or antagonizes the early steps of spliceosome assembly (Fu and Ares, 2014), one ambitious goal is to determine how cell-, tissue-, and disease-specific variations in the expression of these splicing regulators and their association near splice sites induce specific changes in alternative splicing (Barash et al., 2010; Zhang et al., 2010). This challenge is compounded by the fact that only a fraction of the >1,000 RBPs has been studied (Gerstberger et al., 2014) and that all RBPs have splice variants, usually of undetermined function. Moreover, the function of RBPs is often modulated by posttranslational modifications that occur in response to environmental insults and metabolic cues (Fu and Ares, 2014).
An extra layer of complexity to our view of splicing control is added when we consider that experimentally induced decreases in the levels of core spliceosomal components also affect splice site selection (Saltzman et al., 2011). Indeed, reducing the level of dozens of spliceosomal components, including SF3B1, U2AF, and tri-snRNP components, affects the production of splice variants involved in apoptosis and cell proliferation (Papasaikas et al., 2015). Although it remains unclear whether variation in the levels and activity of generic factors is used to control splicing decisions under normal conditions, deficiencies in tri-snRNP proteins or in proteins involved in snRNP biogenesis are now frequently associated with aberrant splicing in disease (e.g., PRPF proteins in retinitis pigmentosa [Tanackovic et al., 2011], the SMN protein in spinal muscular atrophy [SMA; Zhang et al., 2008], and SF3B1, SRSF2, and U2AF1 in MDS [see Spliceosomal proteins in MDS section]). How mutations in generic splicing factors confer gene- and cell type–specific effects is an intriguing question. The suboptimal features of some introns that dictate this sensitivity may normally be mitigated by the high concentration or activity of generic factors. Consistent with this view, repression of PRPF8 alters the splicing of introns with weak 5′ss (Wickramasinghe et al., 2015). Thus, deficiencies in the activity of generic spliceosome components may compromise the splicing of a subset of introns, contributing to the onset of disease.
As splicing decisions are usually made while the pre-mRNA is still being transcribed (Fig. 1 B), regulatory links with transcription and chromatin structure take place at several levels. First, spliceosome components and regulators are recruited to the transcription machinery (e.g., the C-terminal domain of RNA polymerase II) to facilitate their transfer onto the emerging nascent pre-mRNA (Bentley, 2014). Second, the speed of the elongating polymerase provides a kinetic window for the assembly of enhancer or repressor complexes that influence commitment between competing pairs of splice sites (Bentley, 2014; Naftelberg et al., 2015). Third, posttranslational modifications of histones and chromatin remodeling factors impact the speed of transcription as well as the recruitment of adapters that interact with splicing regulators (Luco et al., 2011; Lee and Rio, 2015; Naftelberg et al., 2015). Notably, histone modifications in specific chromatin regions can be triggered by Argonaute proteins bound to endogenous or exogenously provided small RNAs (Alló et al., 2009; Ameyar-Zazoua et al., 2012). Long noncoding RNAs (lncRNAs), whose expressions vary in human diseases (e.g., MALAT-1 in cancer), may also contribute to splicing control by interacting with splicing factors to regulate their availability, or by orchestrating local epigenetic modifications that impact the speed of transcription or the recruitment of adapters (Zhou et al., 2014a; Gonzalez et al., 2015).
Shooting the pre-messenger by disrupting splicing control elements
More than 200 human diseases, including progeria and some forms of breast cancer and cystic fibrosis, are caused by point mutations that affect pre-mRNA splicing by destroying or weakening splice sites, or activating cryptic ones (Wang et al., 2012), thereby producing mRNAs that encode defective proteins or that are targets for nonsense-mediated mRNA decay (NMD). Splicing defects can also lead to the cotranscriptional degradation of nascent pre-mRNAs (Davidson et al., 2012; Vaz-Drago et al., 2015). A splice site mutation in BRAF is associated with resistance to the anticancer agent vemurafenib, but inhibitors of the generic splicing factor SF3B1 decrease the production of the mutation-induced BRAF variant and inhibit drug-resistant cell proliferation (Poulikakos et al., 2011; Salton et al., 2015). Splice site variations can also have health-positive effects, as shown recently for a variant of LDLR that lowers non–high density lipoprotein cholesterol and protects against coronary artery disease (Gretarsdottir et al., 2015). In addition to mutations at splicing signals themselves, mutations that destroy silencer or enhancer elements form another important group of disease-causing alterations that impact alternative splicing (Sterne-Weiler and Sanford, 2014). Because more than half of the nucleotides in an exon may be part of splicing regulatory motifs (Chasin, 2007), synonymous exon mutations, as well as an undetermined number of intron mutations, may further contribute to splicing misregulation that leads to disease. A recent computational analysis relying on RNA sequencing data from normal and disease samples and using >650,000 single-nucleotide variations (SNVs) identified >10,000 intronic and 70,000 missense and synonymous exonic SNVs occurring in splicing regulatory motifs that linked potential splicing defects with disease (Xiong et al., 2015). Notably, the computational tool developed for this analysis predicted, with tantalizing accuracy, the impact of mutation on the direction and amplitude of splicing shifts associated with SMA and hereditary colorectal cancer and identified several intronic autism-associated SNVs with a high potential of splicing impact (Xiong et al., 2015).
Incapacitating the regulators
Disease-causing mutations in intronic or exonic control elements generally affect splicing by perturbing the binding of regulatory proteins that normally recognize them. The activity of the splicing regulators themselves can also be altered in disease. Changes in the nuclear level of regulators, including RBFOX2, hnRNP, and SR proteins, often occur in cancer (Venables et al., 2009; Zhang and Manley, 2013). Although these changes frequently produce splice variants that affect cell cycle control, apoptosis, cell motility, and invasion, the molecular mechanisms that lead to these alterations and to specific downstream events that promote cancer remain largely unclear (Zhang and Manley, 2013; Shilo et al., 2015). Another way to alter the activity of splicing regulators is through sequestration. This is the case in DM1 and DM2 myotonic dystrophies, where muscleblind-like (MBNL) proteins are recruited to mRNAs carrying expansions of CUG and CCUG repeats, respectively. This sequestration compromises MBNL binding to normal RNA targets, deregulates the expression of CUGBP1, and alters alternative splicing of hundreds of transcripts not only in muscle tissues but also in the brain (Poulos et al., 2011; Charizanis et al., 2012; Echeverria and Cooper, 2012; Batra et al., 2014; Goodwin et al., 2015). The formation of cytoplasmic aggregates, possibly also triggered by mRNAs carrying nucleotide expansions, is frequently associated with neuropathological diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD; see Splicing control defects in neuropathological and muscle-related disease section). In other instances, particularly in cancer, the localization and/or activity of splicing factors are misregulated by posttranslational modifications, e.g., phosphorylation (Naro and Sette, 2013). Mutation of generic spliceosomal components is also becoming a recurrent theme in disease, and recent advances in this area are mentioned throughout our review. Likewise, with the increasing awareness that splicing decisions are coordinated with transcription, and thus with processes that modify chromatin, splicing alterations provoked by disease-associated lncRNAs and chromatin-modifying enzymes are likely to become an emerging focus of inquiry.
Impairing splicing factors to promote cancer
Here, we highlight recent work that has focused on the role of spliceosomal components in MDS, a heterogeneous group of disorders that affect hematopoietic progenitor cells and the production of different types of blood cells. MDS often progress to fully malignant acute myeloid leukemia (AML) with the abnormal accumulation of hematopoietic precursors arrested at an early stage of differentiation. We then provide examples of network restructuring of alternative splicing regulators that have been more solidly associated with carcinogenesis or that may constitute new concepts that link splicing factors with the emergence and maintenance of cancer.
Spliceosomal proteins in MDS
Somatic heterozygous mutations in any of the spliceosomal proteins SF3B1, SRSF2, U2AF1, and the U2AF-related gene ZRSR2 occur in >50% of all MDS patients (Papaemmanuil et al., 2011; Quesada et al., 2011; Yoshida et al., 2011). No homozygous mutations have been described, and almost all mutations are missense, usually occurring at conserved positions. Bone marrow and cancer cells harboring these mutations display splicing abnormalities (Makishima et al., 2012; Przychodzen et al., 2013; Gentien et al., 2014). This may be a direct consequence of the specific mutations because the RNAi-mediated depletion of wild-type SF3B1, SRSF2, and U2AF1 in a variety of cell types or the expression of mutated proteins in nonhematopoietic and cancer cell lines also disrupts alternative splicing (Massiello et al., 2006; Pacheco et al., 2006; Graubert et al., 2011; Yoshida et al., 2011; Venables et al., 2013; Brooks et al., 2014; Shao et al., 2014; Dolatshad et al., 2015; Kfir et al., 2015; Komeno et al., 2015; Papasaikas et al., 2015). Here, we will summarize a set of studies that provide tantalizing insight into how mutated SRSF2, SF3B1, U2AF1, and ZRSF2 affect splicing programs and alter hematopoiesis in mice and MDS patients (Fig. 2).
Figure 2.
Diagram summarizing how splicing factors and telomere deficiencies reproduce hematopoietic defects found in MDS. The impact of each deficiency on alternative splicing is indicated with an emphasis on shared alterations (see Spliceosomal proteins in MDS for details).
SRSF2
Notably, telomerase-negative mice with short telomeres that induce a persistent DNA damage response (DDR) present hematopoietic defects that recapitulate the clinical features of human MDS (Colla et al., 2015). Moreover, this telomere deficiency is associated with a decrease in the level of splicing factors that are frequently mutated in MDS (e.g., SRSF2, U2AF2, SF3B2, and SF3A3). Progenitor cells with deficient telomeres produce defective transcripts encoding components involved in DNA repair and chromatin structure. One splicing change reduces the level of the DNA methyl transferase DNMT3a, whose frequent mutation in MDS patients contributes to rapid progression to AML (Walter et al., 2011; Colla et al., 2015). SRSF2 is a splicing regulator that contributes to both generic and alternative splicing (Long and Caceres, 2009). The impact of short telomeres on the expression of SRSF2 inspired Colla et al. (2015) to create SRSF2-haploinsufficient mice. Remarkably, these mice display impaired erythroid differentiation and express several of the defective alternative splicing events caused by telomere dysfunction. Further, they aberrantly splice transcripts encoding components involved in telomere maintenance, potentially providing a feedback loop that may elicit more splicing defects (Colla et al., 2015).
Importantly, the MDS-associated P95H mutation in SRSF2 shifts the affinity of SRSF2 to a subset of binding sites (Kim et al., 2015; Komeno et al., 2015; Zhang et al., 2015a), thus providing an explanation for the fact that the recapitulation of splicing defects observed in SRSF2-haploinsufficient mice is only partial (Colla et al., 2015). CD34+ hematopoietic stem cells from MDS patients with the P95H mutation have defects in the production of splice variants involved in telomere maintenance, DNA repair, and chromatin remodeling (Colla et al., 2015). Finally, murine bone marrow cells expressing the SRSF2-P95H mutant display features that are characteristic of MDS, including increased proliferation of progenitor cells and impaired differentiation (Kim et al., 2015). One of the P95H-mediated splicing alterations in mice reduces the expression of the histone methyl transferase EZH2, an outcome also occurring in human cells expressing mutant SRSF2. Strikingly, restoring expression of EZH2 in SRSF2 mutant mice partially rescues the hematopoietic defect (Kim et al., 2015). Overall, these studies provide strong evidence that MDS-associated mutations in SRSF2 affect the production of splice variants involved in chromatin structure that in turn elicit hematopoietic defects.
SF3B1
SF3B1 is a U2 snRNP–associated protein involved in branch point selection (Corrionero et al., 2011). The depletion of SF3B1 impairs the growth and the differentiation of myeloid cell lines (Dolatshad et al., 2015). SF3B1 haploinsufficiency in mice compromises the repopulating ability of hematopoietic stem cells, but is not sufficient to induce MDS (Visconte et al., 2012; Matsunawa et al., 2014; Wang et al., 2014a; Dolatshad et al., 2015). Decreasing the levels of SF3B1 in myeloid cell lines alters the alternative splicing of transcripts encoding components involved in apoptosis and cell cycle control. Interestingly, in bone marrow cells and progenitor bone marrow stem cells from SF3B1-mutated MDS patients, the expression and splicing of genes/transcripts associated with mitochondrial and heme-related functions are altered, providing a link with the abnormal iron homeostasis observed in MDS patients (Visconte et al., 2012; Dolatshad et al., 2015). Notably, iron homeostasis influences alternative splicing by modulating the activity of SRSF7 (Tejedor et al., 2015). Interestingly, it has been observed that SRSF7 is itself abnormally spliced in MDS patients carrying the SF3B1 mutation (Dolatshad et al., 2015). We speculate that this defective splicing may be responsible, at least in part, for the noted heme deficiency in MDS patients.
SF3B1 is also part of a complex with BCLAF1, U2AF, and PRPF8 that is recruited to chromatin-bound BRCA1 to stimulate the splicing of transcripts encoding factors involved in DNA repair and the DDR (Savage et al., 2014). In line with this finding, several DNA repair and DDR genes (e.g., ABL1, BIRC2, and NUMA1) produce aberrantly spliced transcripts in cells of patients with SF3B1 mutations (Dolatshad et al., 2015). Interestingly, one splicing alteration in these patient cells occurs in EZH1, a functional homologue of the histone methyl transferase EZH2, which is also defectively spliced in SRSF2-mutated cells and contributes to the MDS phenotype (Visconte et al., 2012; Dolatshad et al., 2015; Kim et al., 2015). Notably, the expression and alternative splicing of transcripts encoding RNA-processing factors, including PRPF8 and U2AF2, are also affected in SF3B1-mutated cells (Dolatshad et al., 2015). This observation is important because mutations in PRPF8 and U2AF2 are found in MDS patients (Boultwood et al., 2014). However, although PRPF8 mutations are associated with alternative splicing defects (Kurtovic-Kozaric et al., 2015), U2AF2 mutations appear neutral (Shao et al., 2014). Overall, these results suggest that SF3B1 mutations alter the splicing of transcripts involved in chromatin structure, DNA repair, and the DDR, thereby possibly providing an explanation for the accumulation of DNA damage in hematopoietic progenitor cells of MDS patients (Zhou et al., 2013). A function for SF3B1 in splice site selection has recently been associated with a specific interaction with histone marks that are enriched in exons (Kfir et al., 2015). Although SF3B1 mutations often occur in the C-terminal HEAT repeats involved in protein–protein interactions, it remains to be shown whether these mutations affect the recruitment of SF3B1 to chromatin. If they do, combining a mutated SF3B1 with chromatin modification defects may amplify splicing alterations, gradually leading to more detrimental hematopoietic deficiencies.
U2AF1
U2AF1 is the smaller of two proteins that make up the U2AF heterodimer implicated in generic 3′ss recognition. Although the U2AF1-S34F mutation elicits hematopoietic abnormalities in mice that compromise the repopulating ability of stem cells, it does not elicit MDS (Shirai et al., 2015). Many splicing defects in MDS patients with U2AF1 mutations occur in transcripts that encode components involved in cell cycle and splicing control (Przychodzen et al., 2013). Expression of mutated U2AF1 proteins in a human erythroleukemic cell line causes thousands of splicing alterations, including some in transcripts encoding components involved in DNA methylation (e.g., DNMT3B, also affected by mutations in SF3B1), DDR, and apoptosis (Ilagan et al., 2015). Different U2AF1 mutations alter its binding to 3′ss in different ways and lead to distinct yet overlapping splicing defects (Brooks et al., 2014; Shao et al., 2014; Ilagan et al., 2015). A meta-transcriptome analysis using samples from U2AF1-S34F mutant mice, AML patients with U2AF1 mutations, and primary bone marrow cells overexpressing U2AF1-S34F uncovered common splicing alterations in transcripts encoding splicing proteins and components that are mutated in MDS and AML, or that are involved in hematopoietic stem cell function. These observations provide strong support to the view that mutated U2AF1 elicits abnormal hematopoiesis (Shirai et al., 2015).
ZRSR2
ZRSR2 has been implicated in the splicing of introns that use the U12-dependent minor spliceosome in transcripts encoding cancer-relevant proteins such as PTEN, MAPK1, MAPK3, BRAF, and E2F2 (Madan et al., 2015). MDS-associated mutations in ZRSR2 are often inactivating, and depleting ZRSR2 reduces the growth and clonogenic potential of leukemia cell lines and alters the differentiation potential of human CD34+ bone marrow cells (Madan et al., 2015).
Overall, the studies mentioned above suggest that alternative splicing likely makes a crucial contribution to the clinical evolution of MDS (Fig. 2). Mutations in SRSF2, U2AF1, and SF3B1 may elicit a shared set of splicing alterations that trigger common hematopoietic defects and predispose stem cells to cancer development. The insight gained by studying the contribution of mutated splicing factors to MDS is likely to benefit our understanding of how mutations in splicing factors lead to cancer in general because mutations in SF3B1, U2AF1, and SRSF2 are also found in a variety of solid tumors (Kandoth et al., 2013; Scott and Rebel, 2013; Maguire et al., 2015). Although a recent compilation indicates that splicing factor genes are frequently mutated in different types of cancer (Sveen et al., 2015), a more extensive characterization of the functional impact of these mutations will be required to determine whether these alterations preferentially contribute to specific types of cancer.
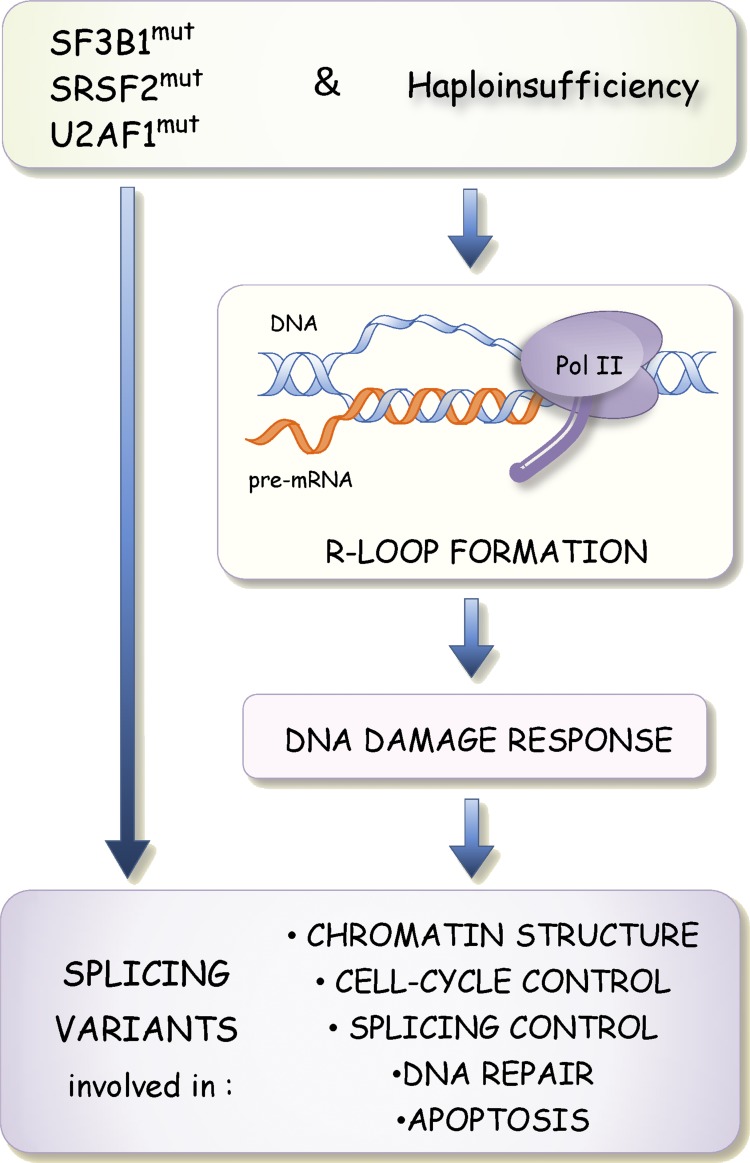
An integrating hypothesis: Altered RBPs cause R loops and persistent DDR signaling that alter splicing
Although mutated SF3B1 and U2AF1 are expected to impact branch site/3′ss selection directly, we speculate that decreases in the level, and changes in the RNA binding specificity of splicing factors may also cause a second wave of alternative splicing changes through activation of the DDR (Fig. 3). This model is based on the observation that when core spliceosomal components are delocalized or when RNA-processing factors such as SRSF1 and RNPS1 are depleted, R loop formation occurs and triggers the DDR to impact alternative splicing (Li and Manley, 2005; Li et al., 2007; Domínguez-Sánchez et al., 2011; Tresini et al., 2015). If we are correct, drops in the level or changes in the activity of SRSF2, SF3B1, and U2AF1 may perturb alternative splicing through persistent R loop–mediated activation of the DDR (Fig. 3), a consequence that would be consistent with the noted accumulation of DNA damage in MDS progenitor cells (Zhou et al., 2013). DNA damage affects the expression, modification, and localization of several splicing regulatory proteins (Shkreta and Chabot, 2015). Likewise, DNA damage caused by deficient telomeres in mice alters the expression of splicing regulators (Colla et al., 2015). Importantly, alternative splicing defects in MDS patients and MDS mouse models affect variants involved in apoptosis, cell cycle control, DNA repair, splicing control, and chromatin structure (Fig. 3), precisely matching the functional categories of transcripts whose alternative splicing is affected by DNA-damaging agents (Shkreta and Chabot, 2015). As drops in the activity of splicing factors are frequently associated with human pathologies, this model may be applicable to a variety of diseases in addition to MDS, including myotonic dystrophies, retinitis pigmentosa, ALS, and FTD.
Figure 3.
Model proposing that changes in alternative splicing triggered by mutations in SF3B1, SRSF2, and U2AF1 also occur through activation of the DDR. Drops in the activity or changes in the binding specificities of splicing factors alter splice site selection directly but are also proposed to promote the formation of R loops when segments of nascent pre-mRNAs are not adequately bound by splicing factors. R loop formation in turn triggers the DDR to elicit splicing changes in transcripts encoding DNA repair, chromatin structure, cell cycle and splicing control, and apoptotic components.
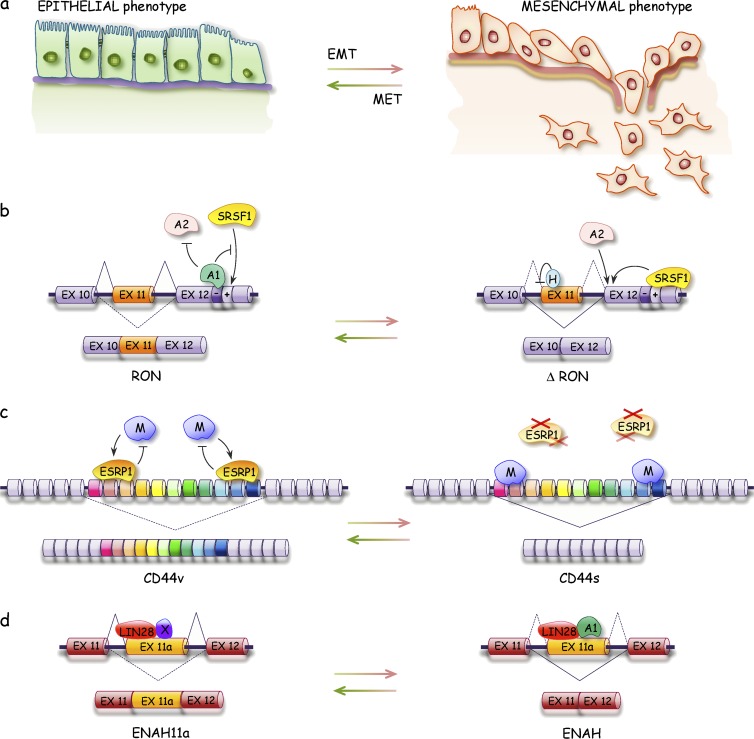
The epithelial–mesenchymal/mesenchymal–epithelial transition connection
Cancer metastasis involves cell migration and tissue invasion through reversible transitions from mesenchymal to epithelial cell types (mesenchymal–epithelial and epithelial–mesenchymal transition [MET and EMT, respectively]; Fig. 4; Yang and Weinberg, 2008). ESRPs and RBFOX2 control the alternative splicing of several transcripts encoding cell adhesion proteins involved in the epithelial or mesenchymal phenotypes (Shapiro et al., 2011; Venables et al., 2013; Braeutigam et al., 2014). A splice variant of the tyrosine kinase receptor RON that promotes cell migration and activates EMT is controlled by antagonistic interactions involving SRSF1 and hnRNP A1, A2, and H proteins (LeFave et al., 2011; Biamonti et al., 2014). Likewise, hnRNP M antagonizes ESRP in the splicing of the cell adhesion molecule CD44 and plays a key role in the metastatic behavior of breast cancer cells in mouse models (Xu et al., 2014). In contrast, RBM47 behaves as a suppressor of breast cancer progression and metastasis (Vanharanta et al., 2014). Consistent with their role in metastasis, the expression of hnRNP M and RBM47 is respectively high and low in aggressive human breast cancer (Vanharanta et al., 2014; Xu et al., 2014). LIN28A, the expression of which increases in the HER2 breast cancer subtype, interacts with hnRNP A1 to modulate the production of splice variants of ENAH that is associated with breast cancer metastasis (Di Modugno et al., 2007; Yang et al., 2015). In addition to the lncRNA MALAT1, which is implicated in metastasis, possibly by controlling alternative splicing (Tripathi et al., 2010; Gutschner et al., 2013), another lncRNA modifies chromatin to prevent the recruitment of a repressive chromatin-splicing adapter complex that normally enforces the mesenchymal-specific splicing of FGFR2 (Gonzalez et al., 2015). lncRNAs may act on opposite functional sides of the oncogenic pathway. On the one hand, the lncRNA INXS, which interacts with Sam68 to favor the production of the proapoptotic Bcl-xS splice variant, is down-regulated in tumors and its overexpression in mouse xenograft models elicits tumor regression (DeOcesano-Pereira et al., 2014). On the other hand, the lncRNA FAS-AS1 interacts with RBM5 to reduce expression of the prosurvival soluble FAS variant (Sehgal et al., 2014). Other lncRNAs that have been implicated in cancer include linc-p21, PANDA, TUG1, and Pint, but their impact on splicing and their contribution to cancer and metastasis are speculative and need to be investigated in more detail (Wang et al., 2014b; Zhang and Peng, 2015).
Figure 4.
Defects in alternative splicing affect EMTs. (A–D) Epithelial and mesenchymal interconversion (A) are affected by splicing factors that control the alternative splicing events associated with these transitions as depicted for the RON (B), CD44 (C), and ENAH (D) transcripts. H, hnRNP H; M, hnRNP M; MET, mesenchymal–epithelial transition; X, unknown protein.
The MYC splicing connection
The overexpression of MYC contributes to malignant transformation and is associated with many cancers. Several studies have established a role for MYC in splicing control (Fig. 5). MYC contributes to cancer metabolism and tumor growth by increasing the levels of splicing regulators PTBP1, hnRNP A1, and hnRNP A2 that shift the production of pyruvate kinase from splice variant PKM1, which drives oxidative phosphorylation, to PKM2, which elicits aerobic glycolysis (Christofk et al., 2008; David et al., 2010). In glioblastoma, the up-regulation of hnRNP A1 promotes the splicing of a transcript encoding the MYC-interacting partner Max to generate ΔMax, producing a feed-forward loop that enhances MYC function and hnRNP A1 expression (Fig. 5; Babic et al., 2013). MYC also stimulates the expression of the SR protein SRSF1, which drives oncogenesis through alternative splicing of a network of transcripts encoding signaling molecules (e.g., RON and MKNK2) and transcription factors (e.g., BIN1; Das and Krainer, 2014). SRSF1 also elicits the production of variants, such as CASC4 with antiapoptotic function, as well as MDM2 and cyclin D1 variants with prooncogenic properties (Olshavsky et al., 2010; Anczuków et al., 2012, 2015; Comiskey et al., 2015). Positive feedback likely occurs because the SRSF1-mediated splice variant BIN1-12a no longer binds to MYC and lacks tumor suppressor activity (Ge et al., 1999; Karni et al., 2007). KRAS mutations that are frequently found in colorectal cancer activate the MAPK–extracellular signal-regulated kinase pathway to increase the level of the transcription factor ELK1 that in turn increases MYC with the expected impact on the production of PKM2 (Hollander, D., and Ast, G., personal communication). The activated MAPK–extracellular signal-regulated kinase pathway also stimulates the expression of Sam68, which increases the level of SRSF1 through alternative splicing (Matter et al., 2002; Valacca et al., 2010). Interestingly, the expression of SRSF1 is also stimulated by the anticancer drug gemcitabine, producing a splice variant of MKNK2 that phosphorylates eIF4E to promote cell growth and drug resistance (Adesso et al., 2013). Gemcitabine resistance is also provided by the expression of PKM2 through the increased production of PTBP1 (Calabretta et al., 2015).
Figure 5.
MYC splicing connections in cancer. As described in the text, MYC acts as master regulator by controlling the expression of splicing factors hnRNP A1, A2, PTBP1, and SRSF1, which promote changes that favor the growth of cancer cells and provide positive feedback loops that further stimulate MYC expression. The KRAS–MAPK pathway, which is relevant to colorectal cancer (KRAS and BRAF mutations) and melanomas (BRAF mutations), converges on the MYC-mediated splicing program through ELK1, which stimulates MYC expression, and Sam68, which improves the production of SRSF1 through alternative splicing. Red arrows are used to indicate impact on alternative splicing.
Splicing factor addiction
The term “oncogene addiction” has been used in the cancer field to describe the increased dependence of cancers on oncogenes for growth and survival (Luo et al., 2009). Recent results suggest that there is an analogous hypersensitivity of cancer cells on splicing factors. This relationship was established when it was noted that MYC-regulated genes and pathways provoke a general increase in pre-mRNA synthesis that imposes a strain on generic splicing (Hsu et al., 2015; Koh et al., 2015). The fact that MYC up-regulates enzymes that modify snRNP proteins in cancer cells is consistent with the high demand for spliceosome components (Koh et al., 2015). Nevertheless, MYC-driven cancer cells are more sensitive to depletions of spliceosome components such as U2AF1 and SF3B1 (Hsu et al., 2015). This splicing stress may also affect the production of functionally important splice variants because decreases in the level or activity of generic spliceosome components also affect alternative splicing. Other cancers may be similarly addicted to splicing factors. For example, PRPF6, a component of the tri-snRNP complex, is overexpressed in a subset of primary and metastatic colon cancers, and its depletion by RNAi in cell lines reduces cell growth and decreases the production of the oncogenic ZAK kinase splice variant (Adler et al., 2014). Likewise, expression of splicing regulator SRSF10 is increased in aggressive colon cancers. The siRNA-mediated depletion of SRSF10 decreases tumor formation in mice, an effect that is mediated, at least in part, by a drop in the production of the oncogenic splice variant of the splicing factor BCLAF1 (Zhou et al., 2014b). Thus, the overall stimulation in gene expression in cancer cells may increase their reliance on splicing factors, hence providing avenues to explore novel anticancer strategies.
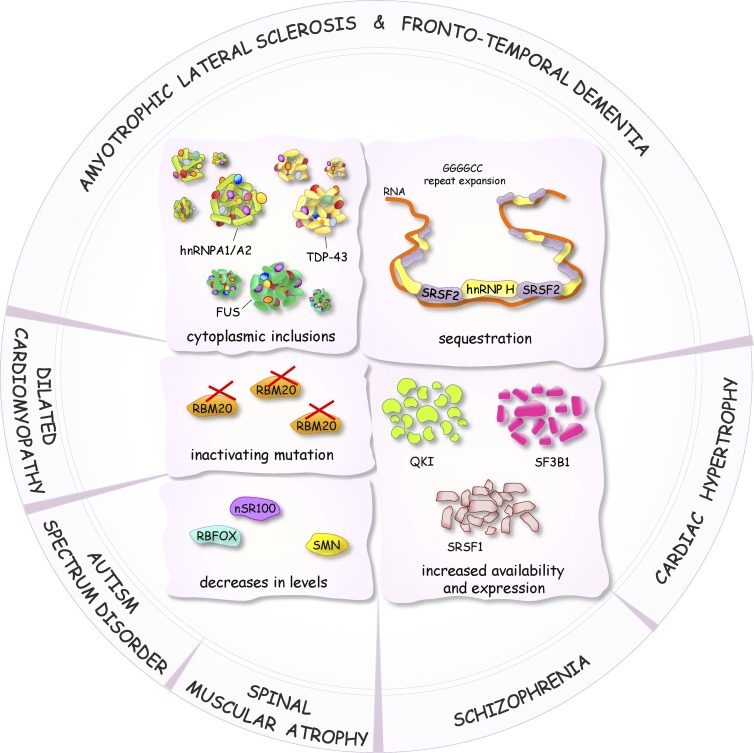
Splicing control defects in neuropathological and muscle-related diseases
As in cancer, pathogenic mechanisms in neurological and muscle-associated diseases can be caused by mutations in genes that affect splicing of their pre-mRNAs, or by mutations that affect the expression and the activity of splicing factors that control splice site utilization. Excellent reviews have recently presented the prevalence of alternative splicing, the role of RBPs, and the functional diversity of splice variants in neuronal systems (Darnell, 2013; Raj and Blencowe, 2015). Here, we present recent advances that solidify the links between splicing control and neuronal and muscular pathologies (Fig. 6). Identifying functionally relevant variants and changes in the expression/activity of regulators remains challenging, particularly in neuropathologies. This is mainly a result of tissue availability and heterogeneity, as well as difficulties in developing adequate animal models that recapitulate human phenotypes.
Figure 6.
Diagram representing some of the defects in the activity, level, and localization of splicing factors linked to the altered production of splice variants in neurological and muscle-associated diseases.
ALS and FTD
Although mutations in the splicing regulatory RBP TDP-43 are found in only a fraction of all cases of ALS and FTD, cytoplasmic inclusions and the nuclear depletion of TDP-43 are hallmarks of these diseases (Janssens and Van Broeckhoven, 2013; Scotter et al., 2015). Decreasing the expression of TDP-43 leads to neuronal defects in mice and affects the alternative splicing of transcripts encoding components important in neuronal development or implicated in neurological diseases (Polymenidou et al., 2011; Tollervey et al., 2011; Yang et al., 2014a). Splicing defects in ALS tissues occur in target TDP-43 transcripts (Arnold et al., 2013; Yang et al., 2014a). A recent study in mice indicates that a decrease in TDP-43 impairs splicing fidelity and leads to the aberrant inclusion of cryptic exons, an effect also seen in brain tissues from ALS-FTD patients (Ling et al., 2015). Similar to TDP-43, mutations and loss of nuclear function of FUS have been linked to alternative splicing changes in ALS, with a few pre-mRNA targets also regulated by TDP-43 (Lagier-Tourenne et al., 2012; Coady and Manley, 2015). Cytoplasmic aggregates of mutated FUS or TDP-43 often sequester other splicing proteins, and this may also contribute to alterations in splicing profiles. For example, the ability of FUS to interact with U1 snRNP is likely responsible for the U1 snRNP cytoplasmic mislocalization in FUS-mutated ALS patient fibroblasts (Yu and Reed, 2015; Yu et al., 2015). ALS-associated mutations in hnRNP A1/A2 proteins also cause cytoplasmic aggregation (Kim et al., 2013). In several ALS-FTD patients, GGGGCC repeat expansion that promotes G-quadruplex formation in the C9ORF72 gene sequester splicing factors such as SRSF2 and hnRNP H, which in turn may promote extensive alternative splicing defects and neurodegeneration (Lee et al., 2013; Prudencio et al., 2015; Zhang et al., 2015b). Further studies should clarify whether the pathogenic impact of aggregates is strictly caused by loss of function or whether toxicity associated with aggregate formation also contributes to the clinical manifestation of ALS and FTD.
Alzheimer’s disease (AD) and Huntington’s disease (HD)
The deposition of oligomeric β-amyloid peptides and the formation of neurofibrillary tangles associated with the hyperphosphorylation of the microtubule-associated TAU protein have been implicated in AD (Ittner and Gotz, 2011). ApoE4 status is one of the strongest genetic risk factors, and it possibly affects both β-amyloid and neurofibrillary tangle pathologies. Many genes involved in these pathways, including ApoE4, sustain splicing mutations that have been linked to AD or present profiles of alternative splicing that are altered in AD tissues (Love et al., 2015). RNA sequencing data suggest considerable alternative splicing abnormalities in AD tissues, including in transcripts encoding presenilin-1 and clusterin (Bai et al., 2013). Several splicing factors whose expression are misregulated in AD have been identified, including RBFOX, SR, and hnRNP A1 proteins, whereas splicing components, such as the U1 snRNP, appear to be depleted from the nucleus to form cytoplasmic aggregates (Bai et al., 2013; Hales et al., 2014). Interestingly, a depletion of U1 snRNP components in HEK293 cells disrupts the expression of splice variants encoding the amyloid precursor protein and increases the level of a β-amyloid peptide (Bai et al., 2013). HD is caused by expanded CAG repeats in the HTT gene that promote missplicing of its transcripts (Sathasivam et al., 2013). The CAG repeats may also sequester splicing factors eliciting alternative splicing defects in other transcripts (Mykowska et al., 2011). Like individuals suffering from FTD, HD subjects display an imbalance in the production of TAU variants that promote deposits. Human HD tissues and a mouse model of HD show alterations in the expression of SRSF6, which may modulate TAU splicing, leading to TAU variants with a greater propensity to form deposits (Yin et al., 2012; Fernández-Nogales et al., 2014).
Schizophrenia (SZ)
SZ is a complex neuronal disease promoting brain dysfunction. A variety of alternative splicing anomalies have been described in the brain or neuronal subtypes of SZ patients, including transcripts encoding a glutamate transporter (EAAT; O’Donovan et al., 2015) and microcephalin (MCPH1; Oldmeadow et al., 2014). A polymorphism associated with an increased risk of SZ occurs in the dopamine receptor gene DRD2 and affects the ability of the splicing regulator ZRANB2 to control alternative splicing of DRD2 transcripts (Cohen et al., 2015). The lncRNA gomafu, which is down-regulated in the gray matter from the superior temporal gyrus of SZ patients, is bound by the splicing regulators QKI and SRSF1 to control the alternative splicing of transcripts implicated in SZ (Barry et al., 2014). Other lncRNAs have been associated with neuronal stem cell differentiation and the control of alternative splicing through interaction with the neuronal splicing factor PTBP1 (Ramos et al., 2015). However, although changes in the expression of lncRNAs involved in epigenetic modifications have been linked to neuronal diseases, their contribution to alternative splicing control remains to be examined (Roberts et al., 2014).
Autism spectrum disorder (ASD)
Mutations in, or altered expression of, >100 genes have been linked to ASD (Devlin and Scherer, 2012; Corominas et al., 2014). The majority of these genes produce splice variants, and recurrent splicing defects in some of them have been noted in autistic individuals (Voineagu et al., 2011; Corominas et al., 2014). RBFOX proteins play a critical role in brain development and function (Gehman et al., 2011, 2012), and RBFOX1 haploinsufficiency has been implicated in a variety of neuropsychiatric disorders including ASD (Voineagu et al., 2011). In the mouse brain, the depletion of RBFOX proteins alters the alternative splicing of transcripts implicated in ASD (Weyn-Vanhentenryck et al., 2014). Identification of a clinically relevant set of splicing events remains challenging because RBFOX proteins affect other pathways in RNA processing and in transcription. Moreover, three highly related RBFOX proteins with partially overlapping functions are expressed in the brain. A recent study has identified a highly dynamic set of microexons (3–15 nucleotides in size) in transcripts of different neurofunctional categories that are misregulated in the brain of autistic individuals. Several neural microexons affect protein–protein interactions that are crucial for neural function, and many are controlled by the splicing regulator nSR100, whose expression is important for normal nervous system development (Quesnel-Vallières et al., 2015) and is reported to be reduced in autistic brain tissues (Irimia et al., 2014). Neural microexon splicing is also regulated by the PTBP1 and RBFOX proteins (Li et al., 2015) that are critical for normal neuronal function (Gehman et al., 2012; Licatalosi et al., 2012; Li et al., 2014). Because microexons have also been linked to SZ and epilepsy, it will be most revealing to characterize the molecular pathways that regulate their inclusion in these neurological disorders.
SMA
Mutations that reduce the level of SMN proteins, which are involved in snRNP biogenesis, cause SMA. Although multiple alternative splicing defects have been noted, it remains unclear which splicing abnormalities cause the human phenotypes (Ule et al., 2005; Zhang et al., 2008, 2013; Fogel et al., 2012; Highley et al., 2014). As the SMN protein deficiency can be rescued by stimulating exon 7 inclusion in the SMN2 pre-mRNA, efforts deployed to achieve this goal in mouse models have produced encouraging results using oligonucleotides that block the activity of an intron splicing silencer (Hua et al., 2015; Staropoli et al., 2015) or small molecules that stimulate exon 7 inclusion with apparent high specificity (Naryshkin et al., 2014; Palacino et al., 2015).
Heart disease
Mutations that truncate the sarcomeric protein titin cause dilated cardiomyopathy (Herman et al., 2012). A loss-of-function mutation in RBM20 affects the alternative splicing of titin, causing dilated cardiomyopathy (Guo et al., 2012). Hypoxic conditions associated with cardiac hypertrophy activate the expression of SF3B1, which in turn induces the production of a splice variant of ketohexokinase associated with contractile dysfunction (Mirtschink et al., 2015).
Advances and challenges in monitoring disease-associated changes in alternative splicing
Today, it is very clear that cells derived from patients with a variety of diseases display splicing defects, with studies relating to cancer and neuropathologies being the most prevalent. These splicing alterations may generate recognizable signatures that can guide diagnostics and may lead to the identification of new therapeutic targets. This important cataloguing effort is now increasing through genome-wide studies that exploit affordable RNA sequencing technologies and access to sequence repositories. Bioinformatic resources designed to interrogate these data are also expanding and are becoming widely available (Tang et al., 2013; Sebestyén et al., 2015; Hollander, D., and Ast, G., personal communication).
The reliable identification of targets that support actionable therapeutic approaches is challenged by the fact that correlations are often derived from heterogeneous clinical samples. Moreover, although documentation of the functional impact of splice variants is accumulating (Kelemen et al., 2013; Pagliarini et al., 2015), the causal contribution of disease-associated splice variants to the disease remains unknown in most cases. The functional assessment of a continuously expanding list of splice variants is an experimentally daunting task, possibly explaining why recent studies have restricted their analysis to mRNA variants encoding proteins with known distinct activities or with premature stop codons that decrease protein production.
To understand the molecular mechanisms that lead to splicing alterations, it will be important to (a) assess the expression, posttranslational modifications, or mutations of splicing regulators and chromatin-modifying components; (b) profile the binding sites of the putative regulatory RBP on target pre-mRNAs in relevant tissue cells as was originally done for NOVA, whose inactivation causes paraneoplastic neurological disorders (Zhang et al., 2010); and (c) sequence the genome of diseased and normal tissues for each patient to identity somatic mutations that may contribute to splicing alterations. This comparison is especially relevant to cancer in which genomes are often intrinsically unstable. Moreover, in light of the model proposed earlier, defects in the activity or levels of splicing factors may lead to R loop–mediated mutations that may have a permanent impact on alternative splicing.
To accommodate the analyses of this vast quantity of data, robust computational methods are being developed to link the production of recurrent variants with changes in RBPs (Sebestyén et al., 2015). Alternatively, combining large-scale collections of molecular interaction datasets (protein–DNA, protein–RNA, and protein–protein) with cancer transcriptome datasets may reveal regulatory pathways relevant to cancer (Hollander, D., and Ast, G., personal communication). Putative connections can then be validated experimentally or confirmed, for example by using The Cancer Genome Atlas. These emerging procedures justify the usefulness of network-based approaches (Yang et al., 2014b) to capture molecular relationships across different regulatory layers that become compromised or that emerge during diseases.
Acknowledgments
We thank Nancy Greenbaum and Raymund Wellinger for thoughtful comments on the manuscript. We thank Gil Ast and Dror Hollander for sharing results before publication. We apologize to colleagues whose work could not be cited because of space constraints.
We acknowledge support from the Canadian Institutes of Health Research. B. Chabot is the Pierre C. Fournier Research Chair in Functional Genomics at Université de Sherbrooke.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AD
- Alzheimer’s disease
- ALS
- amyotrophic lateral sclerosis
- AML
- acute myeloid leukemia
- ASD
- autism spectrum disorder
- DDR
- DNA damage response
- EMT
- epithelial–mesenchymal transition
- FTD
- frontotemporal dementia
- HD
- Huntington’s disease
- hnRNP
- heterogeneous nuclear RNP
- lncRNA
- long noncoding RNA
- MDS
- myelodysplastic syndromes
- RBP
- RNA-binding protein
- SMA
- spinal muscular atrophy
- snRNP
- small nuclear RNP
- SNV
- single-nucleotide variation
- SR
- serine arginine
- SZ
- schizophrenia
References
- Adesso L., Calabretta S., Barbagallo F., Capurso G., Pilozzi E., Geremia R., Delle Fave G., and Sette C.. 2013. Gemcitabine triggers a pro-survival response in pancreatic cancer cells through activation of the MNK2/eIF4E pathway. Oncogene. 32:2848–2857. 10.1038/onc.2012.306 [DOI] [PubMed] [Google Scholar]
- Adler A.S., McCleland M.L., Yee S., Yaylaoglu M., Hussain S., Cosino E., Quinones G., Modrusan Z., Seshagiri S., Torres E., et al. . 2014. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes Dev. 28:1068–1084. 10.1101/gad.237206.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alló M., Buggiano V., Fededa J.P., Petrillo E., Schor I., de la Mata M., Agirre E., Plass M., Eyras E., Elela S.A., et al. . 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16:717–724. 10.1038/nsmb.1620 [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.C., et al. . 2012. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 19:998–1004. 10.1038/nsmb.2373 [DOI] [PubMed] [Google Scholar]
- Anczuków O., Rosenberg A.Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S.K., and Krainer A.R.. 2012. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19:220–228. 10.1038/nsmb.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O., Akerman M., Clery A., Wu J., Shen C., Shirole N.H., Raimer A., Sun S., Jensen M.A., Hua Y., et al. . 2015. SRSF1-regulated alternative splicing in breast cancer. Mol. Cell. 60:105–117. 10.1016/j.molcel.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E.S., Ling S.C., Huelga S.C., Lagier-Tourenne C., Polymenidou M., Ditsworth D., Kordasiewicz H.B., McAlonis-Downes M., Platoshyn O., Parone P.A., et al. . 2013. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. USA. 110:E736–E745. 10.1073/pnas.1222809110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic I., Anderson E.S., Tanaka K., Guo D., Masui K., Li B., Zhu S., Gu Y., Villa G.R., Akhavan D., et al. . 2013. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 17:1000–1008. 10.1016/j.cmet.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B., Hales C.M., Chen P.C., Gozal Y., Dammer E.B., Fritz J.J., Wang X., Xia Q., Duong D.M., Street C., et al. . 2013. U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 110:16562–16567. 10.1073/pnas.1310249110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., and Frey B.J.. 2010. Deciphering the splicing code. Nature. 465:53–59. 10.1038/nature09000 [DOI] [PubMed] [Google Scholar]
- Barry G., Briggs J.A., Vanichkina D.P., Poth E.M., Beveridge N.J., Ratnu V.S., Nayler S.P., Nones K., Hu J., Bredy T.W., et al. . 2014. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry. 19:486–494. 10.1038/mp.2013.45 [DOI] [PubMed] [Google Scholar]
- Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A., and Swanson M.S.. 2014. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 56:311–322. 10.1016/j.molcel.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.L. 2014. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15:163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G., Catillo M., Pignataro D., Montecucco A., and Ghigna C.. 2014. The alternative splicing side of cancer. Semin. Cell Dev. Biol. 32:30–36. 10.1016/j.semcdb.2014.03.016 [DOI] [PubMed] [Google Scholar]
- Boultwood J., Dolatshad H., Varanasi S.S., Yip B.H., and Pellagatti A.. 2014. The role of splicing factor mutations in the pathogenesis of the myelodysplastic syndromes. Adv. Biol. Regul. 54:153–161. 10.1016/j.jbior.2013.09.005 [DOI] [PubMed] [Google Scholar]
- Braeutigam C., Rago L., Rolke A., Waldmeier L., Christofori G., and Winter J.. 2014. The RNA-binding protein Rbfox2: An essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 33:1082–1092. 10.1038/onc.2013.50 [DOI] [PubMed] [Google Scholar]
- Brooks A.N., Choi P.S., de Waal L., Sharifnia T., Imielinski M., Saksena G., Pedamallu C.S., Sivachenko A., Rosenberg M., Chmielecki J., et al. . 2014. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One. 9:e87361 10.1371/journal.pone.0087361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta S., Bielli P., Passacantilli I., Pilozzi E., Fendrich V., Capurso G., Fave G.D., and Sette C.. 2015. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. In press. 10.1038/onc.2015.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K., Lee K.Y., Batra R., Goodwin M., Zhang C., Yuan Y., Shiue L., Cline M., Scotti M.M., Xia G., et al. . 2012. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 75:437–450. 10.1016/j.neuron.2012.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L.A. 2007. Searching for splicing motifs. Adv. Exp. Med. Biol. 623:85–106. 10.1007/978-0-387-77374-2_6 [DOI] [PubMed] [Google Scholar]
- Christofk H.R., Vander Heiden M.G., Harris M.H., Ramanathan A., Gerszten R.E., Wei R., Fleming M.D., Schreiber S.L., and Cantley L.C.. 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 452:230–233. 10.1038/nature06734 [DOI] [PubMed] [Google Scholar]
- Cieply B., and Carstens R.P.. 2015. Functional roles of alternative splicing factors in human disease. Wiley Interdiscip. Rev. RNA. 6:311–326. 10.1002/wrna.1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coady T.H., and Manley J.L.. 2015. ALS mutations in TLS/FUS disrupt target gene expression. Genes Dev. 29:1696–1706. 10.1101/gad.267286.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O.S., Weickert T.W., Hess J.L., Paish L.M., McCoy S.Y., Rothmond D.A., Galletly C., Liu D., Weinberg D.D., Huang X.F., et al. . 2015. A splicing-regulatory polymorphism in DRD2 disrupts ZRANB2 binding, impairs cognitive functioning and increases risk for schizophrenia in six Han Chinese samples. Mol. Psychiatry. In press. 10.1038/mp.2015.137 [DOI] [PubMed] [Google Scholar]
- Colla S., Ong D.S., Ogoti Y., Marchesini M., Mistry N.A., Clise-Dwyer K., Ang S.A., Storti P., Viale A., Giuliani N., et al. . 2015. Telomere dysfunction drives aberrant hematopoietic differentiation and myelodysplastic syndrome. Cancer Cell. 27:644–657. 10.1016/j.ccell.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comiskey D.F. Jr., Jacob A.G., Singh R.K., Tapia-Santos A.S., and Chandler D.S.. 2015. Splicing factor SRSF1 negatively regulates alternative splicing of MDM2 under damage. Nucleic Acids Res. 43:4202–4218. 10.1093/nar/gkv223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas R., Yang X., Lin G.N., Kang S., Shen Y., Ghamsari L., Broly M., Rodriguez M., Tam S., Trigg S.A., et al. . 2014. Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism. Nat. Commun. 5:3650 10.1038/ncomms4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A., Minana B., and Valcarcel J.. 2011. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 25:445–459. 10.1101/gad.2014311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R.B. 2013. RNA protein interaction in neurons. Annu. Rev. Neurosci. 36:243–270. 10.1146/annurev-neuro-062912-114322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., and Krainer A.R.. 2014. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 12:1195–1204. 10.1158/1541-7786.MCR-14-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.J., Chen M., Assanah M., Canoll P., and Manley J.L.. 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 463:364–368. 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L., Kerr A., and West S.. 2012. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 31:2566–2578. 10.1038/emboj.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeOcesano-Pereira C., Amaral M.S., Parreira K.S., Ayupe A.C., Jacysyn J.F., Amarante-Mendes G.P., Reis E.M., and Verjovski-Almeida S.. 2014. Long non-coding RNA INXS is a critical mediator of BCL-XS induced apoptosis. Nucleic Acids Res. 42:8343–8355. 10.1093/nar/gku561 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Devlin B., and Scherer S.W.. 2012. Genetic architecture in autism spectrum disorder. Curr. Opin. Genet. Dev. 22:229–237. 10.1016/j.gde.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Di Modugno F., Demonte L., Balsamo M., Bronzi G., Nicotra M.R., Alessio M., Jager E., Condeelis J.S., Santoni A., Natali P.G., and Nistico P.. 2007. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: Epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res. 67:2657–2665. 10.1158/0008-5472.CAN-06-1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatshad H., Pellagatti A., Fernandez-Mercado M., Yip B.H., Malcovati L., Attwood M., Przychodzen B., Sahgal N., Kanapin A.A., Lockstone H., et al. . 2015. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 29:1092–1103. 10.1038/leu.2014.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Sánchez M.S., Barroso S., Gomez-Gonzalez B., Luna R., and Aguilera A.. 2011. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 7:e1002386 10.1371/journal.pgen.1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverria G.V., and Cooper T.A.. 2012. RNA-binding proteins in microsatellite expansion disorders: Mediators of RNA toxicity. Brain Res. 1462:100–111. 10.1016/j.brainres.2012.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Nogales M., Cabrera J.R., Santos-Galindo M., Hoozemans J.J., Ferrer I., Rozemuller A.J., Hernandez F., Avila J., and Lucas J.J.. 2014. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat. Med. 20:881–885. 10.1038/nm.3617 [DOI] [PubMed] [Google Scholar]
- Fogel B.L., Wexler E., Wahnich A., Friedrich T., Vijayendran C., Gao F., Parikshak N., Konopka G., and Geschwind D.H.. 2012. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet. 21:4171–4186. 10.1093/hmg/dds240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D., and Ares M. Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15:689–701. 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K., DuHadaway J., Du W., Herlyn M., Rodeck U., and Prendergast G.C.. 1999. Mechanism for elimination of a tumor suppressor: Aberrant splicing of a brain-specific exon causes loss of function of Bin1 in melanoma. Proc. Natl. Acad. Sci. USA. 96:9689–9694. 10.1073/pnas.96.17.9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman L.T., Stoilov P., Maguire J., Damianov A., Lin C.H., Shiue L., Ares M. Jr., Mody I., and Black D.L.. 2011. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 43:706–711. 10.1038/ng.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman L.T., Meera P., Stoilov P., Shiue L., O’Brien J.E., Meisler M.H., Ares M. Jr., Otis T.S., and Black D.L.. 2012. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 26:445–460. 10.1101/gad.182477.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentien D., Kosmider O., Nguyen-Khac F., Albaud B., Rapinat A., Dumont A.G., Damm F., Popova T., Marais R., Fontenay M., et al. . 2014. A common alternative splicing signature is associated with SF3B1 mutations in malignancies from different cell lineages. Leukemia. 28:1355–1357. 10.1038/leu.2014.28 [DOI] [PubMed] [Google Scholar]
- Gerstberger S., Hafner M., and Tuschl T.. 2014. A census of human RNA-binding proteins. Nat. Rev. Genet. 15:829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I., Munita R., Agirre E., Dittmer T.A., Gysling K., Misteli T., and Luco R.F.. 2015. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 22:370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin M., Mohan A., Batra R., Lee K.Y., Charizanis K., Gomez F.J., Eddarkaoui S., Sergeant N., Buee L., Kimura T., et al. . 2015. MBNL sequestration by toxic RNAs and RNA misprocessing in the myotonic dystrophy brain. Cell Reports. 12:1159–1168. 10.1016/j.celrep.2015.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert T.A., Shen D., Ding L., Okeyo-Owuor T., Lunn C.L., Shao J., Krysiak K., Harris C.C., Koboldt D.C., Larson D.E., et al. . 2011. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 44:53–57. 10.1038/ng.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretarsdottir S., Helgason H., Helgadottir A., Sigurdsson A., Thorleifsson G., Magnusdottir A., Oddsson A., Steinthorsdottir V., Rafnar T., de Graaf J., et al. . 2015. A splice region variant in LDLR lowers non-high density lipoprotein cholesterol and protects against coronary artery disease. PLoS Genet. 11:e1005379 10.1371/journal.pgen.1005379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Schafer S., Greaser M.L., Radke M.H., Liss M., Govindarajan T., Maatz H., Schulz H., Li S., Parrish A.M., et al. . 2012. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 18:766–773. 10.1038/nm.2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Hammerle M., and Diederichs S.. 2013. MALAT1 — a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 91:791–801. 10.1007/s00109-013-1028-y [DOI] [PubMed] [Google Scholar]
- Hales C.M., Seyfried N.T., Dammer E.B., Duong D., Yi H., Gearing M., Troncoso J.C., Mufson E.J., Thambisetty M., Levey A.I., and Lah J.J.. 2014. U1 small nuclear ribonucleoproteins (snRNPs) aggregate in Alzheimer’s disease due to autosomal dominant genetic mutations and trisomy 21. Mol. Neurodegener. 9:15 10.1186/1750-1326-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman D.S., Lam L., Taylor M.R., Wang L., Teekakirikul P., Christodoulou D., Conner L., DePalma S.R., McDonough B., Sparks E., et al. . 2012. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366:619–628. 10.1056/NEJMoa1110186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highley J.R., Kirby J., Jansweijer J.A., Webb P.S., Hewamadduma C.A., Heath P.R., Higginbottom A., Raman R., Ferraiuolo L., Cooper-Knock J., et al. . 2014. Loss of nuclear TDP-43 in amyotrophic lateral sclerosis (ALS) causes altered expression of splicing machinery and widespread dysregulation of RNA splicing in motor neurones. Neuropathol. Appl. Neurobiol. 40:670–685. 10.1111/nan.12148 [DOI] [PubMed] [Google Scholar]
- Hsu T.Y., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., Echeverria G.V., Sun T., Kurley S.J., Tyagi S., et al. . 2015. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 525:384–388. 10.1038/nature14985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Liu Y.H., Sahashi K., Rigo F., Bennett C.F., and Krainer A.R.. 2015. Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev. 29:288–297. 10.1101/gad.256644.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan J.O., Ramakrishnan A., Hayes B., Murphy M.E., Zebari A.S., Bradley P., and Bradley R.K.. 2015. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 25:14–26. 10.1101/gr.181016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallieres M., Tapial J., Raj B., O’Hanlon D., et al. . 2014. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 159:1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner L.M., and Gotz J.. 2011. Amyloid-β and tau — a toxic pas de deux in Alzheimer’s disease. Nat. Rev. Neurosci. 12:65–72. 10.1038/nrn2967 [DOI] [PubMed] [Google Scholar]
- Janssens J., and Van Broeckhoven C.. 2013. Pathological mechanisms underlying TDP-43 driven neurodegeneration in FTLD-ALS spectrum disorders. Hum. Mol. Genet. 22(R1):R77–R87. 10.1093/hmg/ddt349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A., et al. . 2013. Mutational landscape and significance across 12 major cancer types. Nature. 502:333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., and Krainer A.R.. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185–193. 10.1038/nsmb1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M., Falaleeva M., and Stamm S.. 2013. Function of alternative splicing. Gene. 514:1–30. 10.1016/j.gene.2012.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfir N., Lev-Maor G., Glaich O., Alajem A., Datta A., Sze S.K., Meshorer E., and Ast G.. 2015. SF3B1 association with chromatin determines splicing outcomes. Cell Reports. 11:618–629. 10.1016/j.celrep.2015.03.048 [DOI] [PubMed] [Google Scholar]
- Kim E., Ilagan J.O., Liang Y., Daubner G.M., Lee S.C., Ramakrishnan A., Li Y., Chung Y.R., Micol J.B., Murphy M.E., et al. . 2015. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 27:617–630. 10.1016/j.ccell.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A., et al. . 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 495:467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh C.M., Bezzi M., Low D.H., Ang W.X., Teo S.X., Gay F.P., Al-Haddawi M., Tan S.Y., Osato M., Sabo A., et al. . 2015. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 523:96–100. 10.1038/nature14351 [DOI] [PubMed] [Google Scholar]
- Komeno Y., Huang Y.J., Qiu J., Lin L., Xu Y., Zhou Y., Chen L., Monterroza D.D., Li H., DeKelver R.C., et al. . 2015. SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol. Cell. Biol. 35:3071–3082. 10.1128/MCB.00202-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic-Kozaric A., Przychodzen B., Singh J., Konarska M.M., Clemente M.J., Otrock Z.K., Nakashima M., Hsi E.D., Yoshida K., Shiraishi Y., et al. . 2015. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia. 29:126–136. 10.1038/leu.2014.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C., et al. . 2012. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15:1488–1497. 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., and Rio D.C.. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84:291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C., et al. . 2013. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Reports. 5:1178–1186. 10.1016/j.celrep.2013.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFave C.V., Squatrito M., Vorlova S., Rocco G.L., Brennan C.W., Holland E.C., Pan Y.X., and Cartegni L.. 2011. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 30:4084–4097. 10.1038/emboj.2011.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., and Manley J.L.. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 122:365–378. 10.1016/j.cell.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Li Q., Zheng S., Han A., Lin C.H., Stoilov P., Fu X.D., and Black D.L.. 2014. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife. 3:e01201 10.7554/eLife.01201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Niu T., and Manley J.L.. 2007. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 13:2108–2115. 10.1261/rna.734407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.I., Sanchez-Pulido L., Haerty W., and Ponting C.P.. 2015. RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome Res. 25:1–13. 10.1101/gr.181990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Yano M., Fak J.J., Mele A., Grabinski S.E., Zhang C., and Darnell R.B.. 2012. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 26:1626–1642. 10.1101/gad.191338.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J.P., Pletnikova O., Troncoso J.C., and Wong P.C.. 2015. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 349:650–655. 10.1126/science.aab0983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., and Caceres J.F.. 2009. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 417:15–27. 10.1042/BJ20081501 [DOI] [PubMed] [Google Scholar]
- Love J.E., Hayden E.J., and Rohn T.T.. 2015. Alternative splicing in Alzheimer’s disease. J. Parkinsons Dis. Alzheimers Dis. 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R.F., Allo M., Schor I.E., Kornblihtt A.R., and Misteli T.. 2011. Epigenetics in alternative pre-mRNA splicing. Cell. 144:16–26. 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Solimini N.L., and Elledge S.J.. 2009. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell. 136:823–837. 10.1016/j.cell.2009.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Kanojia D., Li J., Okamoto R., Sato-Otsubo A., Kohlmann A., Sanada M., Grossmann V., Sundaresan J., Shiraishi Y., et al. . 2015. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 6:6042 10.1038/ncomms7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire S.L., Leonidou A., Wai P., Marchio C., Ng C.K., Sapino A., Salomon A.V., Reis-Filho J.S., Weigelt B., and Natrajan R.C.. 2015. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J. Pathol. 235:571–580. 10.1002/path.4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H., Visconte V., Sakaguchi H., Jankowska A.M., Abu Kar S., Jerez A., Przychodzen B., Bupathi M., Guinta K., Afable M.G., et al. . 2012. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 119:3203–3210. 10.1182/blood-2011-12-399774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A., Roesser J.R., and Chalfant C.E.. 2006. SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 20:1680–1682. 10.1096/fj.05-5021fje [DOI] [PubMed] [Google Scholar]
- Matera A.G., and Wang Z.. 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15:108–121. 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunawa M., Yamamoto R., Sanada M., Sato-Otsubo A., Shiozawa Y., Yoshida K., Otsu M., Shiraishi Y., Miyano S., Isono K., et al. . 2014. Haploinsufficiency of Sf3b1 leads to compromised stem cell function but not to myelodysplasia. Leukemia. 28:1844–1850. 10.1038/leu.2014.73 [DOI] [PubMed] [Google Scholar]
- Matter N., Herrlich P., and Konig H.. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 420:691–695. 10.1038/nature01153 [DOI] [PubMed] [Google Scholar]
- Mirtschink P., Krishnan J., Grimm F., Sarre A., Horl M., Kayikci M., Fankhauser N., Christinat Y., Cortijo C., Feehan O., et al. . 2015. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature. 522:444–449. 10.1038/nature14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykowska A., Sobczak K., Wojciechowska M., Kozlowski P., and Krzyzosiak W.J.. 2011. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 39:8938–8951. 10.1093/nar/gkr608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naftelberg S., Schor I.E., Ast G., and Kornblihtt A.R.. 2015. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 84:165–198. 10.1146/annurev-biochem-060614-034242 [DOI] [PubMed] [Google Scholar]
- Naro C., and Sette C.. 2013. Phosphorylation-mediated regulation of alternative splicing in cancer. Int. J. Cell Biol. 2013:151839 10.1155/2013/151839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryshkin N.A., Weetall M., Dakka A., Narasimhan J., Zhao X., Feng Z., Ling K.K., Karp G.M., Qi H., Woll M.G., et al. . 2014. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 345:688–693. 10.1126/science.1250127 [DOI] [PubMed] [Google Scholar]
- Nussbacher J.K., Batra R., Lagier-Tourenne C., and Yeo G.W.. 2015. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 38:226–236. 10.1016/j.tins.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan S.M., Hasselfeld K., Bauer D., Simmons M., Roussos P., Haroutunian V., Meador-Woodruff J.H., and McCullumsmith R.E.. 2015. Glutamate transporter splice variant expression in an enriched pyramidal cell population in schizophrenia. Transl. Psychiatry. 5:e579 10.1038/tp.2015.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldmeadow C., Mossman D., Evans T.J., Holliday E.G., Tooney P.A., Cairns M.J., Wu J., Carr V., Attia J.R., and Scott R.J.. 2014. Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J. Psychiatr. Res. 52:44–49. 10.1016/j.jpsychires.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Olshavsky N.A., Comstock C.E., Schiewer M.J., Augello M.A., Hyslop T., Sette C., Zhang J., Parysek L.M., and Knudsen K.E.. 2010. Identification of ASF/SF2 as a critical, allele-specific effector of the cyclin D1b oncogene. Cancer Res. 70:3975–3984. 10.1158/0008-5472.CAN-09-3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco T.R., Moita L.F., Gomes A.Q., Hacohen N., and Carmo-Fonseca M.. 2006. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell. 17:4187–4199. 10.1091/mbc.E06-01-0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini V., Naro C., and Sette C.. 2015. Splicing regulation: A molecular device to enhance cancer cell adaptation. BioMed Res. Int. 2015:543067 10.1155/2015/543067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino J., Swalley S.E., Song C., Cheung A.K., Shu L., Zhang X., Van Hoosear M., Shin Y., Chin D.N., Keller C.G., et al. . 2015. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 11:511–517. 10.1038/nchembio.1837 [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D., Pellagatti A., Wainscoat J.S., Hellstrom-Lindberg E., Gambacorti-Passerini C., et al. . 2011. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365:1384–1395. 10.1056/NEJMoa1103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasaikas P., Tejedor J.R., Vigevani L., and Valcarcel J.. 2015. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell. 57:7–22. 10.1016/j.molcel.2014.10.030 [DOI] [PubMed] [Google Scholar]
- Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C., et al. . 2011. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14:459–468. 10.1038/nn.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos P.I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M.T., et al. . 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 480:387–390. 10.1038/nature10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos M.G., Batra R., Charizanis K., and Swanson M.S.. 2011. Developments in RNA splicing and disease. Cold Spring Harb. Perspect. Biol. 3:a000778 10.1101/cshperspect.a000778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P., et al. . 2015. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 18:1175–1182. 10.1038/nn.4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przychodzen B., Jerez A., Guinta K., Sekeres M.A., Padgett R., Maciejewski J.P., and Makishima H.. 2013. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 122:999–1006. 10.1182/blood-2013-01-480970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Conde L., Villamor N., Ordonez G.R., Jares P., Bassaganyas L., Ramsay A.J., Bea S., Pinyol M., Martinez-Trillos A., et al. . 2011. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 44:47–52. 10.1038/ng.1032 [DOI] [PubMed] [Google Scholar]
- Quesnel-Vallières M., Irimia M., Cordes S.P., and Blencowe B.J.. 2015. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 29:746–759. 10.1101/gad.256115.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B., and Blencowe B.J.. 2015. Alternative splicing in the mammalian nervous system: Recent insights into mechanisms and functional roles. Neuron. 87:14–27. 10.1016/j.neuron.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Ramos A.D., Andersen R.E., Liu S.J., Nowakowski T.J., Hong S.J., Gertz C.C., Salinas R.D., Zarabi H., Kriegstein A.R., and Lim D.A.. 2015. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 16:439–447. 10.1016/j.stem.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.C., Morris K.V., and Wood M.J.. 2014. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M., Kasprzak W.K., Voss T., Shapiro B.A., Poulikakos P.I., and Misteli T.. 2015. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat. Commun. 6:7103 10.1038/ncomms8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman A.L., Pan Q., and Blencowe B.J.. 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 25:373–384. 10.1101/gad.2004811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D., et al. . 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. USA. 110:2366–2370. 10.1073/pnas.1221891110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage K.I., Gorski J.J., Barros E.M., Irwin G.W., Manti L., Powell A.J., Pellagatti A., Lukashchuk N., McCance D.J., McCluggage W.G., et al. . 2014. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell. 54:445–459. 10.1016/j.molcel.2014.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.M., and Rebel V.I.. 2013. Acquired mutations that affect pre-mRNA splicing in hematologic malignancies and solid tumors. J. Natl. Cancer Inst. 105:1540–1549. 10.1093/jnci/djt257 [DOI] [PubMed] [Google Scholar]
- Scotter E.L., Chen H.J., and Shaw C.E.. 2015. TDP-43 proteinopathy and ALS: Insights into disease mechanisms and therapeutic targets. Neurotherapeutics. 12:352–363. 10.1007/s13311-015-0338-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyén E., Zawisza M., and Eyras E.. 2015. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 43:1345–1356. 10.1093/nar/gku1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal L., Mathur R., Braun F.K., Wise J.F., Berkova Z., Neelapu S., Kwak L.W., and Samaniego F.. 2014. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 28:2376–2387. 10.1038/leu.2014.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C., Yang B., Wu T., Huang J., Tang P., Zhou Y., Zhou J., Qiu J., Jiang L., Li H., et al. . 2014. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 21:997–1005. 10.1038/nsmb.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro I.M., Cheng A.W., Flytzanis N.C., Balsamo M., Condeelis J.S., Oktay M.H., Burge C.B., and Gertler F.B.. 2011. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7:e1002218 10.1371/journal.pgen.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo Y., Siegfried Z., and Karni R.. 2015. The role of splicing factors in deregulation of alternative splicing during oncogenesis and tumor progression. Mol. Cell. Oncol. 2:e970955 10.4161/23723548.2014.970955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai C.L., Ley J.N., White B.S., Kim S., Tibbitts J., Shao J., Ndonwi M., Wadugu B., Duncavage E.J., Okeyo-Owuor T., et al. . 2015. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 27:631–643. 10.1016/j.ccell.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkreta L., and Chabot B.. 2015. The RNA splicing response to DNA damage. Biomolecules. 5:2935–2977. 10.3390/biom5042935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.K., and Cooper T.A.. 2012. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 18:472–482. 10.1016/j.molmed.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staropoli J.F., Li H., Chun S.J., Allaire N., Cullen P., Thai A., Fleet C.M., Hua Y., Bennett C.F., Krainer A.R., et al. . 2015. Rescue of gene-expression changes in an induced mouse model of spinal muscular atrophy by an antisense oligonucleotide that promotes inclusion of SMN2 exon 7. Genomics. 105:220–228. 10.1016/j.ygeno.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Sterne-Weiler T., and Sanford J.R.. 2014. Exon identity crisis: Disease-causing mutations that disrupt the splicing code. Genome Biol. 15:201 10.1186/gb4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A., Kilpinen S., Ruusulehto A., Lothe R.A., and Skotheim R.I.. 2015. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. In press. 10.1038/onc.2015.318 [DOI] [PubMed] [Google Scholar]
- Tanackovic G., Ransijn A., Thibault P., Abou Elela S., Klinck R., Berson E.L., Chabot B., and Rivolta C.. 2011. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 20:2116–2130. 10.1093/hmg/ddr094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.Y., Lee J.C., Hou M.F., Wang C.L., Chen C.C., Huang H.W., and Chang H.W.. 2013. Alternative splicing for diseases, cancers, drugs, and databases. ScientificWorldJournal. 2013:703568 10.1155/2013/703568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor J.R., Papasaikas P., and Valcarcel J.. 2015. Genome-wide identification of Fas/CD95 alternative splicing regulators reveals links with iron homeostasis. Mol. Cell. 57:23–38. 10.1016/j.molcel.2014.10.029 [DOI] [PubMed] [Google Scholar]
- Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V., et al. . 2011. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14:452–458. 10.1038/nn.2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers, J.A. van Ijcken W., Grosveld F.G., Medema R.H., Hoeijmakers J.H., et al. . 2015. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 523:53–58. 10.1038/nature14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. . 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 39:925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen J.J., Niemela E.H., Verma B., and Frilander M.J.. 2013. The significant other: Splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA. 4:61–76. 10.1002/wrna.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Ule A., Spencer J., Williams A., Hu J.S., Cline M., Wang H., Clark T., Fraser C., Ruggiu M., et al. . 2005. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 37:844–852. 10.1038/ng1610 [DOI] [PubMed] [Google Scholar]
- Valacca C., Bonomi S., Buratti E., Pedrotti S., Baralle F.E., Sette C., Ghigna C., and Biamonti G.. 2010. Sam68 regulates EMT through alternative splicing–activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 191:87–99. 10.1083/jcb.201001073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S., Marney C.B., Shu W., Valiente M., Zou Y., Mele A., Darnell R.B., and Massague J.. 2014. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife. 3:e02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz-Drago R., Pinheiro M.T., Martins S., Enguita F.J., Carmo-Fonseca M., and Custodio N.. 2015. Transcription-coupled RNA surveillance in human genetic diseases caused by splice site mutations. Hum. Mol. Genet. 24:2784–2795. 10.1093/hmg/ddv039 [DOI] [PubMed] [Google Scholar]
- Venables J.P., Klinck R., Koh C., Gervais-Bird J., Bramard A., Inkel L., Durand M., Couture S., Froehlich U., Lapointe E., et al. . 2009. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16:670–676. 10.1038/nsmb.1608 [DOI] [PubMed] [Google Scholar]
- Venables J.P., Brosseau J.P., Gadea G., Klinck R., Prinos P., Beaulieu J.F., Lapointe E., Durand M., Thibault P., Tremblay K., et al. . 2013. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell. Biol. 33:396–405. 10.1128/MCB.01174-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconte V., Rogers H.J., Singh J., Barnard J., Bupathi M., Traina F., McMahon J., Makishima H., Szpurka H., Jankowska A., et al. . 2012. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood. 120:3173–3186. 10.1182/blood-2012-05-430876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., and Geschwind D.H.. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 474:380–384. 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., and Luhrmann R.. 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell. 136:701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Walter M.J., Ding L., Shen D., Shao J., Grillot M., McLellan M., Fulton R., Schmidt H., Kalicki-Veizer J., O’Laughlin M., et al. . 2011. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 25:1153–1158. 10.1038/leu.2011.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Sashida G., Saraya A., Ishiga R., Koide S., Oshima M., Isono K., Koseki H., and Iwama A.. 2014a Depletion of Sf3b1 impairs proliferative capacity of hematopoietic stem cells but is not sufficient to induce myelodysplasia. Blood. 123:3336–3343. 10.1182/blood-2013-12-544544 [DOI] [PubMed] [Google Scholar]
- Wang G., Li Z., Zhao Q., Zhu Y., Zhao C., Li X., Ma Z., Li X., and Zhang Y.. 2014b LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol. Rep. 31:1839–1845. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang J., Li K., Zhao W., and Cui Q.. 2012. SpliceDisease database: Linking RNA splicing and disease. Nucleic Acids Res. 40(D1):D1055–D1059. 10.1093/nar/gkr1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck S.M., Mele A., Yan Q., Sun S., Farny N., Zhang Z., Xue C., Herre M., Silver P.A., Zhang M.Q., et al. . 2014. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Reports. 6:1139–1152. 10.1016/j.celrep.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe V.O., Gonzalez-Porta M., Perera D., Bartolozzi A.R., Sibley C.R., Hallegger M., Ule J., Marioni J.C., and Venkitaraman A.R.. 2015. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5′ splice site strength. Genome Biol. 16:201 10.1186/s13059-015-0749-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., et al. . 2015. The human splicing code reveals new insights into the genetic determinants of disease. Science. 347:1254806 10.1126/science.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gao X.D., Lee J.H., Huang H., Tan H., Ahn J., Reinke L.M., Peter M.E., Feng Y., Gius D., et al. . 2014. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 28:1191–1203. 10.1101/gad.241968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., and Weinberg R.A.. 2008. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell. 14:818–829. 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yang C., Wang H., Qiao T., Yang B., Aliaga L., Qiu L., Tan W., Salameh J., McKenna-Yasek D.M., Smith T., et al. . 2014a Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 111:E1121–E1129. 10.1073/pnas.1322641111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bennett B.D., Luo S., Inoue K., Grimm S.A., Schroth G.P., Bushel P.R., Kinyamu H.K., and Archer T.K.. 2015. LIN28A modulates splicing and gene expression programs in breast cancer cells. Mol. Cell. Biol. 35:3225–3243. 10.1128/MCB.00426-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Han L., Yuan Y., Li J., Hei N., and Liang H.. 2014b Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat. Commun. 5:3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Jin N., Gu J., Shi J., Zhou J., Gong C.X., Iqbal K., Grundke-Iqbal I., and Liu F.. 2012. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J. Biol. Chem. 287:30497–30506. 10.1074/jbc.M112.355412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., et al. . 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 478:64–69. 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- Yu Y., and Reed R.. 2015. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc. Natl. Acad. Sci. USA. 112:8608–8613. 10.1073/pnas.1506282112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Chi B., Xia W., Gangopadhyay J., Yamazaki T., Winkelbauer-Hurt M.E., Yin S., Eliasse Y., Adams E., Shaw C.E., and Reed R.. 2015. U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Res. 43:3208–3218. 10.1093/nar/gkv157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., and Peng G.. 2015. Non-coding RNAs: An emerging player in DNA damage response. Mutat. Res. Rev. Mutat. Res. 763:202–211. 10.1016/j.mrrev.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Zhang J., and Manley J.L.. 2013. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 3:1228–1237. 10.1158/2159-8290.CD-13-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Frias M.A., Mele A., Ruggiu M., Eom T., Marney C.B., Wang H., Licatalosi D.D., Fak J.J., and Darnell R.B.. 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 329:439–443. 10.1126/science.1191150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lieu Y.K., Ali A.M., Penson A., Reggio K.S., Rabadan R., Raza A., Mukherjee S., and Manley J.L.. 2015a Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. USA. 112:E4726–E4734. 10.1073/pnas.1514105112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S., et al. . 2015b The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 525:56–61. 10.1038/nature14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., and Dreyfuss G.. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 133:585–600. 10.1016/j.cell.2008.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Pinto A.M., Wan L., Wang W., Berg M.G., Oliva I., Singh L.N., Dengler C., Wei Z., and Dreyfuss G.. 2013. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 110:19348–19353. 10.1073/pnas.1319280110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.L., Luo G., Wise J.A., and Lou H.. 2014a Regulation of alternative splicing by local histone modifications: Potential roles for RNA-guided mechanisms. Nucleic Acids Res. 42:701–713. 10.1093/nar/gkt875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Hasty P., Walter C.A., Bishop A.J., Scott L.M., and Rebel V.I.. 2013. Myelodysplastic syndrome: An inability to appropriately respond to damaged DNA? Exp. Hematol. 41:665–674. 10.1016/j.exphem.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li X., Cheng Y., Wu W., Xie Z., Xi Q., Han J., Wu G., Fang J., and Feng Y.. 2014b BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat. Commun. 5:4581 10.1038/ncomms5581 [DOI] [PubMed] [Google Scholar]