Abstract
Background
Domesticated animals quickly evolve docile and submissive behaviors after isolation from their wild conspecifics. Model organisms reared for prolonged periods in the laboratory also exhibit similar shifts towards these domesticated behaviors. Yet whether this divergence is due to inadvertent selection in the lab or the fixation of deleterious mutations remains unknown.
Results
Here, we compare the genomes of lab-reared and wild-caught Drosophila melanogaster to understand the genetic basis of these recently endowed behaviors common to laboratory models. From reassembled genomes of common lab strains, we identify unique, derived variants not present in global populations (lab-specific SNPs). Decreased selective constraints across low frequency SNPs (unique to one or two lab strains) are different from patterns found in the wild and more similar to neutral expectations, suggesting an overall accumulation of deleterious mutations. However, high-frequency lab SNPs found in most or all lab strains reveal an enrichment of X-linked loci and neuro-sensory genes across large extended haplotypes. Among shared polymorphisms, we also find highly differentiated SNPs, in which the derived allele is higher in frequency in the wild (Fst*wild>lab), enriched for similar neurogenetic ontologies, indicative of relaxed selection on more active wild alleles in the lab.
Conclusions
Among random mutations that continuously accumulate in the laboratory, we detect common adaptive signatures in domesticated lab strains of fruit flies. Our results demonstrate that lab animals can quickly evolve domesticated behaviors via unconscious selection by humans early on a broad pool of disproportionately large neurogenetic targets followed by the fixation of accumulated deleterious mutations on functionally similar targets.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-015-0580-1) contains supplementary material, which is available to authorized users.
Keywords: Behavior, Adaptation, Purifying selection, Positive selection, Extended haplotypes, Domestication genomics, Model organisms, Domestication syndrome
Background
Our recent history of domesticating plants and animals [1] offers a diversity of genetic systems to study evolution in action [2]. Crop and livestock breeders often provoke relatively large phenotypic changes across successive generations conditioned on available standing genetic variation found in wild progenitor populations. Such changes demonstrate the formidable power of directional selection over relatively short periods of time. In fact, Charles Darwin devoted the opening chapter of “On the origins of species” to artificial selection in order to introduce natural selection as the principal driver of evolutionary change [3]. In his two-volume follow-up devoted specifically to domestication, Darwin noted that “selection may be followed either methodically and intentionally, or unconsciously and unintentionally” [4]. These histories can also be modeled as a co-evolutionary framework between humans and the crops and livestock they cultivate [5], whether the selective pressures were intentional or not.
While domesticates usually have reduced effective population sizes relative to their ancestral populations, there still remains ample variation for selection to act upon. Numerous loci involved in animal and crop domestication are found to harbor positive selection coefficients [6] and selectively swept regions [7]. In domestic chickens, genomic sequences from multiple lines reveal the presence of selective sweeps leading to the discovery of causative agents in growth differences between domestic lines [8]. In household pets, selection for certain behavioral and sensory traits [9] have produced signatures of positive selection in vision and hearing genes in domesticated cats [10] and neural development genes in dogs [11]. The growing literature on crop, livestock, and pet domestication reveals that long-term selection by humans can generate strong signals of selection at the genomic level and provides a new lens into the strength and target of selection during our recent domesticated past.
Animals bred in the laboratory as model organisms may also be adapted to human conditions, and over a much shorter time period. Studies comparing laboratory strains of mice and nematodes have identified genetic differences in genes involved in behavior [12] and metabolism [13, 14] suggesting adaptation to novel conditions in the laboratory (e.g., [15]). Over a century ago, Drosophila melanogaster was brought into the laboratory initially as a teaching tool [16], and its fast generation time and relative ease of maintenance quickly made the fruit fly an important genetics research tool [17] in such varied fields such as development, physiology, and evolution. Canton-S(pecial), the oldest known wildtype fly stock, was captured by Calvin Bridges over a century ago from a natural population in Canton, Ohio, and first debuted in his seminal 1916 paper on “Non-disjunction as proof of the chromosome theory of heredity” [18]. Approximately a decade later, Donald Lancefield, another product of Thomas Morgan’s prolific lab at Columbia University, extracted Oregon-R from a population in Roseburg, Oregon [19]. Many commonly used fly stocks eventually coalesce ancestrally to these old North American laboratory stalwarts, Canton-S and Oregon-R (Fig. 1), which were independently extracted from populations in North America and themselves possessing relatively recent African origins ([20]; Fig. 1).
Fig. 1.
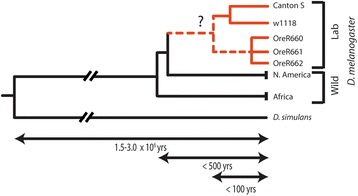
Hypothetical genealogical relationship of laboratory strains in relation to global populations of D. melanogaster and the closely related species, D. simulans. The five lab strains are indicated in red with hypothetical ancestral relationships marked as a dotted line. Estimated time since common ancestors are indicated below
With at least a dozen generations per year, D. melanogaster lab stocks have been isolated from their wild ancestors for over a thousand generations, providing ample time to sufficiently diverge. It is known among fruit fly researchers that behavioral traits of laboratory vs. wild D. melanogaster are distinct, with lab-reared flies far tamer and more manageable than those found in the wild. Although lab-reared flies are generally maintained under relatively standard conditions of temperature, light, and diet, selective pressures in the lab are very different than those found in the wild providing the potential for strong selection for human-accommodating phenotypes and/or the relaxation of selection on traits adapted in nature [21, 22]. On the other hand, domesticates are generally less reproductively fit than their wild relatives [23] and may exhibit similar less optimal behavioral phenotypes. Lab stocks typically experience drastic decreases in population size and higher levels of inbreeding ultimately decreasing the efficacy of selection to rid lines of continually re-occurring deleterious alleles [24]. Whether these phenotypic differences in lab strains are driven by conscious selection, inadvertent selection, a relaxation in selection, or are simply the fixation of deleterious mutations, is not known.
Its relatively recent and well-documented history, access to multiple isolated lines reared in similar environmental settings, well-characterized functional annotations, and the availability of hundreds of genomes from extensively sampled extant populations including a closely related species, make D. melanogaster an ideal model system to study the evolutionary processes that underlie rapid phenotypic change. Here, we investigate genetic differences between common laboratory stains of D. melanogaster to those recently caught from nature to examine whether this important, centuries-old genetic model has evolved convergent domesticated behaviors by adapting to a bottled existence or being inundated by low-fitness alleles. We first document behavioral differences between lab-reared and wild-caught flies with respect to their general activity and locomotory abilities. We then identify SNPs that are differentiated between the genomes of laboratory and wild strains to evaluate the roles of selection vs. drift in flies isolated in bottles. Among uniquely derived alleles found only in lab strains we find patterns of drift and selection across, respectively, low and high frequency classes, when comparing evolutionary parameters such as Grantham distance, missense to silent mutations, positional distribution within codons, and levels of codon bias. We further characterize putative regions under sustained selection among lab strains and find longer than expected haplotypes near high-frequency derived SNPs, also enriched in neuro-sensory genes. Finally, we suggest that this neurogenetic class, residing on a disproportionately large fraction of the genome relative to other functional classes, provides a large mutational target for genetic variation to accumulate and selection to act upon. Thus, the genomics of fly domestication reveal an interplay of evolutionary forces with mutation and selection on a large neurogenetic class of genes playing a pivotal role in D. melanogaster’s brief, but distinguished, history in the laboratory.
Results
Behavior
Although it is well-known among Drosophilists that lab strains are much slower and easier to handle than live flies, literature documenting these differences is lacking. We first confirm anecdotal reports of differences in the overall activity between laboratory stocks (Canton-S, Oregon-R, w1118) and flies from wild populations (Additional file 1: Figure S1). On average, flies from lab strains are significantly less active than wild-caught individuals using different measures of locomotion including standard and angular velocities (Additional file 1: Figure S1A,B; Wilcoxon P < 0.05) and time-spent moving vs. stationary (Additional file 1: Figures S1C, S1D; Wilcoxon P < 0.05). Lab strains also demonstrate less responsiveness in the form of interactions between individuals compared to wild-caught lines where the latter’s movement is more uniformly distributed and gradually increases with proximity to neighbors (Additional file 1: Figure S1E). Flies from laboratory strains do not appear to follow this relationship with motion uncorrelated to the proximity with their nearest neighbor (Additional file 1: Figure S1F). Overall, these results provide general support for a convergence of slow-moving and less responsive behavior in lab strains.
Genetic variation in lab strains
After applying filters for quality and missing data, a total of 98,442,787 base pairs were analyzed across five reassembled laboratory strains of D. melanogaster, 516 genomes from 23 global populations including an extensively sampled population from North America (n = 205), and one closely related species, D. simulans. Among 14,545,645 polymorphic sites, 68.5 % are non-singletons and used for subsequent analyses (Additional file 2: Table S1). To test for signals of domestication, we used three defined categories of SNPs differentiated between lab strains and populations of D. melanogaster from nature: lab-specific SNPs and two types of highly differentiated FST (Fst*) SNPs, depending on whether the derived allele is found at a higher frequency among lab strains (Fst*lab>wild), or in a representative natural population (Fst*wild>lab). While distinct, some overlap exists between the lab-specific and Fst*lab>wild analysis categories: out of a total 50,565 differentiated SNPs, 520 SNPs are common to these two SNP classes (Fig. 2a). In contrast, Fst*wild>lab SNPs are unique and relatively rare (n = 123; Fig. 2a).
Fig. 2.
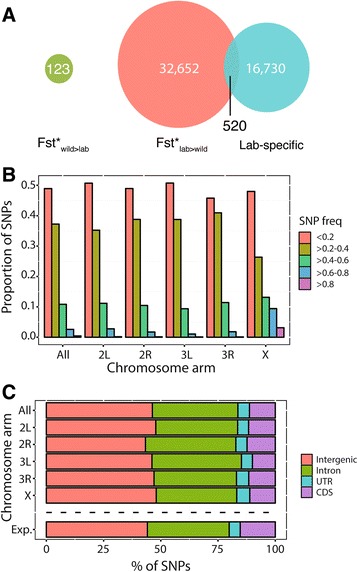
Characterization of derived SNPs differentiated between lab strains and a wild D. melanogaster population. a Number and overlap of lab-specific SNPs (derived and unique among lab strains) with Fst*lab>wild SNPs (derived allele is higher in frequency among lab strains compared to sequenced flies from Raleigh NC). The total number of Fst*wild>lab SNPs (the derived allele is shared and lower in frequency among lab strains) is also shown. b Site frequency distribution of lab-specific SNPs, per chromosome. c Genic characterization of lab-specific SNPs found in the majority (at least 3/5) of laboratory strains, across chromosomal arms
Lab-specific SNPs
The number of lab-specific SNPs is 17,250, with 9,258 and 1,951 SNPs located in genic and CDS regions, respectively (Fig. 2c). The total number of lab-specific SNPs is significantly larger than a subsample of five random genomes from North Carolina and comparison of the number of SNPs specific to this random subset against the entire North Carolinian dataset (performed 1,000 times; Sign test, P < 0.002) as well as the entire global dataset (Sign test, P < 0.001). Lab-specific SNPs represent only a small fraction (0.017 %) of the total SNP variation found among global populations of D. melanogaster, including singletons. Lab-specific SNPs are distributed across all chromosome arms with a slight but significant enrichment across non-coding regions (e.g., intergenic, 1.02x, χ2 = 33.56, P < 0.001; intronic, 1.05x, χ2 = 19.11, P < 0.001; Fig. 2b, c). The X-chromosome is enriched for mid- and high-frequency lab-specific SNPs (χ2 = 201.48, P < 0.001). The number of fixed lab-specific SNPs (i.e., found in all five lab strains, or 4/4 due to missing data) was 75, 46, and 32, respectively, in all genomic regions, genic, and CDS regions (Additional file 2: Table S1). Genes containing fixed lab-specific SNPs are enriched for regulatory and signaling gene ontology (GO) categories (Additional file 3: Table S2; Additional file 4: Table S3) and up-regulated in neural-sensory tissues (Additional file 5: Figure S2).
Fst*lab>wild SNPs
The number of derived SNPs with high FST (i.e., Fst*) that harbor a greater frequency in the lab compared to the wild is 33,172, with 18,284 and 4,116 in genic and CDS regions respectively (Additional file 2: Table S1). Fst*lab>wild SNPs generally describe variants that are highly represented (≥80 %) among lab strains but have a low (<0.35) allele frequency in global populations. Fst*lab>wild are uniformly distributed across chromosome arms and are enriched for genic regions (regulatory, 1.07x, χ2 = 104.09, P < 0.001; Additional file 2: Table S1). The number of Fst*lab>wild SNPs that are fixed (i.e., found in all lab strains) is 30,396 and 3,829, respectively, in all genomic regions and CDS regions (Additional file 2: Table S1). Fst*lab>wild SNPs are functionally enriched in general developmental gene ontology categories (100+ GO categories are significantly enriched; Additional file 3: Table S2; Additional file 4: Table S3). Fst*lab>wild SNPs are upregulated in several tissues including ganglion, larval CNS, and ovary (Additional file 5: Figure S2).
Fst*wild>lab SNPs
A small proportion of Fst* SNPs (n = 123) are highly differentiated in the opposite direction. These Fst*wild>lab SNPs are found at low frequency (≤25 %) among lab strains but have high allele frequencies in global populations (>93 %). Fst*wild>lab SNPs are enriched on the X-chromosome (1.6x, χ2 = 9.45, P < 0.01), across intergenic regions (1.25x, χ2 = 4.14, P < 0.05), and significantly enriched for neurogenetic genes (3.6x, χ2 = 28.73, P < 0.001; Additional file 3: Table S2). Specifically, this SNP category is significantly enriched in nervous system development and photoreceptor development gene ontologies and is upregulated in neural, visual, and ovarian tissues (Additional file 4: Table S3, Additional file 5: Figure S2).
Evolutionary relationships
Phylogenetic analyses on lab-specific SNPs and Fst* SNPs were performed to provide insight on the origin and relationship among isolated lab stocks (Additional file 6: Figure S3). The lab-specific consensus SNP tree reveals Canton-S as ancestral to the three Oregon-R strains and the w1118 strain, as expected (Additional file 6: Figure S3A). The Fst* SNP consensus tree similarly shows a distinct lab monophyletic clade with similar bootstrap support for lab strain topology (Additional file 6: Figure S3B). A distinct origin among lab strains is also seen using a random set of 100,000 polymorphic sites among lab and wild strains (Additional file 6: Figure S3C). The branch lengths of the lab strains dramatically differ between trees indicating each lab strain’s distinctiveness (with the exception of OreR-661 and OreR-662) from each other (Additional file 6: Figure S3A) and the extant North American population (Additional file 6: Figure S3B). The three congruent phylogenetic trees also revealed several surprises including the three Oregon-R strains being paraphyletic, with Ore-661 and Ore-662 more similar to w1118 than Ore-660. Also, while the original Oregon-R strain was independently sampled on the US west coast a few decades later, all lab strains appear to be derived from a single Canton-S common ancestor (Additional file 6: Figure S3C), with the lab clade best supported next to extant populations from France, then North America.
Genome-wide levels of selection
To detect differences in selective constraints in the lab, we estimate a series of evolutionary parameters (Grantham distance, R/S ratio, C1/C2+C3, codon bias) across five frequency bins of lab-specific SNPs located in coding regions. We compare parameter estimates against similarly binned derived SNPs found only in the Raleigh NC population along with simulation estimates expected under strict neutrality. Additional file 2: Table S1 compares the nature and number of lab-differentiated SNPs to those found in the Raleigh NC population across autosomes and X-chromosomes. When not binned according to allele frequency, mean Grantham scores, R/S, and C1/(C2+C3) ratios are similar to random simulations. However, when grouped by frequency class, parameter estimates of medium to high frequency SNPs more closely follow a pattern of similarly binned SNPs from the wild rather than expected neutral patterns based on a random mutational model (Fig. 3).
Fig. 3.
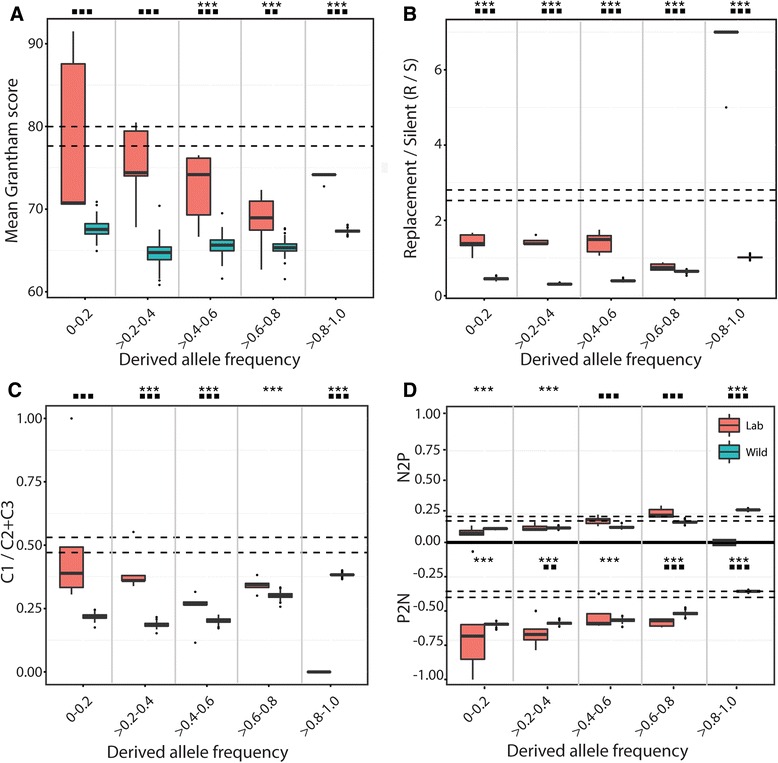
Frequency distribution of lab-specific (red), wild-caught (green), and neutrally simulated derived (between dotted lines) SNPs across various evolutionary parameters. a Mean Grantham score per replacement substitution. b Mean replacement/silent (R/S) ratio per SNP per individual. c Mean ratio of the number of first codon position (C1) SNPs to the number of second and third codon position (C2+C3) SNPs. d Mean proportion of non-preferred to preferred (N2P) codons and preferred to non-preferred (P2N) codons per individual. For all panels, dotted black horizontal lines represent one standard deviation from the mean of 1000 random simulations using D. simulans sequence data (matched to the number of lab-specific SNPs). Statistical significance determined using a Wilcoxon rank-sum test. Asterisks indicate significant differences between lab-specific SNPs and SNPs derived from neutral simulations (***P-value < 0.001, ** P-value < 0.05). Squares indicate significant differences between lab-specific SNPs and SNPs derived from the Raleigh NC population (■■■P-value < 0.001, ■■P-value < 0.05)
Effective population size
The effective population size is a strong determinant of the effectiveness of selection and drift [25, 26]. Watterson’s theta, Θs [27], was estimated across non-overlapping 50 kb windows to compare the amount of genetic variation from an extant population to that found among all lab strains. Like previous genome-wide estimates of nucleotide diversity from the Raleigh NC population [28–30], our estimates of Θ similarly fluctuate across genomic regions with a genome-wide average of ΘNC = 0.0053 (Additional file 7: Figure S4). Although laboratory strains do not collectively comprise a true interbreeding population, we estimate Θlab to provide a relative measure of ancestral Ne. Laboratory nucleotide diversity varies along the genome in a similar fashion as the Raleigh NC population with a genome-wide mean of Θlab = 0.00312 (Additional file 7: Figure S4), indicating at least a two-fold reduction in ancestral Ne.
Extended haplotype blocks
Regions of extended homozygosity present potential signals of positive selection. We observe a mean haplotype length for all lab vs. wild differentiated classes of SNPs (lab-specific, Fst*lab>wild, Fst*wild>lab) of 653 bp (SD = 801 bp). Among lab-specific SNPs, long outlier haplotypes were only found within the high frequency class ≥0.8 and were significantly larger than all other frequency classes (Wilcoxon rank sum test, P = 3.41 × 10−4, Fig. 4). Under a neutral model, we would expect similar haplotype lengths across all frequency classes. In addition, we have run simulations in which we choose sites at random and estimate haplotype lengths. Using 1000 replicates, the random site haplotype lengths are at least two standard errors lower than the observed 5/5 haplotype lengths. A total of 457 (lab-specific: 112, Fst*lab>wild: 342, Fst*wild>lab: 3) large haplotype block outliers (Zhap >2.5) were identified ranging in length from 2,622 bp to 11,985 bp and were significantly enriched (by nearly four times the expected amount) on the X-chromosome (Fig. 5). Candidate lab-specific, Fst*lab>wild, and Fst*wild>lab haplotype blocks contain, respectively, 135, 334, and 4 genes. Genes found within these lab-specific candidate haplotype blocks are enriched for neurogenetic gene ontology categories along with functional classes related to regulation and behavior responses (Table 1) as well as other significant functional categories (Additional file 8: Table S4). However, when normalized by gene length, significant GO category enrichments in large lab-specific haplotype blocks disappear. Within large Fst*lab>wild haplotype blocks, genes are enriched for GO classes involved with axon guidance, post-embryonic system development, and regulation (Table 1), even after normalizing for gene length. No significant GO enrichment is found in the four genes contained within large Fst*wild>lab haplotype blocks.
Fig. 4.
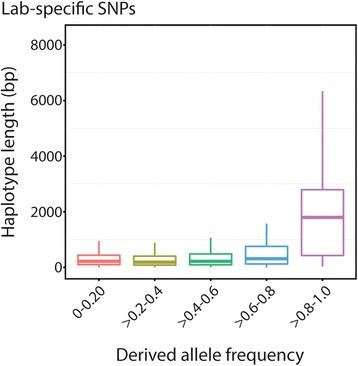
Lab-specific SNP haplotype length across frequency class. Boxplot of extended haplotype length surrounding lab-specific SNPs across derived frequency allele classes
Fig. 5.
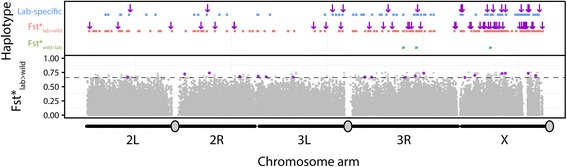
Neurogenetic enrichment of extended haplotype blocks and Fst*wild>lab SNP genomic distribution. In the top panel, colored rectangles indicates haplotype blocks surrounding lab-differentiated SNPs (lab-specific: blue, Fst*lab>wild: red, Fst*wild>lab: green). Size distribution of haplotype blocks per frequency class is found in Fig. 4. Presence of known neurogenetic genes in each block is indicated by purple arrows. In the bottom panel, FST scan identifies derived SNPs in genic regions with higher allele frequency in the Raleigh NC population relative to the laboratory strains (Fst*wild>lab). Dotted line indicates cutoff for Fst*wild>lab SNPs (Z-score > 2.5, above dotted line). Of the 62 Fst* SNPs found in genic regions, SNPs are significantly enriched for neurogenetic genes (P < 0.001). SNPs found within neurogenetic genes are shaded in purple
Table 1.
Enriched gene ontologies (GO) on extended haplotype block outliers for two categories of differentiated SNPs: unique among laboratory strains (lab-specific) or higher in allele frequency in the lab (Fst*lab>wild)
| SNP Type | GO: Biological Process | GO: ID | # of Genes in GOa | **P-value | Count |
|---|---|---|---|---|---|
| Lab-specific | regulation of cellular component size | 32535 | 305 | 0.028412 | 8 |
| Lab-specific | regulation of biological quality | 65008 | 1994 | 0.038348 | 23 |
| Lab-specific | generation of neurons | 48699 | 992 | 0.039188 | 22 |
| Lab-specific | nervous system development | 7399 | 1744 | 0.040839 | 32 |
| Lab-specific | neurogenesis | 22008 | 1525 | 0.041256 | 30 |
| Lab-specific | regulation of anatomical structure size | 90066 | 374 | 0.042371 | 10 |
| Lab-specific | regulation of cell size | 8361 | 89 | 0.042507 | 6 |
| Lab-specific | cellular component organization | 16043 | 5855 | 0.042708 | 43 |
| Lab-specific | proboscis extension reflex | 7637 | 9 | 0.043535 | 3 |
| Lab-specific | reflex | 60004 | 9 | 0.043535 | 3 |
| Lab-specific | cell differentiation | 30154 | 2770 | 0.043641 | 41 |
| Lab-specific | regulation of cell morphogenesis | 22604 | 184 | 0.044224 | 9 |
| Lab-specific | axon development | 61564 | 341 | 0.047764 | 12 |
| Lab-specific | behavioral response to nutrient | 51780 | 10 | 0.048507 | 3 |
| Fst*lab>wild | motor neuron axon guidance | 8045 | 62 | 0.007739 | 9 |
| Fst*lab>wild | system development | 48731 | 2850 | 0.00912 | 88 |
| Fst*lab>wild | post-embryonic organ development | 48569 | 583 | 0.009155 | 29 |
| Fst*lab>wild | multicellular organismal development | 7275 | 3977 | 0.009225 | 106 |
| Fst*lab>wild | biological regulation | 65007 | 13920 | 0.009452 | 112 |
| Fst*lab>wild | single-organism cellular process | 44763 | 23874 | 0.014023 | 162 |
| Fst*lab>wild | regulation of biological process | 50789 | 13269 | 0.015888 | 106 |
| Fst*lab>wild | regulation of cellular process | 50794 | 12824 | 0.023125 | 96 |
| Fst*lab>wild | single-multicellular organism process | 44707 | 5981 | 0.024058 | 118 |
| Fst*lab>wild | post-embryonic appendage morphogenesis | 35120 | 452 | 0.026085 | 24 |
| Fst*lab>wild | appendage development | 48736 | 470 | 0.026896 | 24 |
aNumber of genes from Gene Ontology (Biological Process) from FlyBase FB2015_05
**P-value calculated using Benjamini-Hochberg correction
Gene ontologies within each SNP type are ranked by P-value. No significant gene ontology class was found for haplotypes classified for significantly differentiated SNPs in which the derived SNP is found in lower frequency in the wild (i.e., Fst*wild>lab)
Discussion
Phenotypic and genetic differences in laboratory stocks
The captive genetic model, Drosophila melanogaster, reveals hallmark features consistent with domestication. Laboratory strains show significant differences in behavior from their relatively recently isolated wild progenitors (Additional file 1: Figure S1). Previous studies comparing lab and wild-caught lines of D. melanogaster also report differences in egg and larval survival [31] and life history traits such as pre-adult development, early fecundity, and remating frequency [32, 33]. These traits comprise a suite of behavioral, physiological, and reproductive characters [34] that have converged across multiple strains evolving independently under similar laboratory conditions. Furthermore, it previously has been demonstrated that these traits can quickly evolve significant differences in as little as 8–10 generations [35].
It is paradoxical that domesticates, typically derived from small founder populations and maintained at very low effective population sizes, can effectively adapt to human conditions. From a population genetics perspective, we may expect the opposite: that small captive populations and lower Ne propagate the segregation and eventual fixation of deleterious mutations, thus, imposing a potentially large mutational burden on laboratory strains [36, 37]. Our evolutionary parameter analysis reveals such drift-like signatures at the low end of the site frequency spectrum. Estimates of Grantham distances, R/S ratios, codon positional fraction, and codon bias support a general genome-wide reduction in selection on low-frequency derived alleles in the lab (Fig. 3) where mildly deleterious alleles may persist for longer periods of time [38]. With a decrease in effective population size across each strain, inbreeding and drift dominate, which in turn, can quickly fix allelic and associated phenotypic changes across successive generations of captivity. Short-term isolation studies in Drosophila have shown similar rapid changes across a variety of phenotypes [35, 39] with reductions in performance levels [40]). Thus, the accumulation and fixation of mildly deleterious alleles, particularly on larger neurogenetic genes (see below), and subsequent inbreeding depression [41, 42] may promote both the rapid nature of domestication and its associated convergence of common behavioral traits. These docile and non-aggressive traits may alternatively be described as “lethargic” (at least relative to their wild-caught conspecifics), a term commonly applied to inbred, and often sickly, Drosophila stocks.
When did these changes occur? Our results suggest that these changes towards a domesticated phenotype likely began very early on based on available standing genetic variation of the progenitor population. While heterozygosity in each contemporary isofemale lab strain is virtually zero (data not shown), as a whole, these five lab strains collectively only harbor a two-fold genome-wide decrease in nucleotide diversity, Θlab, relative to a large contemporary population from Raleigh NC (Additional file 7: Figure S4). This diversity estimate primarily reflects the amount of genetic variation captured in each of the five lines from an ancestral population(s). The five lab strains share ~800,000 derived SNPs with extant global populations (a total of ~10 million non-singleton D. melanogaster SNPs pass our data filters) indicating that laboratory stocks collectively extracted a significant fraction D. melanogaster genetic variation at their time of capture.
The importance of ancestral standing genetic variation can also be seen in the 17,250 lab-specific SNPs. In theory, these SNPs represent any of the following: i) de novo mutations that arose in the lab, ii) genetic variation that previously existed in an extinct North American population, or iii) a subset of genetic variation that has been completely lost in extant global populations. We estimate that only a small fraction of these SNPs can be generated de novo (3.5 × 10−9 mutations/bp/generation x 120 × 106 bp x 20 generations/yr x ~75 years x 5 strains ≈ 3,150 lab-specific SNPs [43]). The remainder of the lab-specific SNPs was probably lost in extant wild populations during the last century. Strong evidence supports a recent global sweep in D. melanogaster that dramatically reduced species-wide genetic variation after these particular lab strains were collected [44]. Thus, from the large pool of available genetic variation from their North American progenitor populations, Canton-S and Oregon-R likely experienced similar selection pressures on common genetic variants (see below) during the earliest generations of lab domestication [45]. A phylogenetic analysis of shared lab/wild SNPs also supports a distinct origin of all lab strains (Additional file 6: Figure S3). However, whether the lab strain monophyly is the result of an extinct progenitor population or the loss of global variation is unknown. The inclusion of more sequenced lab strains may differentiate between these two hypotheses.
Adaptation in the lab
Caenorhabditis elegans, like D. melanogaster, has been lab-cultured for over 50 years, and harbors pronounced differences in longevity and fertility when compared to wild isolates [46]. Genome sequencing in the nematode identified SNPs differentiated between wild and long-term laboratory strains enriched for cell cycle and metabolic/growth genes [13]. Their results suggest the presence of strong selection early in nematode domestication for optimal growth under rich nutrient conditions similar to the significant GO term, “behavioral response to nutrients”, found among lab-specific SNPs in fruit flies (Table 1). Laboratory mice have similarly been shown to converge certain phenotypes including melatonin deficiency [15], and a lack of aggression and tameness [47, 48]. The results from our evolutionary parameter analysis, when limited to lab-specific SNPs in the mid- to high frequency range, support that common phenotypic signals of domestication observed in the captive fruit fly have been strongly shaped by selection. These derived SNPs, found in the majority of lab strains, show similar evolutionary patterns to high frequency SNPs from the wild and not to neutral expectations, unlike low-frequency SNPs (Fig. 3). The laboratory setting presents an immediate change in the fitness landscape, permitting rapid and significant changes in phenotype that would be detrimental to their fitness in the wild, across relatively few generations [35, 36].
Inadvertent human habituation and unintentional conditioning may be the primary selective agent for such known differences among lab strains as faster development and reproductive time [13, 14]. For instance, flies that rarely escape the bottle or benchtop may be selected due to human carelessness while fly stocks are transferred to new vials/bottles, or “flipped”. Our behavioral results support such a convergent shift towards less active and responsive flies (Additional file 1: Figure S1). From our genomic analysis, we find that neurogenetic genes, involved in such biological processes as neurogenesis and axon development, are enriched in extended haplotype blocks common to differentiated SNPs (both lab-specific and highly differentiated Fst*) found at high allele frequencies (≥0.8), with an overrepresentation of fixations on the X-chromosome. These genes affect locomotion and visual cues suggesting lab selection on genes involved in behavioral responses.
While an excess of long extended haplotypes on high frequency lab-specific SNPs support an adaptive shift towards domesticated phenotypes, a relaxation of selection on certain loci involved in behavior may have co-occurred in the lab. Conditions in the laboratory are often optimized for growth and reproduction, reducing the natural ability of flies to escape predators or compete for food and mates. Thus, a relaxation of selection on activity levels, aggressiveness, and responsiveness, critical in the wild, may also drive the behavioral differences that converged across lab strains. Characters involved in mating, driven by the sparsity of mates in the vicinity, is a key difference between domestic and wild species [49]. Our behavioral results, showing a reduction in interactive activity in lab flies, is consistent with this hypothesis. In addition, our GO analysis of shared Fst*wild>lab SNPs, in which the derived allele is more frequent in the Raleigh NC population than lab strains, finds a significant enrichment of the neurogenetic functional class, even when corrected for gene size. These SNPs are found in different genes than the SNPs harbored in long extended haplotype blocks, suggesting an extensive cache of genes involved in behavioral differences between flies reared in the lab and those found in the wild. A similar decrease in behavioral activity was observed in lab strains of mice [50], with backcrossing to wild mice isolates allowing them to regain these previously lost behavioral functions [47].
The use of inbred laboratory strains of D. melanogaster presented unique analytical challenges that differ from other domesticated studies. Due to initial and recurrent inbreeding and the lack of an interbreeding population, laboratory strains violate most population genetic models used to infer selection. In this study, the site frequency spectrum is only applied across isolated lab strains as a framework to bin our observed data and could not be used to infer population genetic parameters. Despite these difficulties, our results reveal an interplay of drift and selection at work in the lab. First, we find genome-wide levels of selective constraints in the lab that are significantly lower than a sampled North American population. This pattern is likely caused by low effective population sizes in bottles that promote the accumulation of mildly deleterious mutations under drift-like conditions, which we also observe in low frequency alleles. Second, we observe derived SNPs that are highly differentiated between the lab and a North American population to be significantly enriched in neurogenetic genes, suggesting a differential fitness landscape in behavior. This functional enrichment takes into account the number of genes in each functional class as well as their size. Third, we find signatures of positive selection on extended haplotypes in both lab-specific and highly differentiated SNPs. These, too, are significantly enriched in neurogenetic genes. Fourth, there’s an enrichment of these changes on the X-chromosome (Fig. 2; Fig. 5; Additional file 2: Table S1). The preferential role of the X-chromosome is seen in marked differences in the site frequency spectrum between the X-chromosome and autosomes (Fig. 2b) and in the enrichment of long haplotype blocks on the X-chromosome (Fig. 4). Since many of these long X-linked haplotype blocks are fixed in all five lab strains, the fixation of hemizygous loci likely occurred early in fly domestication.
Preferential role for neurogenetic genes
Our results suggest a central role for neurogenetic genes in domestication. Lab-specific SNPs found in the majority of lab strains are strongly enriched for this functional class, as are highly differentiated SNPs found in high frequency in the wild (Fst*wild>lab SNPs; Additional file 4: Table S3). Large outlier haplotype blocks also contain an overrepresentation of neurogenetic genes (Fig. 5; Additional file 8: Table S4). In most fly labs, inadvertent selection is inevitable: more active, reactive, and sensory-prone flies (and their alleles) have a higher probability of escaping during routine stock transfers. Hence, fly researchers may have unconsciously selected for lethargic flies over thousands of generations in the lab. Selected genes, enriched for sensory functions in eye photoreceptors and peripheral nervous system, can explain these behavioral shifts seen in lab strains. The significance of neurogenetic genes in changing activity and response behaviors across a relatively short evolutionary time period may also relate to how behaviors involved in premating isolation [51–53] can swiftly and easily develop in a population by drift and selection.
Recently, Wilkins et al. [54] proposed a general hypothesis to explain the convergence of various phenotypic traits that differentiate mammalian domesticates from their wild progenitors. These traits are collectively known as the “domestication syndrome” [55, 56] and, in mammals, include such morphological modifications as depigmentation, facial skeletal, and floppy ears as well as behavioral shifts towards docility and tameness [57]. Wilkins et al. [54] argue that a developmental deficit in neural crest genes can generate each of these differences, thus, explaining the commonality of these traits across domesticated mammals. Our results extend the behavioral component of the domestication syndrome to non-vertebrates but through a more general genomic mechanism based on the predominance of mutations on neurogenetic genes affecting overall locomotion and activity. In Drosophila, genes from this ontological category are among the largest in gene number and gene size, providing a large mutational target for rapid behavioral change (Additional file 9: Figure S5). Currently, 1,708 out of 17,716 genes are characterized as “neurogenetic” (according to FlyBase R6.05), and 24 % of known fly genes are expressed in the brain and nervous system [58]. We propose that a large mutational target [85] of neurogenetic genes can explain the rapid evolution of behavior in animal taxa., These neurogenomic loci collectively provide a large genomic substrate for variation to accumulate, and then selection and drift to act, to quickly transform behavior within a relatively short time frame.
Conclusions
Tameness and docility are hallmark features of domestication and the product of artificial selection by breeders. Our results challenge the traditional notion that relatively submissive laboratory animals are solely the product of cumulating deleterious mutations and demonstrate how unconscious selection for human-favored traits plays an important role in driving rapid phenotypic change in the lab. Selection on a large pool of available genetic variation during the early stages of fly domestication, followed by strong and recurrent inbreeding, allow for the successive roles of adaptation and drift in shaping the genetic architecture of domesticated phenotypic traits in a bottle. Our study finds that the genes and phenotypes in fruit fly domestication are enriched in, respectively, neurogenetic and behavioral function, providing a starting point to decode the genomic basis of domestication and promoting its study in genetic model systems such as Drosophila. A detailed mapping of these genes and their SNPs to specific behaviors will not only be informative about the selective pressures that we have inadvertently applied to our immediate biotic environment, but may also provide new general insight on the divergence and isolation of populations.
Methods
Behavioral assays
Locomotory assays were performed separately on five lines of adult D. melanogaster (6–8 days post-eclosion), each of North American origin. Three lines (Canton-S, w1118, and Oregon-R) represent common laboratory stocks originally extracted from nature at least seventy years ago (Fig. 1). Two wild-caught lines were collected from Linvilla PA and Lancaster MA and reared under normal laboratory conditions for less than one year without specific selective regimes (courtesy of the Schmidt lab, University of Pennsylvania). Stocks were maintained in the laboratory at ~24 C, at ~40 % relative humidity, kept in standard 250 ml bottles on Lewis food medium [59], and exposed to a 12 h light–dark cycle. Prior to behavioral assays, flies are anesthetized with light CO2 sedation (<15 s) for transfer and identification, and allowed to acclimatize in the arena setting for 30 min, post-sedation. Assays are conducted in Delrin arenas (McMaster-Carr) following specifications outlined in Simon and Dickinson [60] to optimize mobility and provide an effective environment for automated tracking. To prevent locomotion on the ceilings, glass covers are coated with Sigmacote (Sigma Aldrich). Between assays, arenas are rinsed with ethanol and allowed to dry for a minimum of 15 min to ensure no residues remain from previous behavioral experiments.
Assays are conducted during active afternoon periods across successive days. Each line is recorded using three independent replicates per line tested on different days and randomized to reduce experimental bias. Fly activity, post-acclimation, is recorded for 30 min. Individual and interactive activities are tracked using CTRAX and MATLAB [61]. Errors in the initial tracking are corrected using CTRAX’s Fix errors scripts. All output measurements are analyzed using MATLAB and statistics implemented using custom R scripts.
Genomic data sources
Whole genomic sequencing (125 bp paired-end) reads from three separate Oregon-R lines were downloaded from NCBI (SRX671605, SRX671606, SRX671607). Illumina 150 bp paired-end reads from Canton-S and w1118 were obtained from the Hawley lab (Stowers Institute). Reference assemblies were generated by aligning filtered reads against the D. melanogaster genome following methods described by Lack et al. [62], an assembly pipeline that adds an intermediate realignment step for the purpose of aligning reads around insertion and deletion sites. Briefly, paired-end reads were aligned using BWA v0.7.12 [63] against the complete D. melanogaster reference genome (Dmel Release 5) obtained from FlyBase (flybase.org). Post-alignment files were transformed using SAMtools [64] and Picard v1.79 (broadinstitute.github.io/picard/). Bases are filtered for a minimum quality score of 30 and a minimum read depth of 15x from VCF files generated by the Genome Analysis Toolkit [65]. Additional file 10: Table S5 includes a brief summary of the raw data.
To identify mutational states in the lab, 516 full genome assemblies from natural populations of D. melanogaster were downloaded from the Drosophila Genome Nexus [62] representing 23 countries from Africa, Europe, and North America [20, 66, 67]. Genome-genome alignment of D. simulans R2 assembly [68] against the D. melanogaster R5 assembly was performed using Progressive Mauve [69] using default parameters. To validate the quality of our alignment, the average number of nucleotide substitutions (Dxy; [70]) was estimated for 100 kb non-overlapping windows (Additional file 11: Figure S6) and genome-wide patterns compared to previous literature [71].
Genomic filters and annotations
All genomic analyses were restricted to euchromatic chromosome arms (2L, 2R, 3L, 3R, X). To minimize sampling biases, the combined dataset was subjected to coverage filters for missing data. For a particular site to be used, a minimum 75 % of the laboratory strains (n > 3) and a minimum of 75 % of the population samples (n ≥ 388) must contain a non-ambiguous nucleotide, with no more than two alleles present (i.e., only monoallelic and diallelic sites were included). SNPs are identified across all filtered base pairs, with singletons from global populations excluded to conservatively reduce the effects of sequencing error from low-coverage global samples. Data were also filtered for the presence of a D. simulans allele to infer ancestral state. After filtering for data quality, coverage, and ancestral state, 98,442,787 eligible sites (Additional file 12: Table S6) were used to identify SNPs differentiated between the lab and wild.
We identify several types of derived mutations: SNPs unique to lab strains and SNPs significantly differentiated between lab stains and the wild. Derived SNPs uniquely found in labs (i.e., the lab strain(s) possess a base neither present in known global populations of D. melanogaster nor D. simulans) are classified as “lab-specific” SNPs. Highly differentiated SNPs, often shared across both lab and wild samples, were identified via Hudson’s FST estimator [72, 73] with FST scores Z-transformed as follows: Z-FST = (FST - μFST)/σFST. SNPs with FST estimates harboring a Z-score > 2.5 were considered highly differentiated (Fst*) SNPs. Fst* SNPs are further classified as either: i) “Fst*lab>wild” if the derived allele is found at a higher frequency in the lab, or ii) “Fst*wild>lab” if the derived allele is higher in frequency in the wild. Both lab-specific and Fst* SNPs can be further categorized as polymorphic (1/5,…4/5) or fixed (5/5, or 4/4 in the case of missing data) with respect to their frequency among the five sequenced lab strains.
SNPs are annotated by genomic location (e.g., genic vs. inter-genic) using FlyBase R5.9. SNPs located within a gene model, represented by their longest transcript, are further classified according to their annotated position within a gene model (5’UTR, exon, intron, 3’UTR), and their codon position (C1, C2, C3) if found within an exon. SNPs found within exonic regions are also classified as non-synonymous or synonymous. The relative fitness of each amino acid substitution is estimated using a Grantham score [74], which evaluates biochemical dissimilarity (based on polarity, amino acid size, and side chain composition) between ancestral and derived states, with lower Grantham scores indicating a greater biochemical similarity. Synonymous codon shifts are categorized into four separate classes (P2P, preferred codon > preferred codon; P2N, preferred codon > non-preferred codon; N2P, non-preferred codon > preferred codon; N2N, non-preferred codon > non-preferred codon) according to the classification of Vicario et al. [75].
Detecting selective signals
To determine whether selection is acting in the lab, we compared evolutionary patterns of coding region variation in lab-specific SNPs against: i) a North American population comprising of 205 DGRP genomes from Raleigh NC [28, 62] and ii) neutral expectations. Since negative and positive selection differentially affects the site frequency spectrum, we bin these differentiated SNPs according to their shared frequency among lab strains: with five strains, the site frequency spectrum is divided into fifths.
To generate neutrally simulated data, n mutations, based on the number of SNPs found in the CDS of lab strains, are randomly assigned to D. melanogaster coding regions. SNPs within CDS regions are labeled according to their codon position (C1, C2, C3) and classified as a non-synonymous or synonymous substitution with synonymous SNPs classified as preferred or non-preferred codons. A simple model of equal probability of changing any position within the codon to another nucleotide is applied. 1000 simulations are performed. After binning these simulated data into the five allele frequency classes, we estimate basic evolutionary parameters including mean Grantham score of amino acid substitutions, proportion of non-synonymous SNPs, fraction of polymorphisms in 1st codon position, and shifts in codon preference. Wilcoxon rank-sum tests [76] are used to compare these neutral estimates, as well as those from the North Carolina population (n = 205), against parameter estimates from the lab strains (n = 5).
Recent domestication studies have surveyed genomic regions for significantly reduced heterozygosity [8, 77, 78] to identify selectively swept candidate genes. However, heterozygosity is rare, if not absent, in isogenic strains of D. melanogaster (data not shown). For each SNP, we estimate the mean population (i.e., laboratory) haplotype length conditioned on frequency class. Haplotype length analyses are performed using custom perl scripts allowing for non-congruent haplotypes to extend from each lab-specific SNP. Due to the lower sequencing coverage of lab strains, a maximum of one individual per site is permitted to have missing data. Large outlier haplotype blocks are identified by a Z-hap score > 2.5.
Phylogenetic and functional enrichment analyses
To understand the topological relationship among lab strains, neighbor-joining trees [79] are generated using p-distance [70] and bootstrapped 1,000 times [80] for laboratory-specific and high Fst* (Fst*lab>wild + Fst*wild>lab) SNP sets. To evaluate the topologies between lab strains and extant populations, 100,000 SNPs that are shared in lab and nature were randomly chosen for NJ tree analysis and bootstrapped 1,000 times using MEGA6 [81]. Overrepresented gene ontologies for differentiated SNPs are identified using DAVID [82] and FlyMine [83]. Gene sets are weighted according to the size of their categories and a False Discovery Rate (FDR) is used to correct for the deployment of multiple tests. For selected enrichment analyses, gene lengths were used to normalize the potential impact of genes from certain GO categories covering a disproportionate fraction of the genome. Gene annotation data for tissue specificity and ontogenetic stages are characterized using FlyAtlas [84].
Acknowledgements
Special thanks to Jody Hey, Alexander Platt, Yujin Chung, Ravi Patel, and members of Temple University’s Center for Computational Genetics and Genomics (CCGG) and the Institute for Genomics and Evolutionary Medicine (iGEM) for valuable suggestions to improve our manuscript. We would also like to thank Emily Berhman for wild-caught flies, Danny Miller for wild-type stocks and access to WGS reads, John Williams for assistance with the fly behavioral assays, Justin Lack for assistance with genome assembly, and Matt Hansen for his work on aligning D. melanogaster and D. simulans. This work is partly funded by NSF grant 1407006.
Abbreviations
- bp
base pairs
- CDS
coding sequence
- Fst*
highly differentiated Fst
- Fst*lab>wild
high Fst with larger allele frequency in lab
- Fst*wild>lab
high Fst with larger allele frequency in wild
- GO
Gene Ontology
- CNS
Central Nervous System
- R/S
ratio of replacement to synonymous substitutions
- C1/C2+C3
ratio of the first position codon to 2nd and 3rd position codons
- Θ
nucleotide diversity
- SNP
single nucleotide polymorphism
- Zhap
Z-score haplotype
- SD
standard deviation
- NC
North Carolina
- NCBI
National Center for Biotechnology Information
- UTR
untranslated region
- P2N (P2P
N2P, N2N), preferred to non-preferred codons
- DGRP
Drosophila Genome Reference Panel
Additional files
Differences in activity between laboratory and wild-caught Drosophila. (A) Mean fraction of time spent moving per angular velocity bin (radians/sec) for laboratory (red) and wild-caught (green) flies. Shading indicates standard error across replicates (B) Mean fraction of time spent moving per forward velocity bin. Laboratory flies spend significantly greater proportion of time at lower angular and forward velocity then their wild conspecifics (P-value < 0.05, Mann–Whitney U Test). (C) Mean fraction of time per fly spent walking during a 30 min assay. (D) Fraction of time per fly spent stationary (velocity = 0 m/s). (E-F) Relationship of distance to nearest neighboring fly and its velocity. Heatmap colors denote velocity gradient for wild (E) and lab (F) flies. Wild-caught flies are generally more active with greater velocity when in closer proximity to other flies. (TIF 32946 kb)
SNP characterization. Number and genomic location of laboratory-specific, high-FST/high lab (Fst*lab>wild), and high-FST/low lab SNPs (Fst*wild>lab). Expected proportions based on genic fractions garnered from FlyBase Dmel R5.9. Random simulations capped using the number of laboratory-specific SNPs. (XLSX 43 kb)
Gene characterization. Characterization of genes containing highly differentiated lab SNPs. (XLSX 244 kb)
Gene ontology. Significant Gene Ontology (GO) classes for genes containing highly differentiated SNPs. Enrichments were normalized by gene lengths among gene ontology categories. Significance adjusted for multiple tests via Benjamini-Hochberg correction (Benjamini and Hochberg 1995). (XLSX 48 kb)
Tissue expression. Distribution of tissue expression for coding region SNPs highly differentiated between lab strains and wild-caught lines. (A) Lab-specific SNPs, (B) Fst*lab>wild SNPs, (C) Fst*wild>lab SNPs. (TIF 10759 kb)
Neighbor-joining trees. Phylogenetic trees for (A) lab-specific SNPs (B) highly differentiated (Fst*) SNPs, and (C) random 100,000 polymorphic sites. Bootstrap values for 1000 replicates are placed at each node. Numeric node labels represent individuals from lab strains (1–5), Raleigh, NC (6–210), France (211–219), and Africa (220–521). Location of the laboratory strains are highlighted using red branches. (TIF 38440 kb)
Genome-wide nucleotide diversity. Genome-wide distribution of nucleotide diversity (θs) across 50,000 bp non-overlapping windows in (A) a Raleigh NC population and (B) all lab strains. Centromeres are denoted as ovals. (TIF 15720 kb)
Large extended haplotype gene characterization. Characterization of genes found within large extended haplotype blocks surrounding SNPs (Lab-specific, Fst*lab>wild, and Fst*wild>lab). (XLSX 29 kb)
Genomic coverage of functional classes. (A) Genes are functionally classified by gene ontology and may overlap multiple classes. Neural functional class is highlighted in blue. (B) Comparison between average size of neural vs. non-neural functional classes across gene regions. (TIF 7433 kb)
Summary of sequenced reads, strain origin, and distribution of lab-specific SNPs for each assembled laboratory strain. Counts within parentheses denote the number of lab-specific SNPs found among the five laboratory strains for each of the frequency classes, 1/5, 2/5, 3/5, 4/5, and 5/5. Each assembly used paired-end Illumina reads. All reads were filtered for quality and assembled against the D. melanogaster R5 genome (see Materials and Methods for further details). (XLSX 8 kb)
Genome-wide nucleotide substitution (Dxy) plot for D. simulans and D. melanogaster. Dxy was calculated using 100,000 bp non-overlapping windows. Ovals denote centromeres. (TIF 14250 kb)
Base call quality control. Genome quality control filters use sequence coverage and the presence of a D. simulans R2 base call across chromosome arms. The number of lab-specific SNPs are indicated per chromosome arm. (XLSX 42 kb)
Footnotes
Competing interests
The authors have neither financial nor non-financial competing interests.
Authors’ contributions
CES and RJK both conceived and were involved in the design of the study, performed the genomic analyses, and drafted the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Craig E. Stanley, Jr, Email: craig.stanley@temple.edu.
Rob J. Kulathinal, Email: robkulathinal@temple.edu
References
- 1.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–7. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 2.Trut L, Oskina I, Kharlamova A. Animal evolution during domestication: the domesticated fox as a model. Bioessays. 2009;31:349–60. doi: 10.1002/bies.200800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darwin CR. On the origin of species by means of natural selection. London: Murray; 1859. [Google Scholar]
- 4.Darwin CR. The variation of animals and plants under domestication. London: Murray; 1868. [PMC free article] [PubMed] [Google Scholar]
- 5.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–8. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 6.Hufford MB, Xu X, van Heerwaarden J, Pyhaejaervi T, Chia J-M, Cartwright RA, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44:808–11. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, et al. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 2014;46:1220–6. doi: 10.1038/ng.3117. [DOI] [PubMed] [Google Scholar]
- 8.Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–91. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll CA, Clutton-Brock J, Kitchener AC, O'Brien SJ. The taming of the cat. Sci Am. 2009;300:68–75. doi: 10.1038/scientificamerican0609-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SMJ, et al. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci U S A. 2014;111:17230–5. doi: 10.1073/pnas.1410083111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Axelsson E, Ratnakumar A, Arendt ML, Maqbool K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–4. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 12.Goto T, Tanave A, Moriwaki K, Shiroishi T, Koide T. Selection for reluctance to avoid humans during the domestication of mice. Genes Brain Behav. 2013;12:760–70. doi: 10.1111/gbb.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber KP, De S, Kozarewa I, Turner DJ, Babu MM, de Bono M. Whole genome sequencing highlights genetic changes associated with laboratory domestication of C. elegans. PLoS ONE. 2010;5:e13922. doi: 10.1371/journal.pone.0013922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sterken MG, Snoek LB, Kammenga JE, Andersen EC. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 2015;31:224–31. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara T, Abe K, Mekada K, Yoshiki A, Kato T. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc Natl Acad Sci U S A. 2010;107:6412–7. doi: 10.1073/pnas.0914399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler RE. Drosophila: A life in the laboratory. J Hist Biol. 1993;26:281–310. doi: 10.1007/BF01061971. [DOI] [PubMed] [Google Scholar]
- 17.Allen GE. The introduction of Drosophila into the study of heredity and evolution: 1900–1910. Isis. 1975;66:322–33. doi: 10.1086/351472. [DOI] [PubMed] [Google Scholar]
- 18.Bridges CB. Non-Disjunction as proof of the chromosome theory of heredity. Genetics. 1916;1:107–63. doi: 10.1093/genetics/1.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsley DL, Grell EH. Genetic variants of Drosophila melanogaster. Washington DC: Carnegie Institute; 1968. [Google Scholar]
- 20.Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, et al. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latter BD, Mulley JC. Genetic adaptation to captivity and inbreeding depression in small laboratory populations of Drosophila melanogaster. Genetics. 1995;139:255–66. doi: 10.1093/genetics/139.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sgro CM, Geddes G, Fowler K, Partridge L. Selection on age at reproduction in Drosophila melanogaster: Female mating frequency as a correlated response. Evolution. 2000;54:2152–5. doi: 10.1111/j.0014-3820.2000.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 23.Shabalina SA, Yampolsky LY, Kondrashov AS. Rapid decline of fitness in panmictic populations of Drosophila melanogaster maintained under relaxed natural selection. Proc Natl Acad Sci U S A. 1997;94:13034–9. doi: 10.1073/pnas.94.24.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, Tang T, Tang H, Huang JZ, Shi SH, Wu CI. The accumulation of deleterious mutations in rice genomes: A hypothesis on the cost of domestication. Trends Genet. 2006;22:126–31. doi: 10.1016/j.tig.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Kimura M. Rare variant alleles in the light of the neutral theory. Mol Biol Evol. 1983;1:84–93. doi: 10.1093/oxfordjournals.molbev.a040305. [DOI] [PubMed] [Google Scholar]
- 26.Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 27.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–76. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 28.Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, Tarone AM, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24:1193–208. doi: 10.1101/gr.171546.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sackton TB, Kulathinal RJ, Bergman CM, Quinlan AR, Dopman EB, Carneiro M, et al. Population genomic inferences from sparse high-throughput sequencing of two populations of Drosophila melanogaster. Genome Biol Evol. 2009;1:449–65. doi: 10.1093/gbe/evp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andolfatto P, Przeworski M. Regions of lower crossing over harbor more rare variants in African populations of Drosophila melanogaster. Genetics. 2001;158:657–65. doi: 10.1093/genetics/158.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohane MJ, Parsons PA. Environment-dependent fitness differences in Drosophila melanogaster - Temperature, domestication and the alcohol-dehydrogenase locus. Heredity. 1986;57:289–304. doi: 10.1038/hdy.1986.126. [DOI] [Google Scholar]
- 32.Sgro CM, Partridge L. Evolutionary responses of the life history of wild‐caught Drosophila melanogaster to two standard methods of laboratory culture. Am Nat. 2000;156:341–53. doi: 10.1086/303394. [DOI] [Google Scholar]
- 33.Houle D, Rowe L. Natural selection in a bottle. Am Nat. 2003;161:50–67. doi: 10.1086/345480. [DOI] [PubMed] [Google Scholar]
- 34.Larson G, Piperno DR, Allaby RG. Current perspectives and the future of domestication studies. 2014:341–53. [DOI] [PMC free article] [PubMed]
- 35.Frankham R, Loebel DA. Modeling problems in conservation genetics using captive Drosophila populations - Rapid genetic adaptation to captivity. Zoo Biol. 1992;11:333–42. doi: 10.1002/zoo.1430110505. [DOI] [Google Scholar]
- 36.Lynch M, O'Hely M. Captive breeding and the genetic fitness of natural populations. Conserv Genet. 2001;2:363–78. doi: 10.1023/A:1012550620717. [DOI] [Google Scholar]
- 37.Renaut S, Rieseberg LH. The accumulation of deleterious mutations as a consequence of domestication and improvement in sunflowers and other Compositae crops. Mol Biol Evol. 2015;32:2273–83. doi: 10.1093/molbev/msv106. [DOI] [PubMed] [Google Scholar]
- 38.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–8. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 39.Briscoe DA, Malpica JM, Robertson A, Smith GJ, Frankham R, Banks RG, et al. Rapid loss of genetic-variation in large captive populations of Drosophila flies - Implications for the genetic management of captive populations. Conserv Biol. 1992;6:416–25. doi: 10.1046/j.1523-1739.1992.06030416.x. [DOI] [Google Scholar]
- 40.Giesel JT. Genetic correlation structure of life history variables in outbred, wild Drosophila melanogaster: effects of photoperiod regimen. Am Nat. 1986;128:593–603. doi: 10.1086/284590. [DOI] [Google Scholar]
- 41.Miller PS, Hedrick PW. Inbreeding and fitness in captive populations - Lessons from Drosophila. Zoo Biol. 1993;12:333–51. doi: 10.1002/zoo.1430120404. [DOI] [Google Scholar]
- 42.Vaiserman AM, Zabuga OG, Kolyada AK, Pisaruk AV, Kozeretska IA. Reciprocal cross differences in Drosophila melanogaster longevity: An evidence for non-genomic effects in heterosis phenomenon? Biogerontology. 2013;14:153–63. doi: 10.1007/s10522-013-9419-6. [DOI] [PubMed] [Google Scholar]
- 43.Keightley PD, Trivedi U, Thomson M, Oliver F. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;190:1195–201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kidwell MG. The evolutionary history of the P-family of transposable elements. J Hered. 1994;85:339–46. doi: 10.1093/oxfordjournals.jhered.a111478. [DOI] [PubMed] [Google Scholar]
- 45.Dyck VA, Hendrichs J, Robinson AS. Sterile insect technique. Netherlands: Springer; 2005. [Google Scholar]
- 46.Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chalfin L, Dayan M, Levy DR, Austad SN, Miller RA, Iraqi FA, et al. Mapping ecologically relevant social behaviours by gene knockout in wild mice. Nat Commun. 2014;5:4569. doi: 10.1038/ncomms5569. [DOI] [PubMed] [Google Scholar]
- 48.Goto K, Kurashima R, Watanabe S. Delayed matching-to-position performance in C57BL/6N mice. Behav Processes. 2010;84:591–7. doi: 10.1016/j.beproc.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Kokko H, Brooks R, Jennions MD, Morley J. The evolution of mate choice and mating biases. Proc R Soc Lond B. 2003;270:653–64. doi: 10.1098/rspb.2002.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshiki A, Ike F, Mekada K, Kitaura Y, Nakata H, Hiraiwa N, et al. The mouse resources at the RIKEN BioResource center. Exp Anim. 2009;58:85–96. doi: 10.1538/expanim.58.85. [DOI] [PubMed] [Google Scholar]
- 51.Wu CI, Hollocher H, Begun DJ. Sexual isolation in Drosophila melanogaster: A possible case of incipient speciation. Proc Natl Acad Sci U S A. 1995;92:2519–23. doi: 10.1073/pnas.92.7.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362. doi: 10.2307/2409213. [DOI] [PubMed] [Google Scholar]
- 53.Coyne JA, Orr HA. Speciation. Sunderland MA: Sinauer Associates; 2004. [Google Scholar]
- 54.Wilkins AS, Wrangham RW, Fitch WT. The “Domestication Syndrome” in mammals: A unified explanation based on neural crest cell behavior and genetics. Genetics. 2014;197:795–808. doi: 10.1534/genetics.114.165423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer K. Das domestikationssyndrom. Die Kulturpflanze. 1984;32:11–34. doi: 10.1007/BF02098682. [DOI] [Google Scholar]
- 56.Broom DM. Domestication - The decline of environmental appreciation. Nature. 1990;348:402. doi: 10.1038/348402a0. [DOI] [Google Scholar]
- 57.Belyaev DK. Domestication of animals. Sci J. 1969;5:47–52. [Google Scholar]
- 58.Graveley BR, Brooks AN, Carlson J, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–9. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis EB. A new standard food medium. Drosophila Information Service. 1960;34:117–8. [Google Scholar]
- 60.Simon JC, Dickinson MH. A new chamber for studying the behavior of Drosophila. PLoS One. 2010;5(e8793). [DOI] [PMC free article] [PubMed]
- 61.Branson K, Robie AA, Bender J, Perona P, Dickinson MH. High-throughput ethomics in large groups of Drosophila. Nat Methods. 2009;6:451–7. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, et al. The Drosophila genome nexus: A population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics. 2015;199:1229–41. doi: 10.1534/genetics.115.174664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, et al. Genomic variation in natural populations of Drosophila melanogaster. Genetics. 2012;192:533–98. doi: 10.1534/genetics.112.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482:173–8. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu TT, Eisen MB, Thornton KR, Andolfatto P. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013;23:89–98. doi: 10.1101/gr.141689.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darling AE, Mau B, Perna NT. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5. [DOI] [PMC free article] [PubMed]
- 70.Nei M, Kumar S. Molecular evolution and phylogenetics. USA: Oxford University Press; 2000. [Google Scholar]
- 71.Nolte V, Pandey RV, Kofler R, Schlotterer C. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res. 2013;23:99–110. doi: 10.1101/gr.139873.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: The impact of rare variants. Genome Res. 2013;23:1514–21. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–9. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grantham R. Amino-acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 75.Vicario S, Moriyama EN, Powell JR. Codon usage in twelve species of Drosophila. BMC Evol Biol. 2007;7:226. doi: 10.1186/1471-2148-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilcoxon F. Individual comparisons by ranking methods. Biometrics Bulletin. 1945;1:80–3. doi: 10.2307/3001968. [DOI] [Google Scholar]
- 77.Rubin CJ, Megens HJ, Barrio AM. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA. 2012;109:19529–36. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carneiro M, Rubin C-J, Di Palma F, Albert FW, Alföldi J, Barrio AM, et al. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345:1074–9. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saitou N, Nei M. The Neighbor-Joining method - A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 80.Felsenstein J. Confidence-limits on phylogenies - An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 81.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, et al. FlyMine: An integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson SW, Herzyk P, Dow JAT, Leader DP. FlyAtlas: Database of gene expression in the tissues of Drosophila melanogaster. Nucl Acids Res. 2013;41:D744–50. doi: 10.1093/nar/gks1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Houle D. How should we explain variation in the genetic variance of traits? Genetica. 1998;102:241–52. doi: 10.1023/A:1017034925212. [DOI] [PubMed] [Google Scholar]


