Abstract
In this study, we describe the mitochondrial genome of the excavate flagellate Euglena gracilis. Its gene complement is reduced as compared with the well-studied sister groups Diplonemea and Kinetoplastea. We have identified seven protein-coding genes: Three subunits of respiratory complex I (nad1, nad4, and nad5), one subunit of complex III (cob), and three subunits of complex IV (cox1, cox2, and a highly divergent cox3). Moreover, fragments of ribosomal RNA genes have also been identified. Genes encoding subunits of complex V, ribosomal proteins and tRNAs were missing, and are likely located in the nuclear genome. Although mitochondrial genomes of diplonemids and kinetoplastids possess the most complex RNA processing machineries known, including trans-splicing and editing of the uridine insertion/deletion type, respectively, our transcriptomic data suggest their total absence in E. gracilis. This finding supports a scenario in which the complex mitochondrial processing machineries of both sister groups evolved relatively late in evolution from a streamlined genome and transcriptome of their common predecessor.
Keywords: Euglena gracilis, mitochondrial genome, transcription, RNA editing
Introduction
The mitochondrion and its DNA are hallmarks of most eukaryotic cells, with few exceptions: Derived mitochondrial organelles, mitosomes, and hydrogenosomes usually lack a genome (Maguire and Richards 2014). The content of known mitochondrial genomes varies from 66 protein-coding genes in Andalucia godoyi (Burger et al. 2013) to just 2 in another protist, Chromera velia (Flegontov et al. 2015), although less than two dozen genes are present in a “standard” mitochondrial genome. However, despite its limited size, the architecture of this organellar genome is extremely variable across eukaryotes (Burger et al. 2003; Smith and Keeling 2015). Although it usually occurs in monomeric circular form or as a linear concatemer, with the fast growing number of mitochondrial genomes sequenced, unconventional forms are frequently encountered. These include repeat-rich genomes of large size (Sloan et al. 2012), “scrambled” recombining pools of linear fragments (Waller and Jackson 2009; Flegontov et al. 2015), or thousands of free or mutually interlocked circular molecules (Gray 2012).
Although complex mitochondrial genomes have been described in land plants (Sloan et al. 2012) and some multicellular opisthokonts (Shao et al. 2012), most diversity seems to be hidden in protists. Although unconventional gene sets, gene arrangements and/or post-transcriptional modification of transcripts of mitochondrion-encoded genes were described, for example from jacobids, dinoflagellates and apicomplexans (Gray 2012; Smith and Keeling 2015), one protist group stands out if complexity of its mitochondrial genome and transcriptome is considered, namely the phylum Euglenozoa. Historically, this diverse and ecologically highly significant group is subdivided into classes Euglenida, Kinetoplastea, and Diplonemea, well defined on the basis of both morphological and molecular features (Adl et al. 2012).
Not surprisingly, most attention was devoted to the studies of the mitochondrial genomes of parasitic kinetoplastids, especially the human pathogens Trypanosoma and Leishmania (Povelones 2014; Verner et al. 2015). Their mitochondrial DNA, termed kinetoplast (k) DNA, is composed of dozens of maxicircles and thousands of minicircles, mutually interlocked into a single giant network (Liu et al. 2005). By carrying a dozen of protein-coding genes, maxicircles represent homologs of the classical mitochondrial genome, yet most of the genes are present in an “encrypted” form, which means their transcripts have to undergo extensive RNA editing. In all kinetoplastids studied so far, editing proceeds through insertions and deletions of uridines within a subset of mitochondrial transcripts (Stuart et al. 2005; Aphasizhev and Aphasizheva 2014). Templates for these editing events are provided by hundreds of guide RNAs, which are almost invariably encoded by the mutually interlocked kDNA minicircles (Alfonzo et al. 1999). The highly sophisticated process of uridine insertions/deletions (indel) editing requires a multitude of enzymatic activities, which are provided by several dedicated protein complexes (Hashimi et al. 2013; Aphasizhev and Aphasizheva 2014). Only correctly edited transcripts are translatable on mitochondrial ribosomes that, not surprisingly, are the most complex organellar ribosomes known (Maslov et al. 2006; Zíková et al. 2008).
Due to the extensive heterogeneity of the kDNA minicircles that carry indispensable guide RNA genes, the replication of a single kDNA network has to be highly accurate, with each of the approximately 5,000 minicircles dividing once and only once, and segregating into the daughter cells. This is achieved by a well-studied machinery composed of more than a hundred proteins, involved in de- and re-catenation, topological changes, replication, and maintenance of the kDNA structure (Jensen and Englund 2012; Povelones 2014; Verner et al. 2015). Most data are available from Trypanosoma brucei, which thanks to its tractability through genetic manipulations such as RNA interference, homologous recombination and gene tagging became a model protist, with its mitochondrion belonging to the best-studied organelles.
The situation is quite different in the case of diplonemids, which were considered to be a rather small obscure group. This has changed recently, as on the global scale they emerged as possibly the third most diverse eukaryotic clade in ocean plankton (Lukeš et al. 2015). Their mitochondrial genome is also highly unconventional, yet different from that of the sister kinetoplastid lineage. It is composed of noncatenated minicircles, with each encoding a single fragment of a protein-coding gene (Marande et al. 2005; Vlček et al. 2011; Kiethega et al. 2013). Hence, multiple transcript fragments have to be spliced together, in a stepwise orderly fashion, to generate a translatable molecule. This trans-splicing is further complicated by the insertion of stretches of uridines between the spliced fragments (Marande and Burger 2007; Kiethega et al. 2013; Valach et al. 2014). Detailed mapping of the mitochondrial genome of Diplonema papillatum revealed that all mitochondrial-encoded genes, including ribosomal (r)RNAs, are fragmented and have to undergo trans-splicing (Vlček et al. 2011; Kiethega et al. 2013; Valach et al. 2014). The combination of RNA editing and trans-splicing qualifies the posttranscriptional processing in the D. papillatum mitochondrion as one of the most complex known.
The discovery of RNA editing in the mitochondrion of T. brucei, as the first case of its kind (Benne et al. 1986), was followed by an intense discussion on the origin of this complex process. Although a long list of possible raisons d’etre have been considered, it is now widely accepted that no particular selective advantage is provided by this mechanism, which likely evolved through a neutral evolutionary “ratchet” building complex cellular processes (Gray et al. 2010; Lukeš et al. 2011). However, major insights into the emergence of extensive RNA editing and trans-splicing in the mitochondria of kinetoplastids and diplonemids, respectively, can be provided by comparative analysis with the only known sister clade of both protist groups, the euglenids (Flegontov et al. 2011).
Surprisingly, despite the widespread presence and ecological significance of euglenids, we knew close to nothing about their mitochondrial genomes (Roy et al. 2007), so their comparative potential could not be explored. This is even more surprising as the Euglena gracilis chloroplast genome was among the first plastid genomes ever sequenced (Hallick et al. 1993). As Euglena is also known as an important biotechnological platform (Krajčovič et al. 2015), there is a growing interest in its metabolic capacity. Recent transcriptome sequencing presented Euglena as an organism with high metabolic potential, capable of producing a range of substances with biotechnological importance: vitamins (A, C, and E), paramylon, wax esters, polyunsaturated fatty acids, biotin, and amino acids (Krajčovič et al. 2015; O’Neill et al. 2015). Moreover, E. gracilis mitochondrion is capable of facultative anaerobic metabolism, namely wax ester fermentation (Müller et al. 2012).
Despite significant efforts, only three protein-coding genes were so far detected in the mitochondrial genome of E. gracilis, namely subunits 1 through 3 of cytochrome c oxidase (cox1, cox2, and cox3), the latter subunit initially annotated as nad6 (Tessier et al. 1997; Yasuhira and Simpson 1997). Only recently, the small rRNA (encoded in two separate pieces) from the same species was sequenced (Spencer and Gray 2011). The latter work established the general architecture of the E. gracilis mitochondrial genome: Linear fragments of heterogeneous size (modal size 4 kb) bearing terminal repeats and many small, likely nonfunctional, protein and rRNA gene fragments, testifying of extensive ongoing recombination within the genome (Spencer and Gray 2011). It was proposed that antisense transcripts of these short gene fragments could have given rise to guide RNAs that mediate uridine indel RNA editing (Flegontov et al. 2011; Spencer and Gray 2011). The recombination-prone, scrambled, structure of the genome resembles mitochondrial genomes of dinoflagellates (Waller and Jackson 2009) and C. velia (Flegontov et al. 2015), representing yet another putative case of evolutionary convergence between these unrelated groups (Lukeš et al. 2009).
Here, we have performed an extensive analysis of the E. gracilis mitochondrial genome and transcriptome and report that it encodes only a very limited set of protein-coding genes. Unexpectedly, their transcripts do not undergo any of the complex posttranscriptional modifications used by their cousins, and thus seem to be rather mundane. We consider this simplicity particularly telling from an evolutionary perspective.
Materials and Methods
Cell Culture
Euglena gracilis var. bacillaris cells were axenically grown in total volume of 600 ml in Hutner liquid medium (Hutner et al. 1966) at 27 °C under the permanent light conditions (10 µm/m−2s−1) and constant shaking till they reached the exponential growth phase (1.5–2 × 106 cells/ml).
Isolation of Mitochondrial DNA and RNA
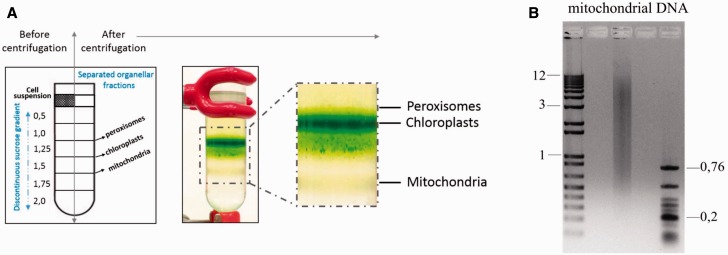
The procedures of mitochondrial isolation reported previously (Davis and Merrett 1973; Moreno-Sánchez et al. 2000) were performed here with several minor modifications. Cells of E. gracilis were harvested by centrifugation at 800 × g for 10 min. The pellet was resuspended in SHE buffer (250 mM sucrose, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1 mM ethylenediaminetetraacetic acid, pH 7.3) supplemented with 0.4% fatty acid-free bovine serum. All the following steps were performed on ice. Sonication of the cell suspension was performed using a thick 19.5-mm probe (Ultrasonic homogenizer model 3,000; Biologics, Inc.) in six cycles consisting of 10 s pulses with 2-min breaks between them. The power of the sonicator was set at 80% and cell disruption was checked under light microscope. If necessary, an additional sonication cycle was added. The sonicate was transferred to new tubes and centrifuged for 15 min at 800 × g and 4 °C; the supernatant was again transferred to new tubes and centrifuged for 15 min at 8,500 × g and 4 °C. The pellet was resuspended in 3 ml of STM buffer (250 mM sucrose, 20 mM Tris–HCl, 2 mM MgCl2, pH 8.0) with 40 U of DNase I (Thermo Scientific) and incubated 30–60 min on ice. Sucrose density gradient centrifugation of the cell lysate allowed separation of the mitochondrial fraction from other organelles as chloroplasts and peroxisomes (Davis and Merrett 1973). The lysate (5 ml) was loaded on top of a sucrose discontinuous density gradient in ultracentrifuge tubes (Cat. No. 344058, Beckman). The gradient consisted of the following layers, bottom to top: 2.0, 1.75, 1.5, 1.25, 1, 0.5 M sucrose, 5 ml each (fig. 1A), and was centrifuged in the SW-28 rotor at 87,041 × g (22,000 rpm) at 4 °C for 4.5 h (L8-M Ultracentrifuge, Beckman). After centrifugation, the mitochondrial fraction situated at the interface of the 1.75 and 1.5 M sucrose layers was collected by a syringe. Mitochondria were washed two times in SHE buffer to remove excessive sucrose, and the final pellet was gently resuspended with a cut-off pipette tip in 500–1,000 μl of SHE buffer supplemented with 0.4% Bovine Serum Albumin (BSA), and then spun for 30 min at 16,000 × g at 4 °C.
Fig. 1.—
Isolation of Euglena mitochondrial fraction in sucrose gradient. (A) The Euglena gracilis cell lysate was loaded onto a discontinuous sucrose gradient. The individual sucrose layers were loaded, from the most dense (2 M sucrose) layer to the least dense (0.5 M sucrose) layer, into a centrifugation tube as depicted in the left part of the figure. Upon centrifugation, few bands of separated cell fractions were visible. The mitochondrial fraction was located at the interface of the 1.75 and 1.5 M sucrose layers. The chloroplast fraction was located between the 1.5 and 1.25 M sucrose layers. The third band corresponded to peroxisomes lying at the interface of the 1.25 and 1.0 M sucrose layers. (B) Isolated mitochondrial DNA loaded on a 1% agarose gel. The sizes of marker DNA fragments are indicated in kilobase.
Mitochondrial DNA was isolated from purified mitochondrial vesicles using DNeasy Blood & Tissue Kit (Qiagen). RNA was isolated using TRIzol Reagent (Cat. No. 15596-026, Life Technologies) following the manufacturer’s recommendation. Concentration of the isolated genetic material was measured using the Qubit fluorimeter (Thermo Fisher Scientific). Total cell DNA was isolated as described previously (Vesteg et al. 2010), whereas total RNA was isolated following the same protocol used for mitochondrial RNA isolation.
Mitochondrial Fraction Purity Control with Quantitative Polymerase Chain Reaction
Purified total or mitochondrial RNA was subjected to cDNA synthesis. Reverse transcription was performed with random hexamer primers using Superscript III (Life Technologies) according to the manufacturer’s instructions. Specific primers (supplementary table S1, Supplementary Material online) were designed for representative genes from each organellar DNA using the Primer3 software (version 0.4.0) (http://bioinfo.ut.ee/primer3-0.4.0/primer3/input.htm, last accessed January 15, 2014). Quantitative polymerase chain reaction (qPCR) was performed using Power SYBR Green PCR Master Mix (Life Technologies) on a microwell plate-based cycler platform (LightCycler 480 System, Roche) and the data were analyzed using the LightCycler 480 software v. 1.5 (Roche). Each experiment had three technical replicates. Specificity of the amplification reactions was confirmed by electrophoresis in a 1% agarose gel.
DNA and RNA Library Preparation and Sequencing
The isolated mitochondrial RNA was partially depleted of rRNA using RiboMinus Eukaryote System v2 (Ambion). A strand-specific RNA-seq library with an insert size ranging from 120 to 300 bp was prepared using NEXTflex Directional dUTP-Based RNA-Seq Kit (Bioo Scientific). The library was sequenced with 100-nt paired reads on the Illumina HiSeq platform (Macrogen, Korea). For mitochondrial DNA, mate pair (MP) and paired-end (PE) libraries of the following insert sizes were prepared: 1,300–4,300 and 280–980 bp, respectively. The MP library was sequenced with Illumina HiSeq obtaining approximately 27 million reads with a read length of 100 nt. The PE library was sequenced with 250-nt paired reads on the Illumina MiSeq platform, producing approximately 25 million reads (supplementary table S2, Supplementary Material online). Sequence reads and assemblies are deposited in the National Center for Biotechnology Information (NCBI) database under accession number PRJNA294935.
Mitochondrial Genome and Transcriptome Assembly
All raw DNA and RNA-seq reads went through quality filtering and adapter trimming in CLC Genomics Workbench v. 6.5 (CLC Inc, Aarhus, Denmark). For PE reads, regions with Phred quality less than 20 were trimmed, no more than one N was allowed in the remaining sequence, than TruSeq adapter trimming and a minimum length threshold of 50 nt were applied. For MP reads, a more complex adapter trimming procedure was implemented to remove internal adapters and pairs with incorrect read orientation. The PE reads were merged in a pairwise fashion using CLC Genomics Workbench v. 6.5 with default settings. The mitochondrial genome of E. gracilis was assembled with the Newbler assembler (GS De Novo Assembler v. 2.9) from MP and PE (merged and not merged) reads. A number of assembly parameters were tested to maximize the length of mitochondrion-encoded genes. Draft RNA-seq assembly was performed with CLC Genomics Workbench v. 6.5 using standard de novo assembly settings.
Identification of E. gracilis Mitochondrial Genes
BLASTn with an E-value cut-off of 10−10 against the E. gracilis chloroplast genome sequence (NC_001603.2) (Hallick et al. 1993) was used to filter out contaminating plastid contigs. Contigs matching the plastid genome with greater than 95% identity were removed. Searching with tBLASTx against mitochondrial genomes of Malawimonas californiana (accession number NC_026311.1), Jakoba libera (NC_021127.1), and Reclinomonas americana (NC_001823.1), we identified putative E. gracilis mitochondrial contigs. As the enrichment of the mitochondrial fraction was confirmed by qPCR, the nucleus-encoded mitochondrial proteins were identified based on low coverage of corresponding contigs (supplementary table S3, Supplementary Material online), and were not considered further. Later on, manual analysis of the graph of alternative contig connections produced by Newbler was performed with an in-house software, in order to close gaps in mitochondrial contigs and assemble long repetitive regions.
Protein Phylogenetic Analysis
All protein alignments were performed using MUSCLE 3.8.31 with default settings, and manually checked for misalignments. Site selection was performed manually, keeping all aligned blocks longer than six amino acids, with no taxon missing in the block. Maximum-likelihood (ML) trees for all mitochondrion-encoded proteins were constructed with RAxML v. 8.0.0 (Stamatakis 2014) with the LG + Г model and 100 bootstrap replicates (supplementary text S1, Supplementary Material online). GenBank accession numbers of sequences used for alignments and phylogenic analysis are available in supplementary text S2, Supplementary Material online. The alignments with selected sites used for building of the phylogenetic trees are available in supplementary text S3, Supplementary Material online.
Detection of RNA Editing
To look for possible U-indel editing events in RNA-seq reads, we used a modification of Bowtie2 v.2.0.2 with base-specific indel penalties (Langmead and Salzberg 2012; David et al. 2015). Edited reads of the mitochondrial transcriptomes of kinetoplastids have U-indels only and can therefore be aligned correctly when gap penalties for T (corresponding to U in RNA) are different from those for A, G, and C (David et al. 2015). The following set of options was routinely used: 1) High gap opening and extension penalties of 10 for A, G, C in the reference and in sequencing reads (–rfg 10,10 –rdg 10,10); 2) minimal gap opening and extension penalties of 1 for T or A (depending on transcript orientation) in the reference and in reads (–rfg-T 1,1 –rdg-T 1,1 or –rfg-A 1,1 –rdg-A 1,1); 3) high mismatch penalty equal to 18 (–mp 18); 4) options allowing terminal mismatches (–gbar 0 –dpad 50), and 5) other options (–end-to-end -D 20 -R 3 -N 1 -L 14 -i S,1,0.50 –score-min L,0,-2). Poor-quality read alignments, especially those introducing large gaps, were removed. Indel and substitution polymorphisms detected in both DNA and RNA-seq read mappings were not considered further as they might represent mitochondrial genome heteroplasmy or alignment artefacts. To accommodate possible biases of this read mapping approach, additional DNA and RNA-seq read mappings were performed with CLC Genomics Workbench v 6.5 (mapping parameters: minimum 90% of read aligned and minimum alignment identity 95%), and resulting polymorphisms compared.
Results
For mitochondrial nucleic acids extraction, we used the organellar mitochondrial fraction instead of total cell lysate (fig. 1A). The purification on a sucrose gradient (see Materials and Methods for details) produced mitochondrial vesicles, the intactness of which was confirmed by an oxygen consumption measurement (data not shown). Afterwards, the mitochondrial DNA was isolated (total amount about 5 µg), and an aliquot (200 ng) was separated on a 1% agarose gel (fig. 1B). The mitochondrial DNA appears as a pool of heterogeneous molecules, with a length distribution similar to that reported previously (ranging from ∼1.0 to ∼9.0 kb and with a peak around 4.0 kb, according to Spencer and Gray 2011).
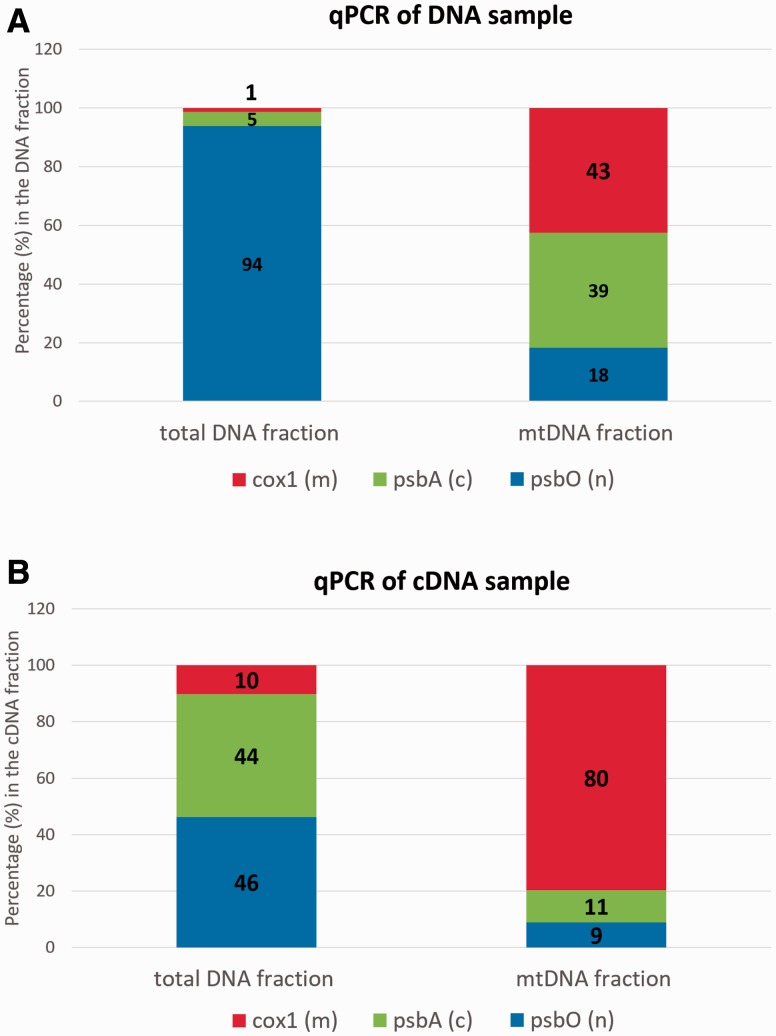
To estimate whether the enrichment method was successful, we the relative abundance of mitochondrial, plastid and nuclear marker genes/transcripts in the total and mitochondrial fractions was analyzed with qPCR (fig. 2). The relative abundance values do not reflect the proportion of mitochondrial, plastid, and nuclear genomes in the fractions (fig. 2), as the genome sizes hugely vary, but rather reflect the coverage of a single gene or transcript representing these genomes. Upon the purification of mitochondria, relative abundance of the mitochondrial marker cox1 increased from 1% to 43% in DNA and from 10% to 80% in cDNA. In the mitochondrial DNA sample, the mitochondrial cox1 and plastid psbA markers had an almost equal abundance, whereas the abundance of the nuclear marker gene psbO was approximately two times lower (fig. 2A). Using purified mitochondrial DNA and RNA, we prepared PE and MP genomic libraries and a strand-specific rRNA-depleted transcriptomic library, and sequenced them with the Illumina technology (supplementary table S2, Supplementary Material online).
Fig. 2.—
Control of mitochondrial fraction enrichment by qPCR. qPCR was performed on DNA and cDNA isolated from mitochondria purified in the sucrose gradient and, as a control, on total DNA and cDNA of Euglena gracilis. Marker genes for organellar genomes were as follows: cox1 (red), a mitochondrial-encoded subunit of complex IV; psbO (blue), a nucleus-encoded subunit of photosystem II; and psbA (green), a chloroplast-encoded subunit of photosystem II. All samples were run in triplicate. (A) qPCR on the DNA samples. The chloroplast DNA represents major contamination of the mitochondrial fraction, most likely due to proximity of the mitochondrial and plastid bands in the sucrose gradient. (B) qPCR on the cDNA samples. Chloroplast transcripts are of low abundance in the mitochondrial fraction cDNA.
DNA reads of the mitochondrial fraction (100-250 nt in length, supplementary table S2, Supplementary Material online) were assembled with Newbler (GS De Novo Assembler v.2.9). We identified mitochondrial-encoded genes through a tBLASTx search: Translated mitochondrial contigs were searched against translated publicly available mitochondrial genomes from M. californiana, R. americana, and J. libera. We chose these organisms for two reasons: At least two of them belong, together with E. gracilis, to the phylum Excavata, and their mitochondrial genomes are the biggest known in terms of gene content (Burger et al. 2013). Nuclear genes were pruned out based on low coverage (supplementary table S3, Supplementary Material online) of the respective contigs in the mitochondrial fraction. Then we tested various assembler settings (data not shown) in order to obtain full-length sequences of mitochondrial-encoded genes, as inferred from alignments with orthologous proteins of other Excavata species (supplementary text S3, Supplementary Material online). The best assembly, that is, the one containing a maximum number of full-length mitochondrial genes, was further improved manually, using an in-house software for visualization and analysis of a graph of alternative contig connections produced by Newbler. Chloroplast sequences were eliminated using a Basic Local Alignment Search Tool (BLAST) search against the chloroplast genome (Hallick et al. 1993).
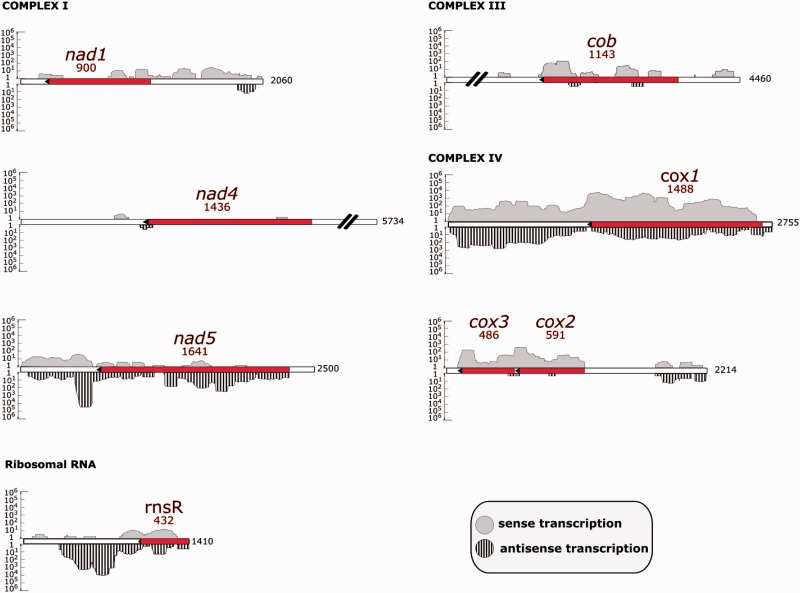
The mitochondrial genome of E. gracilis emerged with a reduced gene complement of just seven protein-coding genes (fig. 3): Subunits of respiratory chain complexes I (NADH dehydrogenase, nad1, nad4, and nad5), III (cytochrome c reductase, cob), and IV (cytochrome c oxidase, cox1, cox2, and cox3). Notably, only three of seven genes were sequenced previously (Spencer and Gray 2011). These seven genes were located on six contigs: One gene per contig, and cox2 and cox3 were closely spaced on a single contig (Spencer and Gray 2011). In agreement with published results (Spencer and Gray 2011), short fragments of all protein-coding genes were scattered throughout mitochondrial contigs, and it was not possible to assemble the mitochondrial genome as a single contig. In summary, our data and published results (Spencer and Gray 2011) allow the reconstruction of the E. gracilis mitochondrial genome as a heterogeneous array of recombination-prone linear fragments with low gene content.
Fig. 3.—
Euglena gracilis mitochondrial genes and their transcription. The mitochondrial contigs contain genes encoding proteins of the respiratory chain: Complex I (nad1, nad4, nad5), complex III (cob), complex IV (cox1, cox2, cox3), and a fragment of SSU rRNA (rnsR). Striped peaks represent antisense transcription, and gray peaks represent sense transcription. Transcript coverage is represented in logarithmic scale on the y axis. Gene lengths are shown in red font, whereas contig lengths are shown in black font.
In order to find actual translation start sites, the predicted protein sequences of E. gracilis were aligned with orthologs from selected Excavata species (supplementary text S3, Supplementary Material online): Diplonemids Diplonema and Rhynchopus; kinetoplastids Bodo, Leishmania, and Trypanosoma; a heterolobosean Naegleria; jakobids Jakoba and Reclinomonas; and a malawimonadid Malawimonas. In ML phylogenetic trees based on these protein alignments, Euglena either branched as a sister-group for the Diplonemea/Kinetoplastea clade, as expected (cox2, see supplementary text S1, Supplementary Material online), or the position of Euglena within Euglenozoa remained unresolved with a bootstrap support cut-off of 80 (in the cases of cox1 and cob). All mitochondrial genes of E. gracilis were represented by full-length sequences in our assembly, except for the nad5 gene, with its 3′-end presumably missing. As compared with aligned protein orthologs, about 100 amino acids of nad5 are missing (supplementary text S3, Supplementary Material online). Numerous short fragments of the nad5 gene scattered throughout mitochondrial contigs apparently prevent unambiguous assembly of the full-length copy, even upon manual contig graph analysis.
In mitochondrial RNA extracts from E. gracilis, there are four major, stoichiometrically equivalent bands corresponding to two fragments of the small subunit (SSU) rRNA, SSU-R and SSU-L, and two fragments of the large subunit (LSU) rRNA, LSU-R and LSU-L (Spencer and Gray 2011). Only the SSU-R and SSU-L fragments were sequenced (Spencer and Gray 2011). In this study, we identified a 432 bp sequence of the SSU-R rRNA gene (fig. 3), and only short highly divergent sequences that could represent LSU rRNA. Protein components of the LSU of the mitochondrial ribosome (rpl proteins) are encoded in the E. gracilis nuclear contigs (data not shown), and are probably targeted to mitochondria.
The transcription of seven mitochondrial contigs was investigated by mapping strand-specific RNA-seq reads (fig. 3): Paired Illumina MiSeq reads (up to 250 nt in length) and longer pseudoreads (up to 400 nt in length) produced from merged overlapping paired reads. As the polyadenylation status of E. gracilis mitochondrial transcripts was not known a priori, we used a total RNA-seq library with partial rRNA depletion. We observed that the cox1, cox2, and cox3 genes were transcribed, whereas the others had very low transcription levels (fig. 3). A relatively high level of antisense transcription has been revealed for the cox1 and nad5 genes (fig. 3).
Most importantly, comparing alignments of genomic and transcriptomic reads to the coding sequences, we found no signs of substitution or insertion/deletion RNA editing in E. gracilis mitochondrial transcripts. However, we cannot exclude the possibility that RNA editing is restricted to the low-coverage transcripts nad1, nad4, nad5, and cob.
Discussion
In the past decades, thousands of mitochondrial genomes were subject to targeted sequencing or were assembled from total genome sequencing efforts. Therefore, it was proposed recently that sequencing additional mitochondrial genomes is unnecessary as it brings neither new insights nor makes any impact on mitochondrial genomics (Smith 2015). However, protists were named as the only exception since when compared with the “standard” metazoan mitochondrial DNAs, their mitochondrial DNAs vary enormously in size, structure, coding capacity and even in posttranscriptional processing (Smith and Keeling 2015). Indeed, some of the most extreme mitochondrial genomes have been described in kinetoplastids and diplonemids. Although the former evolved their organellar genomes into thousands of (non-) catenated circles (Lukeš et al. 2002), the latter fragmented their mitochondrial genes into a jigsaw puzzle (Marande and Burger 2007). Hence, euglenozoans carry a highly diverse array of mitochondrial genomes.
The architecture of the E. gracilis mitochondrial genome has remained elusive for a long time (Spencer and Gray 2011). Initially, it was thought that this was due to the fragile nature of the mitochondrial DNA and the isolation procedure itself (Buetow 1989). Several further attempts led to detection of only a single cox1-bearing fragment (Tessier et al. 1997; Yasuhira and Simpson 1997), and it was suggested that the genome may be fragmented (Flegontov et al. 2011). In this study, we decided to isolate mitochondria first and extract their genetic material from that fraction afterwards. Using this protocol, relatively intact metabolically active mitochondrial vesicles were obtained, as confirmed by oxygen consumption measurements indicating intact respiration. Despite gentle sample handling, we only found genomic molecules 5–8.0 kb in length, in agreement with a previous study (Spencer and Gray 2011). Many of these relatively short fragments carry pseudogenes and short gene fragments, but full-length apparently functional gene copies are also present.
Sequencing of the E. gracilis mitochondrial DNA revealed a remarkably small complement of seven protein-coding genes and a single SSU rRNA gene fragment. In this study, we augmented the data set of E. gracilis mitochondrial genes with four newly identified ones: cob, nad1, nad4, and nad5. Given previous data and the depth of sequencing reported herein (supplementary table S3, Supplementary Material online), we predict that this set of protein-coding genes is final. Thus, the genome encodes subunits of three respiratory complexes (NADH:ubiquinone oxidoreductase, ubiquinone:cytochrome c oxidoreductase, and cytochrome c oxidase) and fragments of the mito-rRNA. No genes encoding subunits of succinate:ubiquinone oxidoreductase (complex II) were found, similar to most animal and fungal mitochondrial DNAs (Gray and Doolittle 1982; Tzagoloff and Myers 1986; Attardi and Schatz 1988; Burger et al. 1996).
Surprisingly, not a single subunit of ATP synthase (complex V) is encoded in the mitochondrial genome of E. gracilis, including atp6, which is almost universally present in mitochondrial genomes except for the most reduced ones (Vázquez-Acevedo et al. 2006). For example, flagellated protists belonging to Kinetoplastea and Diplonemea, the other major euglenozoan groups, invariably encode atp6 in their mitochondria. However, all subunits of complex V are missing from the mitochondrial genomes of chlorophycean algae, including the well-studied Chlamydomonas reinhardtii (Vázquez-Acevedo et al. 2006; van Lis et al. 2007). In this protist, all genes encoding subunits of ATP synthase were relocated to the nucleus. Components of this complex are highly hydrophobic membrane proteins (van Lis et al. 2007), and in order to overcome the problems associated with their targeting back to the organelle, some structural changes must have occurred (Funes et al. 2002; Figueroa-Martínez et al. 2011). In summary, E. gracilis together with green algae, Myzozoa (Alveolata) and a marine invertebrate Paraspadella gotoi, exhibits the remarkable property of lacking all atp genes in its mitochondrial DNA (Feagin 2000; Vázquez-Acevedo et al. 2006). The loss of mitochondrial-encoded subunits of complex V may be connected with the recruitment of new subunits of the respiratory chain. Recent study revealed that E. gracilis shares some unique subunits of the respiratory chain with kinetoplastids (Perez et al. 2014). Apparently, their presence cannot be explained by the parasitic lifestyle of most kinetoplastids, as many euglenozoans are nonparasitic, including the photosynthetic euglenids and free-living diplonemids. At the same time, these subunits are not present outside Euglenozoa and may thus be a consequence of biochemical uniqueness of the kinetoplastids and euglenids (Perez et al. 2014).
We were able to find only one of two fragments of the split SSU rRNA (Spencer and Gray 2011). We suggest that a highly divergent mitochondrial LSU rRNA in E. gracilis escapes detection by BLAST, similar to its counterpart in the sister clade Diplonemea (Valach et al. 2014). The fragmentation of rRNA genes is not an unusual feature of mitochondrial genomes (Smith and Keeling 2015). For example, eight and four scrambled fragments of LSU and SSU rRNA, respectively, are encoded by the linear mitochondrial DNA of Chl. reinhardtii. It is likely that corresponding transcripts come together through base-pairing interactions (Boer and Gray 1988). An even more extreme case of scrambled rRNA genes has been observed in the parasitic protist Plasmodium falciparum: Its LSU and SSU rRNA genes are encoded in at least 27 distinct modules (Feagin et al. 2012).
We speculate that the availability of glucose in the medium in which E. gracilis was cultivated (2% concentration) leads to inhibition of the respiratory chain activity and hence to silencing of at least some of its subunits (cob, nad1, nad4, and nad5). We cannot ignore the increasing evidence for molecular crosstalk between mitochondria and plastids (Leister 2005). As E. gracilis was grown under light, photosynthesis might have taken control over the cell energy metabolism, and there was no need for high expression of mitochondrial genes. However, a recent study of E. gracilis did not reveal any significant changes in enzymatic activities of the respiratory complexes or oxygen consumption in the presence or absence of light (Krnáčová et al. 2015).
RNA editing encompasses a range of transcript-processing mechanisms, including insertions and deletions of single or multiple uridine residues (=U-indel), as well as base modifications and replacements, occurring in both noncoding and protein-coding sequences (Gott and Emeson 2000; Gray 2003). The U-indels are characteristic and specific for kinetoplastids (Verner et al. 2015), one of three major euglenozoan clades, although a potentially similar type of RNA editing was recently found in their sister clade, the diplonemids (Marande and Burger 2007; Valach et al. 2014). Despite the abundance of short gene fragments and antisense transcripts, both previously hypothesized as pre-requisites for the emergence of U-indel editing through a neutral evolution mechanism (Flegontov et al. 2011; Spencer and Gray 2011), our results suggest, rather surprisingly, that RNA editing does not operate in the mitochondrion of E. gracilis. After the complete nuclear genome of E. gracilis becomes available, it will be insightful to check whether any of the key enzymatic activities needed for U-indel editing exist in this organism.
One of the features typical for organellar DNAs is their enrichment in A + T content (Smith et al. 2011). Euglena is not an exception when it comes to A + T content, with both mitochondrial and plastid genomes containing just 25% of G + C (Fonty et al. 1975; Buetow 2011). Despite this general trend, few species have GC-rich mitochondrial genomes: A parasitic fungus (Fricova et al. 2010), lycophyte (Smith 2009; Hecht et al. 2011), some fishes of the teleost group (Clare et al. 2008), and also Chlorophyta, comprising the majority of green algae (Lewis and McCourt 2004). A recent study suggested that the nucleotide composition skewed toward G + C may be a result of nonadaptive forces (Smith et al. 2011).
Having remained intractable for decades, the mitochondrial genome of E. gracilis finally emerges in its almost complete form. With its complement of just seven protein-coding genes, it represents one of the smallest mitochondrial genomes in terms of gene content. Remarkably, many other gene-poor mitochondrial genomes, namely those of dinoflagellates and alveolates, exhibit a genome architecture very similar to that of Euglena: An array of recombination-prone relatively short linear molecules, in which repeats and nonfunctional gene fragments have accumulated (Flegontov et al. 2015). Recently, a mitochondrial genome of the same type has also been revealed in a parasitic plant, Viscum scurruloideum: It lost all subunits of complex I, has an extremely active recombination and a very high rate of protein divergence (Skippington et al. 2015). It is rather difficult to imagine how such disordered systems are propagated in a stable manner for millions of years, and it is also not entirely clear how they emerged in the first place. One possibility is that an initial gene loss was followed by relaxed selective constraints (Smith and Keeling 2015), which drive the accumulation of “molecular chaos.”
Euglena is a plastid-bearing alga, and plastid and mitochondrial genomes demonstrate similar evolutionary trends in some organisms. Emergence of unorthodox genome structures and transcript processing mechanisms is frequently correlated in both organelles within one cell, and may be driven by unusually high or low mutation rates, determined by nuclear-encoded proteins targeted into both mitochondria and plastids (Smith and Keeling 2015). Notably, both mitochondrial and plastid genomes of E. gracilis have accumulated odd molecular features. Its plastid genome has an unusually high number of introns (∼150 introns taking up 40% of the genome), including 15 introns within introns called twintrons (Hallick et al. 1993). Determining mutation rates in both organellar genomes of Euglena, which are now finally available, is an important direction of future research.
Supplementary Material
Supplementary tables S1–S3 and texts S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Vojtěch David (Institute of Parasitology, České Budějovice) for technical assistance and Evgeny S. Gerasimov (Moscow State University, Russia) for providing unpublished assembly-finishing software. This work was supported by the Grant Agency of the Czech Republic 15-21974S and Praemium Academiae award (both to J.L.). P.F. was supported by the Moravian-Silesian region projects MSK2013-DT1, MSK2013-DT2, and MSK2014-DT1, and by the Institution Development Program of the University of Ostrava.
Literature Cited
- Adl SM, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonzo JD, Blanc V, Estévez AM, Rubio MAT, Simpson L. 1999. C to U editing of the anticodon of imported mitochondrial tRNA (Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 18:7056–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I. 2014. Mitochondrial RNA editing in trypanosomes: small RNAs in control. Biochimie. 100:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi G, Schatz G. 1988. Biogenesis of mitochondria. Annu Rev Cell Biol. 4:289–333. [DOI] [PubMed] [Google Scholar]
- Benne R, et al. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46:819–826. [DOI] [PubMed] [Google Scholar]
- Boer PH, Gray MW. 1988. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell 55:399–411. [DOI] [PubMed] [Google Scholar]
- Buetow DE, editor. 1989. The mitochondrion. In: The biology of Euglena. Vol. 4. San Diego (CA): Academic Press; p. 247–314. [Google Scholar]
- Buetow DE. 2011. Euglena. In: Battista J, editor. Encyclopedia of life sciences (ELS). Chichester (United Kingdom): John Wiley & Sons, Ltd; 1–5. [Google Scholar]
- Burger G, Gray MW, Forget L, Lang BF. 2013. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 5:418–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709–716. [DOI] [PubMed] [Google Scholar]
- Burger G, Lang BF, Reith M, Gray MW. 1996. Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc Natl Acad Sci U S A. 93:2328–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare EL, Kerr KCR, von Konigslow TE, Wilson JJ, Hebert PDN. 2008. Diagnosing mitochondrial DNA diversity: applications of a sentinel gene approach. J Mol Evol. 66:362–367. [DOI] [PubMed] [Google Scholar]
- David V, et al. 2015. Gene loss and error-prone RNA editing in the mitochondrion of Perkinsela, an endosymbiotic kinetoplastid. MBio; 6(6). pii: e01498-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B, Merrett MJ. 1973. Malate dehydrogenase isoenzymes in division synchronized cultures of Euglena. Plant Physiol. 51:1127–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE. 2000. Mitochondrial genome diversity in parasites. Int J Parasitol. 30:371–390. [DOI] [PubMed] [Google Scholar]
- Feagin JE, et al. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One 7:e38320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Martínez F, et al. 2011. What limits the allotopic expression of nucleus-encoded mitochondrial genes? The case of the chimeric cox3 and atp6 genes. Mitochondrion 11:147–154. [DOI] [PubMed] [Google Scholar]
- Flegontov P, et al. 2015. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol Biol Evol. 32:1115–1131. [DOI] [PubMed] [Google Scholar]
- Flegontov P, Gray MW, Burger G, Lukeš J. 2011. Gene fragmentation: a key to mitochondrial genome evolution in Euglenozoa? Curr Genet. 57:225–232. [DOI] [PubMed] [Google Scholar]
- Fonty G, Crouse EJ, Stutz E, Bernardi G. 1975. The mitochondrial genome of Euglena gracilis. Eur J Biochem. 54:367–372. [DOI] [PubMed] [Google Scholar]
- Fricova D, et al. 2010. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology 156:2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S, et al. 2002. The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0-ATPase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J Biol Chem. 277:6051–6058. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. 2000. Functions and mechanisms of RNA editing. Annu Rev Genet. 34:499–531. [DOI] [PubMed] [Google Scholar]
- Gray MW. 2003. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 55:227–233. [DOI] [PubMed] [Google Scholar]
- Gray MW. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 4(9):a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Doolittle WF. 1982. Has the endosymbiont hypothesis been proven? Microbiol Rev. 46:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lukeš J, Archibald JM., Keeling PJ, Doolittle PF. 2010. Irremediable complexity? Science 330:929–921. [DOI] [PubMed] [Google Scholar]
- Hallick RB, et al. 1993. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 21:3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Zimmer SL, Ammerman ML, Read LK, Lukeš J. 2013. Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex. Trends Parasitol. 29:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V. 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 3:344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutner SH, Zahalsky AC, Aaronson SA, Baker H, Frank O. 1966. Culture media for Euglena gracilispter. Methods Cell Biol. 2:217–228. [Google Scholar]
- Jensen RE, Englund PT. 2012. Network News: the replication of kinetoplast DNA. Annu Rev Microbiol. 66:473–491. [DOI] [PubMed] [Google Scholar]
- Kiethega GN, Yan Y, Turcotte M, Burger G. 2013. RNA-level unscrambling of fragmented genes in Diplonema mitochondria. RNA Biol. 10:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajčovič J, Vesteg M, Schwartzbach SD. 2015. Euglenoid flagellates: a multifaceted biotechnology platform. J Biotechnol. 202:135–145. [DOI] [PubMed] [Google Scholar]
- Krnáčová K, et al. 2015. Characterization of oxidative phosphorylation enzymes in Euglena gracilis and its white mutant strain WgmZOflL. FEBS Lett. 589:687–694. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. 2005. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354:110–116. [DOI] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM. 2004. Green algae and the origin of land plants. Am J Bot. 91:1535–1556. [DOI] [PubMed] [Google Scholar]
- Liu B, Shawn YL, Motyka A, Agbo EEC, Englund PT. 2005. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 21:363–369. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW. 2011. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 63:528–537. [DOI] [PubMed] [Google Scholar]
- Lukeš J, et al. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 1:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J, Flegontova O, Horák A. 2015. Diplonemids. Curr Biol. 25:R702–R704. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Leander BS, Keeling PJ. 2009. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc Natl Acad Sci U S A. 106:9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire F, Richards TA. 2014. Organelle evolution: a mosaic of “mitochondrial” functions. Curr Biol. 24:R518–R520. [DOI] [PubMed] [Google Scholar]
- Marande W, Burger G. 2007. Mitochondrial DNA as a genomic jigsaw puzzle. Science 318:415. [DOI] [PubMed] [Google Scholar]
- Marande W, Lukeš J., Burger G. 2005. Unique mitochondrial genome structure in Diplonemids, the sister group of Kinetoplastids. Eukaryot Cell. 4:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov D, et al. 2006. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol Biochem Parasitol. 148:69–78. [DOI] [PubMed] [Google Scholar]
- Moreno-Sánchez R, et al. 2000. Oxidative phosphorylation supported by an alternative respiratory pathway in mitochondria from Euglena. Biochim Biophys Acta. 1457:200–210. [DOI] [PubMed] [Google Scholar]
- Müller M, et al. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 76:444–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EC, et al. 2015. The transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry. Mol BioSyst. 11:2808–2820. [DOI] [PubMed] [Google Scholar]
- Perez E, et al. 2014. The mitochondrial respiratory chain of the secondary green alga Euglena gracilis shares many additional subunits with parasitic Trypanosomatidae. Mitochondrion 19:338–349. [DOI] [PubMed] [Google Scholar]
- Povelones ML. 2014. Beyond replication: division and segregation of mitochondrial DNA in Kinetoplastids. Mol Biochem Parasitol. 196:53–60. [DOI] [PubMed] [Google Scholar]
- Roy J, Faktorová D, Lukeš J, Burger G. 2007. Unusual mitochondrial genome structures throughout the Euglenozoa. Protist 158:385–396. [DOI] [PubMed] [Google Scholar]
- Shao R, Zhu XQ, Barker SC, Herd K. 2012. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol Evol. 4:1088–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD. 2015. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci U S A. 112:E3515–E3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10:e1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, et al. 2011. The GC-rich mitochondrial and plastid genomes of the green alga Coccomyxa give insight into the evolution of organelle DNA nucleotide landscape. PLoS One 6:e23624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 12:10177–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2015. The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief Funct Genomics.. Advance Access published June 27, 2015; doi:10.1093/bfgp/elv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2009. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 71:627–639. [DOI] [PubMed] [Google Scholar]
- Spencer DF, Gray MW. 2011. Ribosomal RNA genes in Euglena gracilis mitochondrial DNA: fragmented genes in a seemingly fragmented genome. Mol Genet Genomics. 285:19–31. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 30:97–105. [DOI] [PubMed] [Google Scholar]
- Tessier LH, van der Speck H, Gualberto JM, Grienenberger JM. 1997. The cox1 gene from Euglena gracilis: a protist mitochondrial gene without introns and genetic code modifications. Curr Genet. 31:208–213. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Myers AM. 1986. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 55:249–285. [DOI] [PubMed] [Google Scholar]
- Valach M, Moreira S, Kiethega GN, Burger G. 2014. Trans-splicing and RNA editing of LSU rRNA in Diplonema mitochondria. Nucleic Acids Res. 42:2660–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lis R, Mendoza-Hernández G, Groth G, Atteia A. 2007. New insights into the unique structure of the F0F1-ATP synthase from the chlamydomonad algae Polytomella sp. and Chlamydomonas reinhardtii. Plant Physiol. 144:1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Acevedo M, et al. 2006. The mitochondrial ATP synthase of chlorophycean algae contains eight subunits of unknown origin involved in the formation of an atypical stator-stalk and in the dimerization of the complex. J Bioenerg Biomembr. 38:271–282. [DOI] [PubMed] [Google Scholar]
- Verner Z, et al. 2015. Malleable mitochondrion of Trypanosoma brucei. Int Rev Cell Mol Biol. 315:73–151. [DOI] [PubMed] [Google Scholar]
- Vesteg M, et al. 2010. A possible role for short introns in the acquisition of stroma-targeting peptides in the flagellate Euglena gracilis. DNA Res. 17:223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlček C, Marande W, Teijeiro S, Lukeš J, Burger G. 2011. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 39:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Jackson CJ. 2009. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. BioEssays 31:237–245. [DOI] [PubMed] [Google Scholar]
- Yasuhira S, Simpson L. 1997. Phylogenetic affinity of mitochondria of Euglena gracilis and kinetoplastids using cytochrome oxidase I and hsp60. J Mol Evol. 44:341–347. [DOI] [PubMed] [Google Scholar]
- Zíková A, et al. 2008. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 7:1286–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.