Abstract
Genome-wide studies of African populations have the potential to reveal powerful insights into the evolution of our species, as these diverse populations have been exposed to intense selective pressures imposed by infectious diseases, diet, and environmental factors. Within Africa, the Sahel Belt extensively overlaps the geographical center of several endemic infections such as malaria, trypanosomiasis, meningitis, and hemorrhagic fevers. We screened 2.5 million single nucleotide polymorphisms in 161 individuals from 13 Sahelian populations, which together with published data cover Western, Central, and Eastern Sahel, and include both nomadic and sedentary groups. We confirmed the role of this Belt as a main corridor for human migrations across the continent. Strong admixture was observed in both Central and Eastern Sahelian populations, with North Africans and Near Eastern/Arabians, respectively, but it was inexistent in Western Sahelian populations. Genome-wide local ancestry inference in admixed Sahelian populations revealed several candidate regions that were significantly enriched for non-autochthonous haplotypes, and many showed to be under positive selection. The DARC gene region in Arabs and Nubians was enriched for African ancestry, whereas the RAB3GAP1/LCT/MCM6 region in Oromo, the TAS2R gene family in Fulani, and the ALMS1/NAT8 in Turkana and Samburu were enriched for non-African ancestry. Signals of positive selection varied in terms of geographic amplitude. Some genomic regions were selected across the Belt, the most striking example being the malaria-related DARC gene. Others were Western-specific (oxytocin, calcium, and heart pathways), Eastern-specific (lipid pathways), or even population-restricted (TAS2R genes in Fulani, which may reflect sexual selection).
Keywords: genome-wide diversity, admixture, selection, Sahel
Introduction
Africa was the cradle of modern humans and the only inhabited continent for around two-thirds of their history, hence extant African populations harbor the greatest worldwide genetic diversity (Prugnolle et al. 2005; Soares et al. 2012; Rito et al. 2013). This rich genetic pool has been modeled by complex demographic (changes in population size, migration events, and admixture) and genetic (natural selection, recombination, and mutation) events. Genomic studies in African populations are therefore of paramount potential in revealing main aspects of human population history and genetic susceptibility to diseases. Before the advent of genome-wide studies (GW), a large screening in Africa consisting in 1,327 microsatellites showed that the population structure follows quite reasonably the self-described ethnic and linguistic groups, implying overall a large and subdivided population structure in Africa that was opposed in several populations by extensive mixture of ancestries owing to large-scale migration events that occurred throughout history (Tishkoff et al. 2009). First GWs focused on African-Americans and in their Western African ancestors (Smith et al. 2004; Zakharia et al. 2009), and even the 1,000 Genomes project (Abecasis et al. 2012) includes mainly Western African samples (four Western and one Eastern of Bantu/Western ancestry). Other GWs began to describe either population structure of hunter-gatherers as the Pygmies (Patin et al. 2014), Khoisan (Pickrell et al. 2012) and Khoisan-speaking Hadza and Sandawe from Tanzania (Pickrell et al. 2012), or admixture with Eurasians in East and South Africa (Pickrell et al. 2014). But it remains difficult to pursue GW in other African populations across the continent and to evaluate the genomic effects of local demographic and selective pressures taking into account the myriad of environments, climates, diets, lifestyles, and exposure to infectious diseases (Campbell and Tishkoff 2008; Teo et al. 2010).
Among the African regions, the Sahel is of particular importance because of its major role as a migration corridor (Newman 1995; Cerny et al. 2009; Pereira et al. 2010; Cerny et al. 2011; Soares et al. 2012). The Sahel Belt lies between the Sahara desert in the north and the tropical forests in the south (more or less between parallels 20°N and 10°N), lacks high mountains or other barriers, and constitutes a particular and very specific ecosystem (fig. 1A) made of semi-arid grasslands, savannas, steppes, and thorn shrublands. The eastern part of the Belt goes even below the equator, including Ethiopia, South Sudan, Somalia, and Kenya. Despite its overall aridity (UNEP 2008), the Sahel has important water resources, consisting in long river courses (Nile, Niger, and Senegal) and Lake Chad. Climatic changes have transformed Sahel’s aridity along time, from a notoriously arid-uninhabitable desert during the Last Glacial Maximum to a humid-fertile landscape of lakes and savannah from 10 to 6 thousand years ago (ka) in the Holocene Climatic Optimum (Drake et al. 2011). These climatic oscillations and annual cycles imply population expansions and migrations when conditions are favorable, and bottlenecks and isolation in refugia when periods are more difficult.
Fig. 1.—
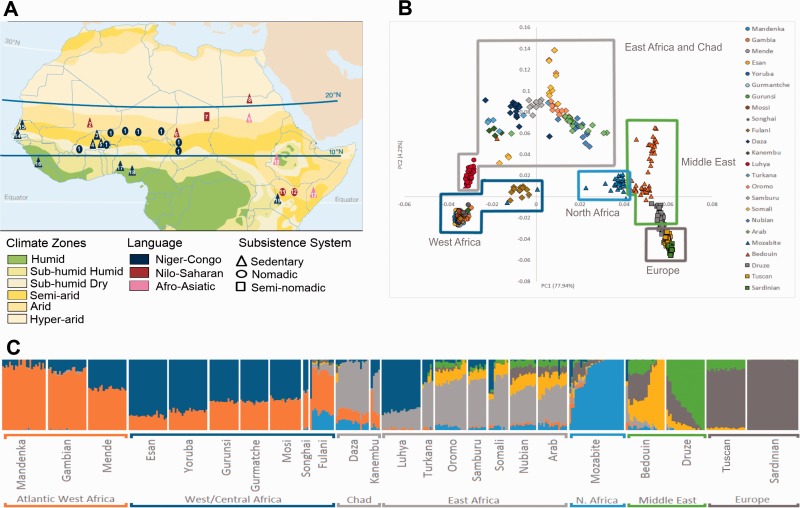
Location of studied samples and population structure across Sahel. (A) Geographic locations of the populations studied here, with subsistence system and family language affiliation identified. The colored zones indicate the current climate zones. Numbers 1 to 13 refer to groups studied here: 1 – Fulani; 2 – Songhai; 3 – Mossi; 4 – Gurunsi; 5 – Gurmantche; 6 – Kanembu; 7 – Daza; 8 – Nubians; 9 – Sudanese Arabs; 10 – Oromo; 11 – Samburu; 12 – Turkana; and 13 – Somali. Numbers 14 to 19 refer to groups from 1000 Genomes project and Li et al. (2008): 14 – Gambian in Western Division; 15 – Mandenka in Senegal; 16 – Mende in Sierra Leone; 17 – Yoruba in Ibadan, Nigeria; 18 – Esan in Nigeria; and 19 – Luhya from Kenya. (B) PCA1 versus PCA2. (C) Admixture analysis for K = 7 ancestral populations (each represented by a color). Each vertical line is an individual.
Two sympatric lifestyles co-exist in the Sahel: nomadic pastoralists who find pasture during the short rainy season in southern Sahara, and sedentary farmers who settled in the more humid areas. Pastoralists raise livestock, which was probably introduced 8-5 ka in Africa from the Near East, following the Nile Valley, and eventually they used the Sahel Belt as a corridor to reach Western Africa (Hanotte et al. 2002), leaving genetic evidences in the human mitochondrial DNA pool (Cerny et al. 2009). Unlike nomadic pastoralism, sedentary farming is a rather recent phenomenon in the area (Marshall and Hildebrand 2002). The largest pastoral nomadic group in the world is the Fulani ethnic group, living today in 17 African countries and amounting to almost 40 million people, of which one-third still preserves the nomadic way of life (Cerny et al. 2006). Their origin is uncertain, with one of the hypothesis stating that in ancient times, the Fulani people moved from Egypt/Ethiopia to Senegal and at the beginning of the 13th century, migrated southwards, where grazing was available (Steverding 2008). Also, northwards of Lake Chad, the seminomadic Daza people live in small hamlets concentrated in oases around the Lakes of Ounianga (Podgorna et al. 2013), where they cultivate dates and grain, and practice transhumance of camels and donkeys. Turkana and Samburu, who originated in Sudan (Spencer 1965; Lamphear 1988) and live currently in North Kenya, are dedicated to cattle pastoralism and a traditional way of life, similar to their neighbor Maasai. Oromo and Somali were also nomadic in the past, but currently they are mostly sedentary (Lewis 1999; Etefa 2012).
Domesticated animals, which are essential to the former and extant economic Sahelian subsistence, also play a significant role as reservoirs of human parasites (Wolfe et al. 2007). Combined with climate and environmental conditions, the close contact between humans and cattle has been contributing to the massive burden of endemic and epidemic diseases in the Sahel. The Sahelian countries have one of the highest under-five years’ mortality rates, with the majority of deaths mainly caused by pneumonia, diarrhea, and malaria. The region experiences recurrent outbreaks of cholera, measles, meningitis, polio, and typhoid (WHO 2012). It is among the worldwide regions with highest burden of malaria, and it has been shown that the vector Anopheles gambiae can survive the long Sahelian dry-season by entering into diapause (Huestis and Lehmann 2014). The Sahel region is known as the meningitis belt, with person-to-person spread epidemics caused by the bacteria Neisseria meningitides (Molesworth et al. 2002). Another highly conditioning disease in the region is the human African trypanosomiasis or sleeping sickness (Steverding 2008), caused by Trypanosoma brucei gambiense (in Western and Central Africa) and T. brucei rhodesiense (in Eastern and Southern Africa). These protozoan parasites are transmitted to mammalian hosts by the bite of infected tsetse flies (Glossina sp.), whose range in Africa overlaps largely the Sahel Belt. Other neglected tropical diseases, including a large majority caused by helminth infections (such as hookworm, schistosomiasis, and lymphatic filariasis), are also endemic in this region (Hotez and Kamath 2009). Thus, the Sahel Belt is a zone of strong selective pressure acting upon the human population.
To better characterize the genetic structure and adaptive history of Sahelian populations, we genotyped 2.5 million single nucleotide polymorphisms (SNPs; Human Omni 2.5 DNA Bead Chip, Illumina) in 13 populations (n = 161 individuals – fig. 1A and supplementary table S1, Supplementary Material 1 online) from the interior western region (Burkina), the Chad Basin, and across the Eastern side of the Sahelian zone. These populations are from diverse linguistic and subsistence system affiliations, and have extensively escaped the influence of the Bantu migration, which contributed greatly to homogenize the population diversity in the bulk of sub-Saharan Africa below the Belt (Gurdasani et al. 2015; Silva et al. 2015). We first evaluated the population structure in the Belt and then inferred candidate selected genomic regions by using two complementary haplotype-based tests, integrated haplotype score (iHS; Voight et al. 2006) and cross population extended haplotype homozygosity (XP-EHH; Sabeti et al. 2007). iHS has good power to detect selective sweeps (consisting in the fixation of a beneficial mutation) at moderate frequency (50–80%), and XP-EHH is most powerful for selective sweeps above 80% frequency (Voight et al. 2006; Sabeti et al. 2007). Notwithstanding the difficulty in proving that these candidate regions in fact reflect the action of positive selection (Hernandez et al. 2011), they display an outlier pattern of diversity that is consistent with positive selection and are enriched in true positives. The level of resolution of our study allowed us to get informative insights into the palimpsest of genome-wide candidate selected regions across the Sahel, one of the worldwide zones most exposed to infection.
Material and Methods
Population Samples, Genome-wide Genotyping, and Published Data
DNA samples analyzed in this study were collected from 13 populations in eight African countries. Further information relative to these populations is provided in supplementary table S1 (Supplementary Material 1 online) and geographic location can be found in fig. 1A. This study obtained ethical approval from the Ethics Committee of the University of Porto, Portugal (17/CEUP/2012). A total of 171 individuals were genotyped for the Illumina Human Omni 2.5 DNA Bead Chip, containing approximately 2.5 million SNPs. Nine samples from the Turkana population were excluded from the analysis, as they were closely related, and another Turkana was a clear ancestry outlier of the population (genetically close to Europeans). We ended up with 161 individuals. Quality control is summed up in supplementary fig. S1 (Supplementary Material 1 online), and a total of 2,247,183 SNPs passed quality control checking. To increase spatial resolution, we included 50 randomly selected unrelated individuals from relevant populations from the 1,000 Genomes Project (Abecasis et al. 2012), and from Li et al. (Li et al. 2008), reported in supplementary table S2 (Supplementary Material 1 online). The extended low-density data set contained 370,470 SNPs and 732 individuals. The build used in all analyses was GRCh37.
Population Structure and Differentiation
Several population genetic analyses assume independent markers, so SNPs were pruned for pairwise linkage disequilibrium (LD) in PLINK (Purcell et al. 2007), by removing any SNP that had a r2 > 0.4 with another SNP, within a 50-SNPs sliding window with step of 20 SNPs. Principal component analysis (PCA), which infers worldwide axes of human genetic variation from the allele frequencies of various populations, was carried out by using the smartpca tool, included in the EIGENSOFT package (Patterson et al. 2006). ADMIXTURE, which provides a maximum likelihood estimation of the population structure (Alexander et al. 2009), was run for several values (from 2 to 7) of clusters or ancestral populations, K. The optimal K was estimated through cross-validation of the logistic regression. Wright’s FST metric was calculated using Vcf tools (Danecek et al. 2011). Details are provided in supplementary fig. S1 (Supplementary Material 1 online).
Local Ancestry Inference
We applied the RFMix algorithm (Maples et al. 2013), which uses a LD model between markers to infer ancestry for each segment of the genome between a mixture of two putative ancestral panels of haplotypes. We used as ancestral data sets the phased data from the 1,000 Genomes Database, the Italian sample representing southern European ancestry and Gambian or Luhya representing the African ancestry (the first when testing for Fulani and the later when analyzing Central and Eastern Sahelian populations). Italians are a good proxy population for the shared Mediterranean ancestry across southern Europe, North Africa, and the Near East/Arabian Peninsula, which is mixed with the sub-Saharan ancestry across the Sahel (Botigue et al. 2013). For comparison purposes, we assayed the effect of using Great Britain (GB) or Northern Europeans from Utah (CEU) as the non-African parental population in the Oromo data set, although by being derived North European populations, these are not geographically, historically, and anthropologically supported as good proxies for non-African admixture in the Sahel (Table S10). Some differences were observed; for instance, the main block on chromosome 2, in the region of the LCT gene, which has been mainly selected in North Europe, was identified in all three analyses, but the size was different: it was larger when using Italians (133,640,409-137,586,694, totaling 3,946,285 bp) than when using GB (133,346,735-135,210,390 – 1,863,655 bp) or CEU (133,346,735-134,727,858 – 1,381,123 bp). So, an input from a non-European population is confirmed in this region, although we do not know if the driver was LCT or another neighbor gene.
Our samples were phased in SHAPEIT v.2 (Delaneau et al. 2012) using HapMap reference panel and fine-scale genetic map. Information on ancestry was obtained for each locus along chromosomes for every individual, and these values were averaged in each population. The null hypothesis equates that the proportion of a certain parental ancestry in an admixed population will be equal across the genome. The test hypothesis is that a certain region of the genome may have a significantly higher proportion of a parental ancestry in comparison with its genome mean value, indicating the action of some event (as positive selection). To identify regions that have a significantly higher proportion of a given parental ancestry, we followed published studies (Bryc et al. 2010) considering only regions outside the range defined by the genome mean value for a certain ancestry ± 3SD. Genes identified as statistically significantly increased in non-African or in African ancestries in the admixed Central and Eastern Sahelian populations were verified for association with complex diseases in the Catalog of Published Genome-Wide Association Studies (downloaded from https://www.genome.gov/26525384 on the 2nd of April 2015) (Welter et al. 2014).
Analysis of Selection
We had to join some of the neighboring samples owing to low sample size, and for that we took into account language affiliation, subsistence system, and the results from population structure, so that reasonably homogeneous sets of populations could be obtained (as also performed by Pickrell et al. 2009, and having in mind the values indicated by them, of a minimum of ∼40 chromosomes for iHS and as few as 20 chromosomes for XP-EHH). Thus, we joined: Gurmantche, Gurunsi, Mossi, and Songhai (recalled Burkina); Daza and Kanembu; Arabs and Nubians; and Turkana and Samburu. We estimated the iHS (Voight et al. 2006) by using the selscan package (Szpiech and Hernandez 2014). SNPs were prunned for minor allele frequency (MAF) >1% in each population. iHS tracks the decay of haplotype homozygosity for both the ancestral and derived haplotypes extending from a tested core SNP. Scores were calculated for each SNP within the population as a whole and standardized within each of 100 bins of allele frequency, using the norm tool. To facilitate comparison of genomic regions between populations and to gain power by detecting selective sweeps affecting the haplotypic structure at surrounding SNPs (Pickrell et al. 2009), we split the genome into non-overlapping segments of 100 kb, so that at least 20 SNPs are present in each segment. For each window, the proportion of absolute standardized iHS scores higher than 2 were calculated and used to order the windows (we confirmed that this measure was more robust in our data set than using simply the highest absolute iHS score, as basically the ordered genes were more similar between neighbor populations when using that measure). Moreover, 100-kb windows were assigned a percentile score based on the proportion of extreme iHS values in the segment. In the 99% percentile range, we checked if significant windows could be collapsed owing to their tandem location, and we did so while maintaining the highest percentile value for the new collapsed window. Percentiles were recalculated for the remaining windows after collapsing. With this strategy, we diverged from others (Voight et al. 2006; Pickrell et al. 2009) because we confirmed that binning windows by their number of SNPs (in bins incremented by 10 each, and excluding windows with <20 SNPs) could decrease the significance of relevant regions; basically, the sharing of top candidate selected regions was higher between Sahelian populations in our strategy than when using bins.
The SelScan tool (Szpiech and Hernandez 2014) was also used to estimate XP-EHH. We calculated XP-EHH for the following pairs of populations: each African population compared with Italians, each Western African population compared with Oromo, and Eastern and Central Sahel populations compared with Gambians. Consistently with other tests for selection, only markers with MAF >1% were used for XP-EHH computation. The obtained XP-EHH values were normalized by subtracting genome-wide mean XP-EHH and dividing by standard deviation (Szpiech and Hernandez 2014). Whole genome was divided into 100-kb windows (same windows used in iHS analysis). In every window, top XP-EHH value was selected and if it falls into top 0.1% of XP-EHH values, the window was selected and was searched for genes in a whole 100-kb window and also in the 5-kb flanking region around the highest XP-EHH value. Previous reports (Pickrell et al. 2009) have shown that the maximum XP-EHH scores are more powerful as a statistic than the fraction of extreme scores, in opposition to what happens for iHS and we followed those indications in our analyses. Details are provided in supplementary fig. S2 (Supplementary Material 1 online).
Pathway Analysis – KEGG Pathway Database
Enrichments in every KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway (http://www.genome.jp/kegg/pathway.html, last accessed April 2, 2015) were tested for all genes identified as positively selected using the three following tests: iHS (genes in 100-kb window, top 300 windows), XP-EHH vs Italians (top 300 windows, only genes in the 5-kb flanking region around the top XP-EHH value were considered), and XP-EHH vs Gambians/Oromo (for Eastern and Central Sahel populations, we used Gambia as a reference population; for West Africans, we used Oromo; also, top 300 windows and genes in the 5-kb flanking region around the top XP-EHH). Every gene that appeared in one of the three selection tests received a score calculated from its percentile in the test (the higher the selection signal, the higher the score). If the gene was flagged in several tests, the individual scores were summed up for that gene within a pathway and then normalized to fit scale from 0 to 10. We produced heat maps for each pathway displaying the genes that appeared in the selection test in at least one population (Supplementary Material 3 online). We also produced a global heat map displaying all pathways, summing up the scores for the genes flagged as selected in that pathway (Supplementary Material 2 online). Details are provided in supplementary fig. S2 (Supplementary Material 1 online).
Results and Discussion
We first studied the genetic structure of Sahelian populations using ADMIXTURE (Alexander et al. 2009). Figure 1C highlights the strong impact of gene flow in this region for K = 7, which includes a Mozabite/North African component that allows to address the hypothesis of a North African ancestry in Fulani (other K plots and cross-validation are presented in supplementary figs. S3 and S4, Supplementary Material 1 online). Western African populations present varying proportions of two clusters, one being more frequent in Atlantic Western populations (orange in fig. 1C; 90% in Mandenka), whereas the other is more frequent in Western/Central populations, especially in Esan and Yoruba of Nigeria (dark blue; reaching 74–81% frequency). This main Western/Central component probably represents groups speaking Niger-Congo languages, including Narrow Bantu speakers, as can be verified by its high frequency (75%) in Luhya, a Bantu-speaking population from Kenya, who also presents a substantial proportion of Eastern African ancestry (23%). In clear contrast with Western Africans, Eastern Africans present a considerable input from Near Eastern (green color), Arabian (yellow color), and North African (light blue color) components (23–49%), with the exception of Turkana (7%), as well as some Somali, who instead show substantial fractions of Western/Central ancestry (27%), possibly received from the Bantu expansions in the region of the African Great Lakes. Both populations from Chad, the Daza and Kanembu, present a high Eastern African component (light gray; 41–61%), mixed with Atlantic Western (14–25%), Western/Central African (10–29%), and North African (5–13%; lower in the sedentary Kanembu and higher in semi-nomadic Daza) backgrounds. The nomadic Fulani present the reverse pattern for the Atlantic Western versus Eastern African components (55% and 11%, respectively), but the proportion of the North African component is even higher (23%) than in Daza (∼10%). These results support the hypothesis of a North African origin and a Western to Central Africa past migration for Fulani. Notice that in ADMIXTURE results for K < 7, the Mozabite/North African component is not identified, being identical to the European component, indicating that these two components are very similar.
PCA of the data confirms these relationships between populations (fig. 1B and supplementary fig. S5, Supplementary Material 1 online), as well as Wright’s FST metrics (supplementary figs. S6 and S7, Supplementary Material 1 online). Altogether, our results support the role of the Sahelian Belt as a main corridor for human migrations across the African continent.
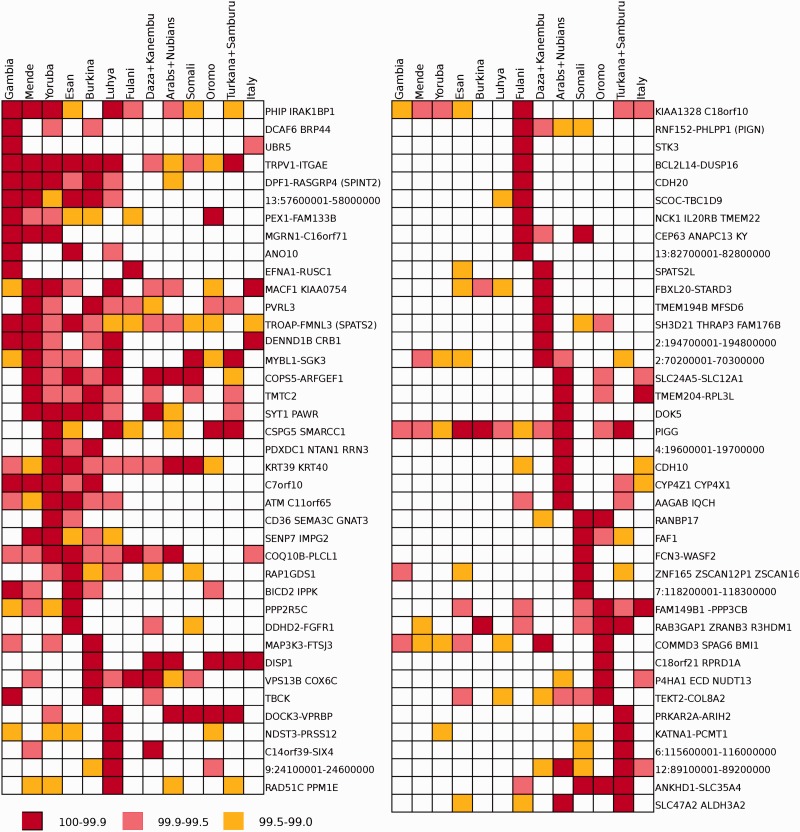
We next sought to identify genomic regions showing excess ancestry from non-Sahelian populations, as well as outliers for informative statistics on positive selection detection, and to describe the fine-scale geographical distribution of these selection signals across the Sahel. Figure 2 illustrates the top-10 iHS candidate selected regions observed in each Sahelian population, and signals of selection in Italians for comparison (supplementary table S11, Supplementary Material 1 online, reports the iHS significant regions in all populations). Western Sahelian samples share a higher amount of significant regions between them than the Eastern group, many of them in the 1% tail of the distribution. Given the North African influences in Fulani and the non-African (via Eastern African) influence in Daza + Kanembu, these Central Sahelian populations have a mixed pattern of selection of the observed in Western and Eastern Sahelian groups. The results for the top-10 XP-EHH, when comparing with Italians (supplementary fig. S16 and S17 and table S12, Supplementary Material 1 online), show a higher sharing of selected genes between Western and Eastern Sahelian pools than in iHS, compatible with the longer time needed for the stronger selective sweeps detected in XP-EHH analysis. When comparing Western with Eastern groups (supplementary fig. S14 and S15 and table S12, Supplementary Material 1 online), the pattern of top XP-EHH-based selected genes was very homogeneous within the Western group, as well as very homogeneous within the Eastern group, but different between them.
Fig. 2.—
Top-10 iHS in each Sahelian population and matching selected genes in Italians. Some of the regions contain many genes, and only the first and last genes are indicated, with interesting genes reported inside brackets.
RFMix software (Maples et al. 2013) was used to deconvolute the genomes of admixed Sahelian populations into segments originating from their two main parental populations. Interestingly, some of the selection signals were associated with a local genomic enrichment of the non-predominant ancestry in the admixed Sahelian populations (supplementary tables S3–S8 and figs. S8–S13, Supplementary Material 1 online), as described below.
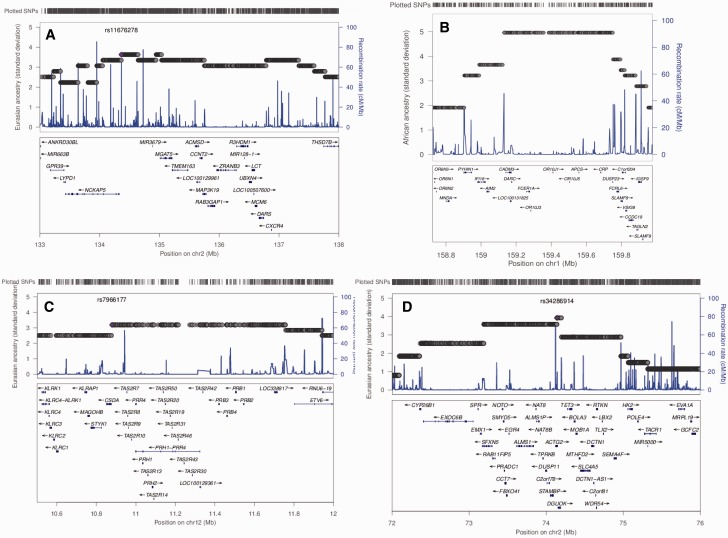
The best example of a widespread candidate selection signal across the Sahel is the malaria-associated DARC gene (fig. 3F), located on chromosome 1. This gene, the Duffy antigen/receptor for chemokines, encodes a membrane-bound chemokine receptor used by Plasmodium vivax for internalization into red blood cells (Demogines et al. 2012), so that Duffy-negative phenotype is thought to confer resistance against malaria caused by this parasite. There is complete fixation of the protective null allele in the Western region and, as we detected here, a local enrichment of African ancestry in the highly admixed Sudanese Arabs and Nubians (fig. 4B), where P. vivax is frequent. This evidence attests to the high selective pressure for DARC gene in the Sahel environmental context, even in its northeastern-most border. Although selection on the DARC gene has been identified a long time ago, our work properly contextualizes its importance in the Sahelian selective landscape.
Fig. 3.—
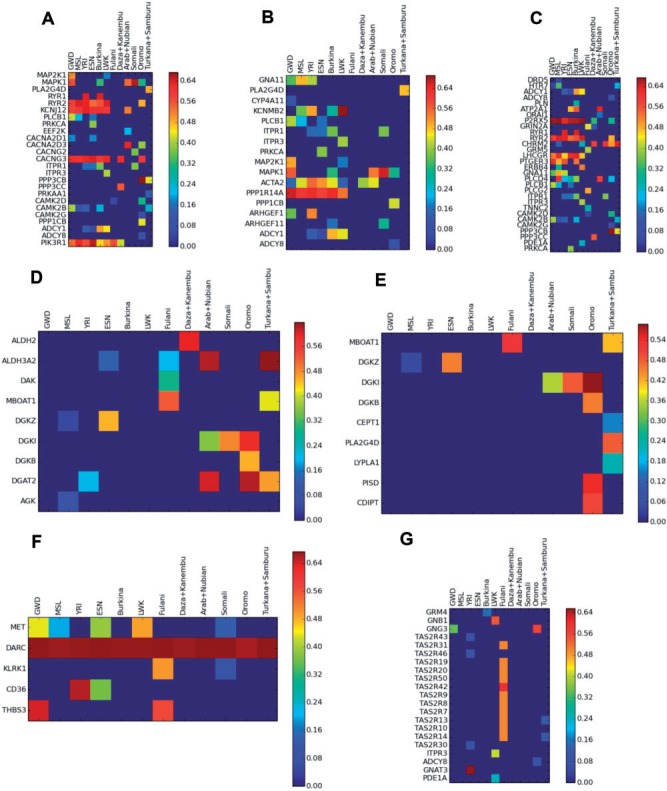
Selected genes in informative metabolic pathways (KEGG database). (A) Oxytocin signaling pathway. (B) Vascular smooth muscle contraction. (C) Calcium signaling pathway. (D) Glycerolipid metabolism. (E) Glycerophospholipid metabolism. (F) Malaria. (G) Taste transduction. GWD – Gambia; MSL – Mende; YRI – Yoruba; ESN – Esan; LWK – Luhya.
Fig. 4.—
Locus zoom of a few enriched ancestry regions. (A) Chromosome 2 in Oromo. (B) Chromosome 1 in Sudanese Arabs + Nubians. (C) Chromosome 12 in Fulani. (D) Chromosome 2 in Turkana + Samburu.
Another long genomic candidate selected region across the Sahel Belt is placed on chromosome 17, but it shows distinct peaks in the iHS measure between Western and Eastern African regions. The iHS peak is located around the ITGAE gene in the Western populations, whereas it is centered on TRPV1/SHPK genes (78,300 bp apart) in the Central and Eastern samples (supplementary figs. S18 and S19, Supplementary Material 1 online). This can be an ongoing process of differentiating selection on a previously large selected genomic region. ITGAE is a receptor for E-cadherin and mediates adhesion of intra-epithelial T-lymphocytes to epithelial cell monolayers (involved in KEGG pathway regulation of actin cytoskeleton), playing a role in the immune system. Grossman et al. (2013) also detected signals of selection for this gene (non-synonymous SNP) in Yoruba, and further confirmed its functional impact through structural homology modeling and conservation analysis. The TRPV1 gene encodes a receptor for capsaicin, the main pungent ingredient in hot chili peppers, which is also activated by noxious increases in temperature (Gavva et al. 2004), and SHPK controls glucose metabolism and acts as a modulator of macrophage activation (Haschemi et al. 2012).
Three other regions show signals of positive selection in almost all populations across Sahel, in the iHS analysis (fig. 2). One is located on chromosome 12 around the SPATS2 gene (supplementary figs. S20 and S21, Supplementary Material 1 online), which plays a role in spermatogenesis (Senoo et al. 2002). Another is on chromosome 17, containing the keratin genes KRT39 and KRT40, the latter being expressed during hair follicle differentiation (Langbein et al. 2007). On chromosome 4, a signal is detected on the PIGG gene, which is involved in the glycosylphosphatidylinositol anchor biosynthesis pathway, allowing the attachment of cell surface proteins to the cell membrane (Stokes et al. 2014).
Notwithstanding the ubiquitous selected genes across the Sahel, there were different profiles of candidate selected genes in the Western and Eastern sides. The main difference resides in strong signals of selection in several genes of the calcium and related heart and oxytocin pathways in the western region (fig. 3A–C), whereas, in the eastern side, glycerolipid and glycerophospholipid metabolism pathways displayed stronger signals of selection (fig. 3D and E). Oxytocin controls a wide variety of central and peripheral effects, especially the stimulation of uterine contractions during parturition and milk release in lactation, which are per se strong selection drivers, but it also influences cardiovascular regulation (Arrowsmith and Wray 2014), redounding effects of the tandem selected genes (PIK3R1, CACNG3, RYR2, and KCNJ12) in pathways related with heart, namely, on cardiac muscle contraction and adrenergic signaling in cardiomyocytes. By opposition, the Eastern trans-region selection signal displayed by the glycerolipid and glycerophospholipid metabolism pathways is mainly owing to the high values of selection in the DGAT2 gene found on top XP-EHH vs West (supplementary fig. S15, Supplementary Material 1 online) and also the DGKI gene. This, together with the high value of selection detected in iHS in Eastern Sahel (except in Arab + Nubian) for the RAB3GAP1 gene that is associated with cholesterol, testifies the importance of lipid metabolism in Eastern Sahel. It was already reported that the Eastern African pastoralist Maasai, whose diet of milk, blood, and meat is rich in lactose, fat, and cholesterol, display selection signals at the RAB3GAP1/LCT/MCM6 region (Wagh et al. 2012). Here we show that this pattern is not only observed in nomads such as Maasai, Turkana, and Samburu, who may share a common/related origin, but also in the sedentary Oromo and Somali. Interestingly, that region in chromosome 2 is not autochthonous, having a significant non-African enrichment in Oromo (fig. 4A) that probably conferred an advantage to these populations, who have milk and blood as important food sources. Another non-African enriched region detected in Turkana + Samburu is located in another region of chromosome 2 (fig. 4D), containing the ALMS1 gene associated with leprosy (Grossman et al. 2013) as well as the NAT8 gene involved in creatinine levels and chronic kidney disease, which is frequent in African descendants. The Eurasian enrichment is probably a protection against chronic kidney disease in these Eastern groups.
The candidate selected regions restricted to Western Sahel have genes playing important roles. This is the case of a region in chromosome 19, rich in many genes, where the highest iHS (supplementary figs. S22 and S23, Supplementary Material 1 online) and XP-EHH vs East values are attained for the gene SPINT2, which has been associated with diarrhea (Heinz-Erian et al. 2009), and neighboring gene CATSPERG, which is required for sperm hyperactivated motility and male fertility (Wang et al. 2009). Other examples are: BTRC, previously associated with HIV (Nazari-Shafti et al. 2011); TLR5, suggested to alter NF-kB signaling in response to bacterial flagellin (Grossman et al. 2013); PSMD8, a member of the 26S proteasome involved in the regulation of transcription initiation (Durairaj and Kaiser 2014); and LHCGR, whose mutations result in disorders of male secondary sexual character development (Jeha et al. 2006).
The most geographically restricted selection signature was detected in Fulani, corresponding to a non-African ancestry enriched region in chromosome 12 (figs. 3G and 4A). This region contains TAS2R genes, which detect natural alkaloids such as quinine and strychnine (Kim et al. 2005; Reed et al. 2010; Ledda et al. 2014), possibly establishing a bridge with a Fulani ritual, of vital importance for some nomadic groups. The ceremony consists in a public flogging named “Sharo,” a test of manhood: all youths must pass this ordeal without flinching to be considered as adults and eligible to get married (Adeola 2014). The courage is fortified by the previous drinking of a beverage (native beer or palm tree) containing seeds of the plant Datura metel, which initiates a stupefying, narcotic effect. The seeds of this plant contain alkaloids (Oliver-Bever 1986) scopolamine or hyoscine, which depress the central nervous system, as well as hyoscyamine, which blocks all the body secretions, including the lachrymal glands, preventing tearing. Could non-African alleles in TAS2R genes on chromosome 12, introduced by the admixed origin of Fulani, allow a more efficient processing of these alkaloids contained in the beverage? This is an interesting hypothesis to be further tested, as it would be a striking example of sexual selection driving a significant excess of non-African ancestry. It must be reinforced that these TAS2R genes are distinct from the TAS2R16 on chromosome 7, which detects salicin, a bitter β-glycoside anti-inflammatory compound, reported previously as having been under selection (through Fay, Wu’s H, and McDonald–Kreitman tests) in Eurasian, East African, and Fulani populations (Li et al. 2011; Campbell et al. 2014). We did not detect signals of selection in TAS2R16.
Conclusion
In summary, by combining a rich population survey, a high-resolution genome-wide chip, and local ancestry and selection tools, we have demonstrated the power to dissect the palimpsest of complex interactions between cross and local selective pressures and demographic factors. We have confirmed independently selection signals described before, found new candidates to be further investigated, and provided a first glimpse of their spatial distribution across a considerable region of the African continent that has been playing a major role in human migrations along millennia. Signals are very strong in certain genes, persisting across Sahel, probably indicating that the selection event occurred in the ancestral African population, before the main Pleistocene migrations that established the bulk of the Sahelian genetic landscape (Soares et al. 2012; Rito et al. 2013). Malaria selection in the DARC gene is most probably an old (before 100,000 years ago) pathogen-driven selection force (Karlsson et al. 2014). But several differences exist between Western and Eastern regions, largely explained by the high admixture with non-African ancestry observed in the Eastern side and by recent pathogens that could have led to local adaptations. One possible example is the non-African enrichment signal detected in Eastern Sahel for ALMS1 gene, which has been associated with leprosy based on GW association analysis in Indian patients (Grossman et al. 2013). Leprosy pathogen dates to around 12,000 years (Karlsson et al. 2014).
It is interesting that many of the enriched-ancestry and candidate selected regions are large and contain several genes that can contribute to adaptation to different selective factors. For instance, the Eastern Sahelian selected RAB3GAP1/LCT/MCM6 region, related with lipid metabolism, also contains the CXCR4, determinant in HIV entrance into cells and tuberculosis resistance. The TAS2R region in Fulani contains PRB3 gene, which acts as a bacterial receptor. The olfactory receptors OR51B5 and OR51B6 have been associated with sickle-cell anemia (Solovieff et al. 2010) but probably owing to their overlap with the HbE gene and neighboring HbB gene. An indirect evidence of their probable role in other non-olfactory sensory functions is their confirmed expression outside the olfactory epithelium, such as in kidney (Pluznick et al. 2009) and sperm (Spehr et al. 2003). OR10J5 gene, located in the region containing DARC and other genes associated with hematology traits, has been said to play a role in angiogenesis and its expression in aorta and coronary artery has been confirmed (Kim et al. 2015).
The modeling of the African genetic diversity by selection driven by infectious diseases, since the origin of modern humans till present times, provides useful insights into natural ways of resistance. These natural strategies can potentially be mimicked pharmacologically, opening new avenues in the combat of infectious and other complex diseases. A good example of this potential is provided by the HIV resistance of CCR5-Δ32 homozygous, which is being used as the basis of stem cell transplantation trial therapies (Novembre and Han 2012).
Supplementary Material
Supplementary data S1–S3, figures S1–S23, and table S1–S12.24 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The research leading to these results has received funding from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013/ under REA grant agreement no. 290344 (EUROTAST). This project was also supported by the Grant Agency of the Czech Republic (13-37998S-P505). P.S. is supported by FCT (the Portuguese Foundation for Science and Technology), through FCT Investigator Programme (IF/01641/2013). IPATIMUP integrates the i3S Research Unit, which is partially supported by FCT. FEDER, COMPETE, and FCT fund IPATIMUP (PEst-C/SAU/LA0003/2013) and CBMA (PEst-OE/BIA/UI4050/2014).
Literature Cited
- Abecasis GR, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeola BS. 2014. Datura Metel L: Analgesic or Hallucinogen? “Sharo” Perspective. Middle East J Sci Res. 21:993–997. [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith S, Wray S. 2014. Oxytocin: its mechanism of action and receptor signalling in the myometrium. J Neuroendocrinol. 26:356–369. [DOI] [PubMed] [Google Scholar]
- Botigue LR, et al. 2013. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci U S A. 110:11791–11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, et al. 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 107:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, et al. 2014. Origin and differential selection of allelic variation at TAS2R16 associated with salicin bitter taste sensitivity in Africa. Mol Biol Evol. 31:288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA. 2008. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 9:403–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny V, et al. 2006. MtDNA of Fulani nomads and their genetic relationships to neighboring sedentary populations. Hum Biol. 78:9–27. [DOI] [PubMed] [Google Scholar]
- Cerny V, et al. 2009. Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evol Biol. 9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerny V, et al. 2011. Genetic structure of pastoral and farmer populations in the African Sahel. Mol Biol Evol. 28:2491–2500. [DOI] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156––2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. 2012. A linear complexity phasing method for thousands of genomes. Nat Methods. 9:179–181. [DOI] [PubMed] [Google Scholar]
- Demogines A, Truong KA, Sawyer SL. 2012. Species-specific features of DARC, the primate receptor for Plasmodium vivax and Plasmodium knowlesi. Mol Biol Evol. 29:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake NA, Blench RM, Armitage SJ, Bristow CS, White KH. 2011. Ancient watercourses and biogeography of the Sahara explain the peopling of the desert. Proc Natl Acad Sci U S A. 108:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durairaj G, Kaiser P. 2014. The 26S proteasome and initiation of gene transcription. Biomolecules 4:827–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etefa T. 2012. Integration and peace in East Africa: a history of the Oromo Nation. New York: Palgrave Macmillan. [Google Scholar]
- Gavva NR, et al. 2004. Molecular determinants of vanilloid sensitivity in TRPV1. J Biol Chem. 279:20283–20295. [DOI] [PubMed] [Google Scholar]
- Grossman SR, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdasani D, et al. 2015. The African Genome Variation Project shapes medical genetics in Africa. Nature 517:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanotte O, et al. 2002. African pastoralism: genetic imprints of origins and migrations. Science 296:336–339. [DOI] [PubMed] [Google Scholar]
- Haschemi A, et al. 2012. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 15:813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz-Erian P, et al. 2009. Mutations in SPINT2 cause a syndromic form of congenital sodium diarrhea. Am J Hum Genet. 84:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, et al. 2011. Classic selective sweeps were rare in recent human evolution. Science 331:920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez PJ, Kamath A. 2009. Neglected tropical diseases in sub-saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Negl Trop Dis. 3:e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huestis DL, Lehmann T. 2014. Ecophysiology of Anopheles gambiae s.l.: persistence in the Sahel. Infect Genet Evol. 28:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeha GS, Lowenthal ED, Chan WY, Wu SM, Karaviti LP. 2006. Variable presentation of precocious puberty associated with the D564G mutation of the LHCGR gene in children with testotoxicosis. J Pediatr. 149:271–274. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Kwiatkowski DP, Sabeti PC. 2014. Natural selection and infectious disease in human populations. Nat Rev Genet. 15:379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, et al. 2015. Expression of human olfactory receptor 10J5 in heart aorta, coronary artery, and endothelial cells and its functional role in angiogenesis. Biochem Biophys Res Commun. 460:404–408. [DOI] [PubMed] [Google Scholar]
- Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. 2005. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat 26:199–204. [DOI] [PubMed] [Google Scholar]
- Lamphear J. 1988. The People of the Grey Bull: the origin and expansion of the Turkana. J Afr Hist. 29:27–39. [Google Scholar]
- Langbein L, et al. 2007. Novel type I hair keratins K39 and K40 are the last to be expressed in differentiation of the hair: completion of the human hair keratin catalog. J Invest Dermatol. 127:1532–1535. [DOI] [PubMed] [Google Scholar]
- Ledda M, et al. 2014. GWAS of human bitter taste perception identifies new loci and reveals additional complexity of bitter taste genetics. Hum Mol Genet. 23:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis IM. 1999. A pastoral democracy: a study of pastoralism and politics among the Northern Somali of the Horn of Africa. Oxford: James Currey Publishers. [Google Scholar]
- Li JZ, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319:1100–1104. [DOI] [PubMed] [Google Scholar]
- Li H, Pakstis AJ, Kidd JR, Kidd KK. 2011. Selection on the human bitter taste gene, TAS2R16, in Eurasian populations. Hum Biol. 83:363–377. [DOI] [PubMed] [Google Scholar]
- Maples BK, Gravel S, Kenny EE, Bustamante CD. 2013. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 93:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall F, Hildebrand E. 2002. Cattle before crops: the Beginnings of Food Production in Africa. J World Prehistory 16:99–143. [Google Scholar]
- Molesworth AM, et al. 2002. Where is the meningitis belt? Defining an area at risk of epidemic meningitis in Africa. Trans R Soc Trop Med Hyg. 96:242–249. [DOI] [PubMed] [Google Scholar]
- Nazari-Shafti TZ, et al. 2011. Mesenchymal stem cell derived hematopoietic cells are permissive to HIV-1 infection. Retrovirology 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JL. 1995. The peopling of Africa: a geographic interpretation. New Haven: Yale University Press. [Google Scholar]
- Novembre J, Han E. 2012. Human population structure and the adaptive response to pathogen-induced selection pressures. Philos Trans R Soc Lond B Biol. Sci. 367:878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Bever B. 1986. Medicinal plants in tropical West Africa. Cambridge: Cambridge University Press. [Google Scholar]
- Patin E, et al. 2014. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat Commun. 5:3163. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, et al. 2010. Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel. Eur J Hum Genet. 18:915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, et al. 2012. The genetic prehistory of southern Africa. Nat Commun. 3:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, et al. 2014. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci U S A. 111:2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluznick JL, et al. 2009. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci U S A. 106:2059–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgorna E, Soares P, Pereira L, Cerny V. 2013. The genetic impact of the lake chad basin population in North Africa as documented by mitochondrial diversity and internal variation of the L3e5 haplogroup. Ann Hum Genet. 77:513–523. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Manica A, Balloux F. 2005. Geography predicts neutral genetic diversity of human populations. Curr Biol. 15:R159–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, et al. 2010. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 19:4278–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rito T, et al. 2013. The first modern human dispersals across Africa. PLoS One 8:e80031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Hoshino S, Mochida N, Matsumura Y, Habu S. 2002. Identification of a novel protein p59(scr), which is expressed at specific stages of mouse spermatogenesis. Biochem Biophys Res Commun. 292:992–998. [DOI] [PubMed] [Google Scholar]
- Silva M, et al. 2015. 60,000 years of interactions between Central and Eastern Africa documented by major African mitochondrial haplogroup L2. Sci Rep. 5:12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, et al. 2004. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 74:1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, et al. 2012. The expansion of mtDNA haplogroup L3 within and out of Africa. Mol Biol Evol. 29:915–927. [DOI] [PubMed] [Google Scholar]
- Solovieff N, et al. 2010. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5' olfactory receptor gene cluster. Blood 115:1815–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M, et al. 2003. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299:2054–2058. [DOI] [PubMed] [Google Scholar]
- Spencer P. 1965. The Samburu: a study of gerontocracy in a nomadic tribe. Oxon: Routledge. [Google Scholar]
- Steverding D. 2008. The history of African trypanosomiasis. Parasit Vectors. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MJ, Murakami Y, Maeda Y, Kinoshita T, Morita YS. 2014. New insights into the functions of PIGF, a protein involved in the ethanolamine phosphate transfer steps of glycosylphosphatidylinositol biosynthesis. Biochem J. 463:249–256. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD. 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 31:2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Small KS, Kwiatkowski DP. 2010. Methodological challenges of genome-wide association analysis in Africa. Nat Rev Genet. 11:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, et al. 2009. The genetic structure and history of Africans and African Americans. Science 324:1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNEP. 2008. AFRICA Atlas of our changing environment. Sioux Falls (SD): United Nations Environment Programme. [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh K, et al. 2012. Lactase persistence and lipid pathway selection in the Maasai. PLoS One 7:e44751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu J, Cho KH, Ren D. 2009. A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol Reprod. 81:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, et al. 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42:D1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2012. Sahel food and health crisis: emergency health strategy. West Africa Regional Health Working Group. p. 28. [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharia F, et al. 2009. Characterizing the admixed African ancestry of African Americans. Genome Biol. 10:R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.