Summary
Predicting host health status based on microbial community structure is a major goal of microbiome research. An implicit assumption of microbiome profiling for diagnostic purposes is that the proportional representation of different taxa determine host phenotypes. To test this assumption, we colonized gnotobiotic zebrafish with zebrafish-derived bacterial isolates and measured bacterial abundance and host neutrophil responses. Surprisingly, combinations of bacteria elicited immune responses that do not reflect the numerically dominant species. These data are consistent with a quantitative model in which the host responses to commensal species are additive, but where various species have different per capita immunostimulatory effects. For example, one species has a high per capita immunosuppression that is mediated through a potent secreted factor. We conclude that the proportional representation of bacteria in a community does not necessarily predict its functional capacities; however, characterizing specific properties of individual species offers predictive insights into multi-species community function.
Introduction
Animals and their resident microbial communities, or microbiota, are a complex ecosystem. These microbes derive nutrients from the host environment, and in turn, they influence normal animal development and health. The gastrointestinal microbiota are critical for nutrient acquisition and immune system development (Bäckhed et al., 2005; Hooper et al., 2012). Metagenomic profiling of gut microbiota has identified deviations from taxonomic compositions associated with health in diseases such as obesity (Turnbaugh et al., 2009), diabetes (Wen et al., 2008), and inflammatory bowel diseases (IBD)(Frank et al., 2007). An implicit assumption in these compositional analyses is that the relative abundances of different taxa can predict pathology; however, application of this assumption to clinical data does not uncover consistent trends. For example, both an increased (Turnbaugh et al., 2009) and decreased ratio (Jumpertz et al., 2011) of Bacteroidetes to Firmicutes have been associated with obesity. Additionally, a meta-analysis of human obesity-associated microbiota concluded that small shifts in many taxa, rather than large differences in a few taxa, are more likely to predict obesity (Walters et al., 2014). Thus, the extent to which microbiota composition can be used to predict community function and human health status remains an open question.
The complexity and variability of vertebrate-associated microbiota presents substantial challenges to unraveling their functional potential. For example, DNA sequence-based surveys of microbiota cannot distinguish between active and inactive or resident and transient members. Another limitation of such surveys is that they only provide information on the proportional representation of taxa but not their per capita contributions to community functions, such as the capacity to induce an inflammatory response. These limitations emphasize the need for simplified, defined model systems to connect the composition of resident bacterial communities with their emergent properties. We created a tractable system to study the impact of microbiota composition on the intestinal innate immune response using the zebrafish, Danio rerio. The zebrafish is an excellent model to examine microbial community function because hundreds of zebrafish can be easily derived and maintained in a germ-free (GF) or gnotobiotic state with defined microbial isolates (Milligan-Myhre et al., 2011). The zebrafish intestinal microbial community is well-characterized; a large number of intestinal microbes that span the phylogenetic diversity observed in the zebrafish microbiota can be maintained in culture and have had their genomes sequenced (Stephens et al., 2015). Furthermore, zebrafish transgenesis and optical transparency allows for high resolution monitoring of host and microbial cells in vivo (Jemielita et al., 2014). We exploited these properties to develop an assay in which we monitor both the composition of the bacterial community and the innate immune response in an individual fish, using GFP-expressing neutrophils as a metric of the host response. Neutrophils are a primary component of the initial inflammatory response and critical for host defense (Harvie and Huttenlocher, 2015). Neutrophil homeostasis is established and maintained by the microbiota, as GF larvae have reduced intestinal (Bates et al., 2007) and systemic neutrophils and reduced neutrophil responses to injury (Kanther et al., 2014). Thus neutrophil dynamics are a sensitive measure of host responses to intestinal microbiota.
Here we use our gnotobiotic zebrafish model to measure the host neutrophil response to individual microbiota constituents and small communities assembled from these members. We show that the per capita immunostimulatory effect of individual species within a community varies widely, such that minor members can exert dominant effects. A simple mathematical model based on additive responses to individual species describes the neutrophil response to these communities by accounting for the per capita effect of each species. Our approach demonstrates the feasibility of predicting the function of a microbial community based on its structure, which in the future may be expanded to more complex systems to improve our understanding of human disease-associated microbial communities and our ability to restore them to a healthy state.
Results
Microbial isolates induce unique neutrophil responses
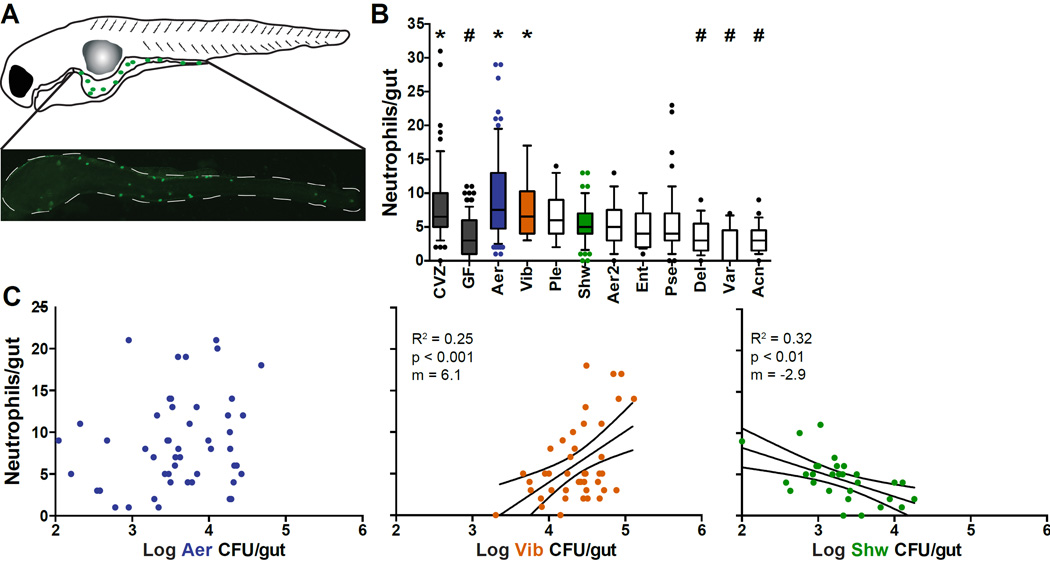
To assay the influence of individual bacterial species on the intestinal innate immune response, we raised GF zebrafish and inoculated their aquatic environment with single bacterial isolates (mono-associations) from our collection of zebrafish intestinal bacteria (Stephens et al., 2015). Bacteria were introduced at 4 days post fertilization (dpf), by which time their intestine had opened, and at 6 dpf we dissected the intestine and assessed neutrophil populations (Fig. 1A) and bacterial colony forming units (CFU) per intestine. All neutrophil responses to the individual strains we tested were within the range observed for GF and conventionalized (CVZ) fish, yet there was a wide range of responses both between and within groups (Fig. 1B). For some strains the variation in neutrophil response correlated with variation in bacterial abundance. For example, for a Vibrio species, the number of neutrophils increased with bacterial abundance and was fit by a linear relationship between neutrophil number and log(CFU) (Fig. 1D). The log(CFU) of two species, Shewanella (Fig. 1D) and Acinetobacter (not shown), were negatively correlated with neutrophil number. A third pattern, characteristic of most isolates, represented by Aeromonas, displayed no clear relationship between neutrophil and bacterial abundance (Fig. 1D). We used representatives of each of these three species-specific neutrophil responses to explore whether we could predict the neutrophil response to more complex communities.
Fig. 1. Resident microbial isolates induce unique neutrophil responses.
A. Dissected mpx:GFP zebrafish intestine. B. Intestinal neutrophil recruitment in conventionalized fish (CVZ), germ free fish (GF), or fish inoculated with ten individual mono-associations. Middle line, median; boxes, quartiles; whiskers, 10th–90th percentile. Aer, Aeromonas; Vib, Vibrio; Ple, Plesiomonas; Shw, Shewanella; Aer2, Aeromonas sp. 2; Ent, Enterobacter; Pse, Pseudomonas; Del, Delftia; Var, Variovorax; Acn, Acinetobacter. * Significantly different from GF; # significantly different from CVZ (ANOVA). C. Correlation between neutrophil influx and logarithm of bacterial abundance per intestine for Aeromonas (left), Vibrio (middle), and Shewanella (right). Linear regression analysis. For all conditions, N ≥ 20 fish, derived from at least three independent experiments.
Complex dynamics in microbial di-associations influence microbial abundance and neutrophil response
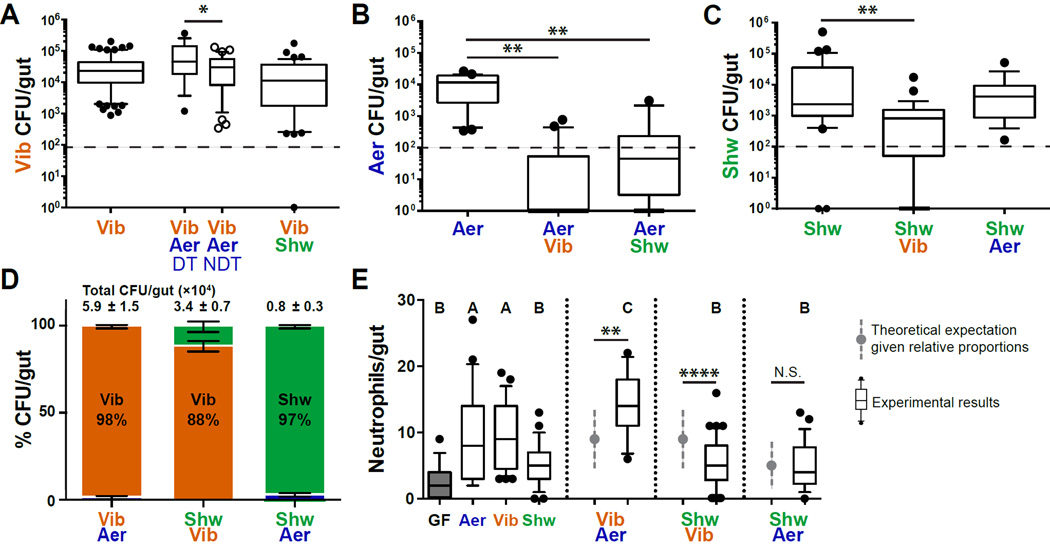
To test whether the relationship between bacterial and neutrophil abundance in mono-association is indicative of their contribution in a complex community to the neutrophil population, we examined every dual species combination (di-association) between Aeromonas, Vibrio, and Shewanella. In a di-association, the two species are added together at the same concentration to the aquatic environment of GF fish at 4 dpf, and the CFU/gut and intestinal neutrophil influx are assayed at 6 dpf. In di-associations between Vibrio and either Shewanella or Aeromonas, Vibrio was the numerically dominant member and its abundance was unchanged or increased, respectively, compared to its abundance in a mono-association (Fig. 2A). In the di-association with Vibrio, Aeromonas was undetectable in 55% of fish and when it did co-colonize with Vibrio, it was present at a significantly lower abundance than in a mono-association (Fig. 2B). Notably, in fish colonized with Aeromonas, Vibrio abundance increased compared to the fish with no detectable Aeromonas (Fig. 2A). In terms of relative abundance, Vibrio dominated the di-associations with Aeromonas (98% ± 1%) and Shewanella (89% ± 3%), and Shewanella dominated the di-association with Aeromonas (97% ± 1%) (Fig. 2C, 2D). The abundance of these species in the water did not change in comparison to mono-associations (Fig. S1A), indicating that the dynamics are host-associated.
Fig. 2. Microbial di-associations reveal complex dynamics between microbes.
A. Vibrio abundance in mono- or di-association with either Aeromonas or Shewanella. The Aeromonas di-association is split into two categories, Aeromonas detected (DT) or not detected (NDT) in the fish. B. Aeromonas abundance in mono- or di-association with either Vibrio or Shewanella. C. Shewanella abundance in mono- or di-association with Vibrio or Aeromonas. Dashed line indicates limit of quantification. For A, B, C *p < 0.05, **p < 0.01, ANOVA. D. Relative proportion of the total number of bacteria in di-associations (avg ± SEM). Total bacterial load is listed above bar graph. E. Intestinal neutrophil influx for each mono- or di- association. Conditions that share a letter are not statistically different, ANOVA. Grey dots and dashed lines represent the expectation for the di-associations given the relative proportion of species. **p < 0.01, **** p < 0.0001, one sample t test with theoretical mean; N.S., not significant. For all conditions, N ≥ 20 fish, derived from at least three independent experiments. See also Figure S1.
In di-associations between Aeromonas and either Shewanella or Vibrio, the neutrophil response reflected the dominant member (Fig. 2D, 2E, S1B, S1C). Notably, in the Vibrio and Aeromonas di-association, neutrophil influx was higher than predicted given the relative proportion of members and a simple expectation of a sum of neutrophil responses (Fig. 2E, grey bars). However, the expectation that the dominant species determines the neutrophil response failed in the Vibrio and Shewanella di-association. In this case, Vibrio was the dominant species (Fig. 2D), yet intestinal neutrophil influx was significantly reduced compared our expectation (Fig. 2E, grey bars). In fact, neutrophil influx was similar to a Shewanella mono-association (Fig. 2E, S1C), which suggests that the minor species Shewanella had a disproportionate impact on the neutrophil response.
A model of additive responses to bacterial species can explain intestinal neutrophil responses in di-associations
Because neutrophil responses to Vibrio di-associations with Aeromonas and Shewanella differed from a simple expectation (Fig. 2E), we explored whether we could construct a mathematical model of the neutrophil response to two-member communities, based on knowledge of the responses to individual species (Fig. 1D) and their abundances in di-associations (Fig. 2A–C). To avoid over fitting the data, we constructed a minimal model that parameterizes key aspects of bacterial growth and interactions between bacterial species and neutrophils. We modeled bacterial growth and competition with Lotka-Volterra equations (eq. 1), which apply to a variety of ecological systems including host-associated microbial communities (Fisher and Mehta, 2014; Marino et al., 2013; Stein et al., 2013):
| (1) |
where Pi, ri, and Ki denote the population, growth rate, and carrying capacity, respectively, of species i, γij characterizes the effect of species j on the dynamics of species i, and bi defines the effect of the neutrophil population on species i. We modeled the neutrophil population (N) with linear influx and exit terms and, importantly, an additive contribution from each bacterial species (eq. 2):
| (2) |
where αN and kN are the influx and exit rate of neutrophils, respectively, and αi is the effect of species i on neutrophil influx. Inspired by the observed form of the mono-association data (Fig. 1D), we modeled αi as being linearly dependent on the logarithm of bacterial abundances (eq. 3):
| (3) |
where Mi characterizes the slope of the bacteria-neutrophil interaction and Ti is the effective threshold for a positive effect. For Vibrio, we constrain α > 0 to specify that no population levels suppress neutrophil numbers. We also considered a sigmoidal model of bacteria-neutrophil interactions, which yields similar behaviors (Supplemental methods). Experiments suppressing the immune response (Fig. 3) and simulations (Supplemental methods) imply that the data can be modeled without incorporating potential influences of neutrophils on bacterial abundance; i.e. bi can be set to zero, and Equation 1 is independent of N. This and other omitted interactions may exist in more complex communities; however, we aimed to determine whether a minimal model could describe our observed di-association data.
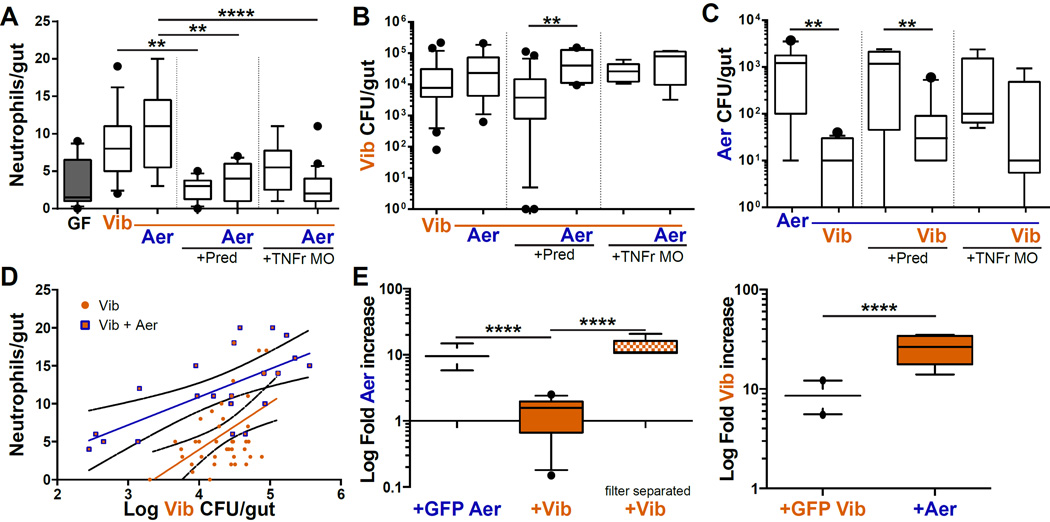
Fig. 3. An interaction between Vibrio and Aeromonas promotes Vibrio abundance.
A. Immunosuppression via prednisolone (Pred) or the tnfr morpholino (TNFr MO) maintains neutrophil influx at GF levels in the presence of Vibrio mono- and di-associations. B. In the absence of a neutrophil response, Vibrio abundance increases in the presence of Aeromonas. C. Aeromonas abundance is reduced in the presence of Vibrio and is unaffected by prednisolone or tnfr morpholino treatment. D. The linear relationships between log(CFU) Vibrio abundance and neutrophil influx in mono- (orange circles) or in di-association with Aeromonas (blue and orange squares). E. Growth of Aeromonas and Vibrio in vitro either in co-culture with a GFP-tagged isogenic strain or mixed together on filter paper. Vibrio reduced Aeromonas growth (left), and Aeromonas increased Vibrio growth (right). Separating the species with filter paper prevented the reduction of Aeromonas. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, T-test. See also Figure S2.
Most parameters (ri, Ki, αN/kN, γij, MVibrio, and TVibrio) are well constrained by experimental data. If the slope and threshold parameters for the influence of Shewanella and Aeromonas mono-associations on neutrophil influx were precisely known, all model parameters would be fixed. The scatter in the data (Fig. 1D) prevent this, but we can examine the Mi/Ti parameter space for regions that are consistent with both the mono-association data and the observed neutrophil number in di-associations of each of these species with Vibrio. For both di-associations with Vibrio, we find such overlapping regions in parameter space (Supplemental methods). Thus, our additive model of bacterial/neutrophil interactions is sufficient to describe the observed data. This indicates that relatively few Shewanella are required to dominate the immune response; their large per capita effect is parameterized by a combination of large slope M and low threshold T. The success of a simple additive model in predicting the host neutrophil response to a two-member bacterial community suggests that 1) neutrophil feedback on bacterial populations is negligible in the context of normal neutrophil responses to commensals and 2) certain disproportionately impactful species, like Shewanella, may use interesting mechanisms to influence neutrophil dynamics in complex, multi-species communities.
An interaction between Vibrio and Aeromonas drives Vibrio growth and neutrophil influx
The model predicted that the higher than expected neutrophil influx in the di-association between Vibrio and Aeromonas was independent of neutrophil feedback, and likely dependent on an increase in Vibrio abundance conferred by the presence of Aeromonas. An alternative explanation, inconsistent with our model, would be that increased Vibrio abundance occurs as a result of positive feedback from the neutrophil influx elicited by Aeromonas, with Vibrio behaving like a pathobiont that thrives in an inflamed environment (Mazmanian et al., 2008). To distinguish between these two possibilities we implemented two independent means of immune suppression, prednisolone (Oehlers et al., 2011; a steroid immunosuppressant) and a tumor necrosis factor receptor (tnfr) morpholino (Bates et al., 2007; which blocks pro-inflammatory TNFα signaling; Fig. 3A). We found that under conditions of low neutrophil influx, Vibrio abundance still increased in the di-association with Aeromonas in comparison to the Vibrio mono-association (Fig. 3B). Prednisolone did not affect the growth of Vibrio or Aeromonas in vitro (Fig. S2) or in mono-association (Fig. 3C). These data support our model’s prediction that neutrophils do not feedback on bacterial abundance and suggest an interaction between Vibrio and Aeromonas. The slope of the relationship between the logarithm of Vibrio abundance and neutrophil influx was unchanged in the di-association compared to the Vibrio mono-association (Fig. 3D), however the intercept is higher, suggesting that either Aeromonas contributes to the neutrophil influx or Vibrio has an increased per capita effect in the presence of Aeromonas.
We further explored the interaction between Aeromonas and Vibrio by asking whether they influence each other’s populations when grown in direct contact or in close vicinity in vitro. Compared to a co-culture with differentially marked isogenic strains cross species co-culture promoted the growth of Vibrio and inhibited growth of Aeromonas in a contact dependent manner (Fig. 3E). These experiments establish that we can recapitulate in vitro an inter-species interaction that occurs in vivo and alters the potential of the community to induce intestinal neutrophil influx.
Shewanella controls the neutrophil response via a secreted anti-inflammatory factor
As a minor member of the di-association with Vibrio, Shewanella directed a lower than expected neutrophil response (Fig. 2E) and abolished the relationship between Vibrio abundance and neutrophil influx (Fig. 4A). Our model posited that Shewanella exerted a large per capita effect on the neutrophil response, which we reasoned could be mediated through a potent secreted product. When we treated Vibrio mono-associated fish with 500-ng/ml concentrated Shewanella cell-free supernatant (CFS), we observed that a secreted factor (or factors) from Shewanella was sufficient to induce a low neutrophil response to Vibrio (Fig. 4B), while Vibrio abundance remained unaltered (Fig. S3A). Heat killing of Shewanella, which inactivates secretion and denatured proteins, eliminated Shewanella’s effect (Fig. 4B). Interestingly, Shewanella CFS did not alter neutrophil influx (Fig. 4C) or abundance (Fig. S3B) in an Aeromonas mono-association, which suggests either that Shewanella’s anti-inflammatory factor specifically inhibits a pro-inflammatory activity of Vibrio or that Aeromonas inactivates the anti-inflammatory factor.
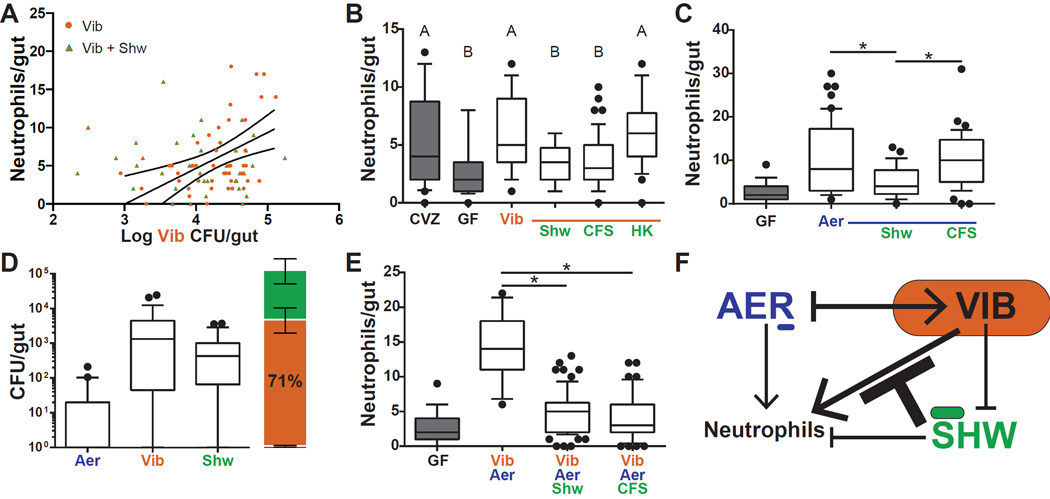
Fig. 4. Shewanella controls the neutrophil response via a secreted anti-inflammatory factor.
A. The presence of Shewanella (green and orange triangles) abolishes the linear relationship between log(CFU) Vibrio (orange circles) abundance and neutrophil influx. Four independent experiments are displayed. B. Intestinal neutrophil influx in response to Vibrio with live Shewanella, Shewanella cell free supernatant (CFS), or heat-killed Shewanella (HK). Conditions that do not share a letter are significantly different (ANOVA, Tukey’s range test). C. Intestinal neutrophil influx in response to a di-association of Aeromonas with either live Shewanella or Shewanella CFS. D. Abundance and percentages (avg ± SEM) of Vibrio, Aeromonas, and Shewanella in the tri-association. E. Neutrophil influx in response to the Vibrio and Aeromonas di-association, the tri-association, or the Vibrio and Aeromonas di-association with Shewanella CFS. *p < 0.05, T-test. F. Model of the inter-bacterial interactions occurring in the intestine that influence bacterial abundance and intestinal neutrophil influx. Arrow thickness denotes interaction strength; oval sizes reflect relative abundance in the tri-association. See also Figure S3.
Finally, we examined the host-microbiota system with the three-member community. The intestines of fish inoculated with equal parts Aeromonas, Vibrio, and Shewanella were dominated by Vibrio (71% ± 7%), with Shewanella contributing 28% ± 7%, and Aeromonas contributing 1% ± 0.3% (Fig. 4D). Despite the numerical dominance of Vibrio, intestinal neutrophil influx was significantly lower than observed in the Vibrio and Aeromonas di-association (Fig. 4E). Furthermore, Shewanella CFS was sufficient to elicit this phenotype when added to a Vibrio and Aeromonas di-association (Fig. 4E). Thus, in a three-member microbial community, a numerically minor member can determine the neutrophil response to the community through the activity of a potent secreted anti-inflammatory factor (Fig. 4F).
Discussion
Two major challenges of microbiome research are to use compositional data to predict the functions of a complex microbial community, such as its inflammatory potential, and to manipulate community membership to promote a specific function. Here, we describe a simple mathematical model that accounts for both competition between microbes and the immunomodulatory effect of each member on the host and predicts the collective immune response elicited by the composite community as the sum of the effects of each individual member, scaled to its particular per capita effect. Our model demonstrates the feasibility of predicting the function of a microbiota based on its composition when specific properties of the individual species are known. Our modeling approach could be expanded to more complex systems, such as the mouse or human gastrointestinal tract, where the mono-association data in our model could be replaced with data based on other individual traits, such as pro- or anti-inflammatory properties measured in a cell based assay (Mastropietro et al., 2015). It will be interesting to see whether other functions of complex microbial communities, such as carbohydrate metabolism in mice (Sonnenburg et al., 2006) and nutrient acquisition in flies (Newell and Douglas, 2014) are consistent with additive contributions from species with different per capita effects or whether a quantitative description of these systems will require evoking non-additive interactions. Finally, our model predicts non-monotonic changes in the neutrophil population over time (Supplemental methods), which may be observable with live imaging of host-bacterial dynamics in real time (Jemielita et al., 2014).
From our model we also gained mechanistic insights into bacterial-bacterial and bacterial-host interactions within the system, which is a step toward manipulating a community to secure a desired function. Our modeling and experimental analysis suggested that in our system, bacterial-bacterial interactions play the dominant role in determining community membership, with no evidence for neutrophil feedback on the bacterial populations. We speculate that this would not be true in a pathologically inflamed intestine, where certain members would likely experience growth inhibition and other inflammation-adapted species would thrive (Winter et al., 2010). We observed a strong bacterial-bacterial interaction between Vibrio and Aeromonas both when co-colonizing the zebrafish intestine and growing in contact in vitro. In the intestine, Vibrio’s impairment of Aeromonas growth correlated with an increase in Vibrio abundance, and thus a corresponding increase in the neutrophil recruiting capacity of the community. Given the contact dependent nature of the in vitro interaction between Vibrio and Aeromonas, it is possible that this interaction involves a type VI secretion system (MacIntyre et al., 2010; Stephens et al., 2015). Our ability to replicate the in vivo dynamic between Aeromonas and Vibrio in vitro highlights a strength of our system and allows us to further interrogate the mechanism of interaction between these species.
The microbiota and the host must maintain a homeostatic relationship both to activate neutrophils for responding to injury and infection (Kanther et al., 2014) and to allow the resident microbes to persist. The range of neutrophils required to establish this relationship is represented in CVZ fish, and all examined mono-associations were within this range. Notably, the average neutrophil response to each bacterial isolate was proportional to that species’ average abundance (Fig. S3C), consistent with the observation that generic bacterial immunostimulatory molecules, such as lipopolysaccharide, contribute to the regulation of neutrophil influx (Bates et al., 2007). However, different isolates exhibited different relationships between neutrophil number and bacterial load across individual mono-associated fish, suggesting that individual bacteria have specialized mechanisms by which they influence the host neutrophil response.
In both two- and three-member communities Shewanella acts as a keystone species (Power et al., 1996) by exerting a disproportionately large effect on the neutrophil population given its low abundance. Shewanella strains are used as probiotics in aquaculture (Tapia-Paniagua et al., 2014), suggesting that they retain immunodominance in complex, natural communities. The human intestinal microbiota contains many low abundance species (Arumugam et al., 2011), and some have a disproportionately large impact on inducing dysbiosis and disease (Hajishengallis et al., 2012) or on promoting health (Sokol et al., 2008). For example, Faecalibacterium prausnitzii, whose absence correlates with IBD (Cao et al., 2014; Sokol et al., 2008, 2009), comprises only 4 – 6% of the mucosa-associated microbiota, yet it reduces pro-inflammatory cytokine signaling and colitis severity through a secreted anti-inflammatory factor (Sokol et al., 2008). Similarly, in our system Shewanella generates a low neutrophil response via a secreted anti-inflammatory factor. We do not know whether this anti-inflammatory factor acts on the host or on Vibrio; however, the abundance of Vibrio is slightly, although not significantly, reduced in the presence of Shewanella and its CFS. This slight reduction in Vibrio may contribute to a reduced neutrophil response, or alternatively it may be the result of a low inflammatory environment elicited by Shewanella. Such an environmental alteration is a characteristic of a keystone species. Given the central role keystone species play in ecosystem function, identifying them will be critical for our ability to engineer microbial communities to promote a required function. Here we have identified one such species and identified two measurable properties—a high per capita effect and a negative relationship between abundance and neutrophil response—that may be used to screen for other such species. Identifying critical players with large per capita effects, like Shewanella, will advance our ability both to predict community functions and to manage community membership.
Experimental procedures
For additional details, see supplemental materials and methods.
Gnotobiotic zebrafish husbandry
All zebrafish experiments were performed following protocols approved by the University of Oregon Institutional Animal Care and Use Committee. Conventionally-raised wild-type (AB × Tu strain) and Tg(BACmpx:GFP)i114 (referred to as mpx:GFP) (Renshaw et al., 2006) were maintained as described (Westerfield, 1993). Zebrafish embryos were derived GF and associated with bacterial isolates as previously described (Bates et al., 2006). At 6 dpf the mpx:GFP zebrafish were anesthetized in Tricaine (Western Chemical, Inc., Ferndale, WA), mounted in 4% methylcellulose (Fisher, Fair Lawn, NJ), and their intestines were dissected using sterile technique. The number of GFP-positive cells was quantified visually for each fish using a fluorescent microscope (SteREO Discovery.V8, Zeiss).
Microbiology
Bacteria used for inoculations were zebrafish isolates ZOR0001 (Aeromonas), ZWU0020 (Vibrio), ZOR0012 (Shewanella), ZNC0006 (Variovorax), ZNC0008 (Delftia), ZOR0008 (Acinetobacter), ZOR0002 (Aeromonas sp. 2), ZWU0006 (Pseudomonas), ZOR0011 (Pleisomonas), and ZOR0014 (Enterobacter) (Stephens et al., 2015). To determine the CFU/intestine, dissected zebrafish intestines were placed in 100-µl sterile EM, homogenized, diluted, and cultured on tryptic soy agar plates (TSA; BD, Sparks MD). For di- and tri-associations, bacterial species were distinguished by colony morphology.
Morpholino injections
Splice-blocking MOs (Gene Tools, Corvallis, OR) were injected into the embryos at the one cell stage. The TR1v1/TR1v2 (1.2 moles and 6 moles, respectively) were used as previously described (Bates et al., 2007).
Prednisolone treatments
The prednisolone solution was prepared and administered as described (Oehlers et al., 2011).
Concentration of CFS
Shewanella was grown over night shaking in TSB. 1 ml of overnight culture was used to inoculate 50-ml TSB, which was kept shaking at 30° C for 2 h. The supernatant was filtered (Corning Inc., Corning NY) and concentrated with a centrifugal device with a 10-kda weight cut off (Pall Life Sciences, Ann Arbor, MI).
In vitro co-culture assay
Vibrio and Aeromonas were grown overnight shaking in TSB (BD, Sparks, MD). 5 × 108 bacterial cells of each strain were mixed together and spotted onto filter paper on brain heart infusion media agar plate (BHI, BD, Sparks, MD). A co-culture of an isogenic fluorescently tagged strain with the wild-type counterpart served as controls. To determine contact dependency, the filter paper was placed between the strains (MacIntyre et al., 2010).
Statistics and modeling
Statistical analysis was performed using Prism (Graphpad Software). Statistical significance was defined as p < 0.05. Modeling details can be found in Supplemental experimental methods.
Supplementary Material
Acknowledgements
We thank Rose Sockol and UO Zebrafish Facility staff for fish husbandry. We thank Guillemin lab members for insightful discussions and Tiffani Jones for critical reading of the manuscript. Research reported in this publication was supported by the NIH: by the NIGMS under award number P50GM098911, by the NIDDK under award number 1F32DK098884-01A1 (to ASR), and by the NICHHD under award P01HD22486, which provided support for the UO Zebrafish Facility. The content is solely the responsibility of the authors and does not represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
Conceptualization, ASR and KG; Methodology, ASR and RP; Formal analysis, ASR, RP, and ARB; Investigation, ASR; Writing – Original Draft, ASR and RP; Writing – Review & Editing, ASR, RP, ARB, BJMB, and KG; Funding Acquisition, ASR and KG; Resources, KG; Supervision, KG and BJMB.
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. Enterotypes of the human gut microbiome. Nature. 2011:1–7. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: A meta-analysis and systematic review of the literature. Gastroenterol. Res. Pract. 2014;2014 doi: 10.1155/2014/872725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CK, Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One. 2014;9:e102451. doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvie EA, Huttenlocher A. Neutrophils in host defense: new insights from zebrafish. J. Leukoc. Biol. 2015;98:1–15. doi: 10.1189/jlb.4MR1114-524R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K. Spatial and Temporal Features of the Growth of a Bacterial Species Colonizing the Zebrafish Gut. MBio. 2014;5:1–8. doi: 10.1128/mBio.01751-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Tomkovich S, Sun X, Grosser MR, Koo J, Flynn EJ, Jobin C, Rawls JF. Commensal microbiota stimulate systemic neutrophil migration through induction of Serum amyloid A. Cell. Microbiol. 2014;16:1053–1067. doi: 10.1111/cmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. USA. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. Mathematical modeling of primary succession of murine intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2013:1–6. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastropietro G, Tiscornia I, Perelmuter K, Astrada S, Bollati-fogolín M. HT-29 and Caco-2 Reporter Cell Lines for Functional Studies of Nuclear Factor Kappa B Activation. Mediat. Inflamm. 2015:1–13. doi: 10.1155/2015/860534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Milligan-Myhre K, Charette JR, Phennicie RT, Stephens WZ, Rawls JF, Guillemin K, Kim CH. Study of Host-Microbe Interactions in Zebrafish. Elsevier Inc.; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Douglas AE. Interspecies Interactions Determine the Impact of the Gut Microbiota on Nutrient Allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 2014;80:788–796. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlers SH, Flores MV, Okuda KS, Hall CJ, Crosier KE, Crosier PS. A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev. Dyn. 2011;240:288–298. doi: 10.1002/dvdy.22519. [DOI] [PubMed] [Google Scholar]
- Power ME, Tilman D, Estes JA, Menge BA, Bond WJ, Mills S, Daily G, Castilla JC, Lubchenco J, Paine RT. Challenges Quest for Keystones. Bioscience. 1996;46:609–620. [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, Blugeon S, Bridonneau C, Furet J-P, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doraé J. Low counts of faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CTL, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, Sander C, Xavier JB. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens W, Burns A, Stagamann K, S W, Rawls J, Guillemin K, Bohannan B. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2015 doi: 10.1038/ismej.2015.140. 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Paniagua S, Vidal S, Lobo C, Prieto-Álamo M, Jurado J, Cordero H, Cerezuela R, García de la Banda I, Esteban M, Balebona M, et al. The treatment with the probiotic Shewanella putrefaciens Pdp11 of specimens of Solea senegalensis exposed to high stocking densities to enhance their resistance to disease. Fish Shellfish Immunol. 2014;41:209–221. doi: 10.1016/j.fsi.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio rerio) Eugene, OR: Institute of Neuroscience University of Oregon; 1993. [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.