Abstract
The identification of genetic variants responsible for behavioral variation is an enduring goal in biology, with wide-scale ramifications, ranging from medical research to evolutionary theory on personality syndromes. Here, we use for the first time a large-scale genetical genomics analysis in the brains of chickens to identify genes affecting anxiety as measured by an open field test. We combine quantitative trait locus (QTL) analysis in 572 individuals and expression QTL (eQTL) analysis in 129 individuals from an advanced intercross between domestic chickens and Red Junglefowl. We identify 10 putative quantitative trait genes affecting anxiety behavior. These genes were tested for an association in the mouse Heterogeneous Stock anxiety (open field) data set and human GWAS data sets for bipolar disorder, major depressive disorder, and schizophrenia. Although comparisons between species are complex, associations were observed for four of the candidate genes in mice and three of the candidate genes in humans. Using a multimodel approach we have therefore identified a number of putative quantitative trait genes affecting anxiety behavior, principally in chickens but also with some potentially translational effects as well. This study demonstrates that chickens are an excellent model organism for the genetic dissection of behavior.
Keywords: anxiety, behavioral genes, eQTL, QTL, causal genes, personality
THE identification of genes that harbor causal mutations underlying a quantitative trait (QTG) is rare for any trait (Flint 2003) and even more so for behavior. Successful examples of behavioral QTG identification are almost exclusively limited to mouse and rat models (Yalcin et al. 2004; Chiavegatto et al. 2008; Kim et al. 2009; Tomida et al. 2009; Gyetvai et al. 2011; Wang et al. 2012; Heyne et al. 2014), Drosophila (Anholt and Mackay 2004; Mackay 2004; Fitzpatrick et al. 2005), and the honeybee (Robinson et al. 2005, 2008). The ramifications for the identification of such behavioral genes are many and varied. For example, mood-based disorders are one of the top 10 causes of disability worldwide (Murray and Lopez 1996; Vos et al. 2015), yet the identification of susceptibility loci for such traits has been severely restricted (Kas et al. 2007), with only a handful identified. This is despite the often high heritability estimates for diseases such as schizophrenia (McGue and Bouchard 1998), bipolar disorder (Burmeister et al. 2008), and major depressive disorder (Burmeister et al. 2008). More generally, very little is known about what polymorphisms affect behavior in a nonmorbid fashion, i.e., alleles that cause natural quantitative variation. Knowledge of such polymorphisms would surely assist in the identification of actual disease loci. With regard to evolutionary theory, behavioral personality studies now exist for a wide range of species (Sih et al. 2004); however, the genes underlying such traits still remain largely unexplored.
It has been suggested that interspecies trait genetics can help reveal alleles influencing susceptibility in humans (Kas et al. 2007), with the mouse model primarily considered for such a role, although others have been suggested (Kaluev and Cachat 2012). Domestic chickens exhibit a wide range of behavioral as well as morphological differences, as compared to their wild-derived progenitor, the Red Junglefowl (RJF). Behaviorally, for example, they differ in anxiety and antipredator behavior, as well as sociality (Schütz et al. 2001). These extreme changes in anxiety are most likely brought about by selection for tameness and reduced fear of humans (Price 1984). Chickens possess a compact genome (∼1.09 Gb) and a high recombination rate (1 cM ∼ 350 kb). Coupled with the extreme diversification brought about by domestication, they lend themselves to analyses designed to identify genes harboring variants underlying anxiety-like behavior. Several quantitative trait locus (QTL) studies of behavior have been performed (Schutz et al. 2002, 2004; Buitenhuis et al. 2004), yet no candidate genes have been reliably identified. Previous studies of anxiety QTG in chickens have been restricted to specific candidate genes or regions (Wiren et al. 2009; Wirén et al. 2013). If gene polymorphisms were identified, it would enable the vast knowledge of the neuroanatomy of chickens to be combined with the genetic mechanisms for a truly holistic examination of these traits.
Assessing anxiety in animals is potentially problematic, and various different tests have been devised to test anxiety in model organisms. Without doubt the most popular test is the open field test (Archer 1973; Ramos and Mormède 1997; Belzung 1999), which was devised by Hall (1934). This is a test of anxiety/emotionality in rats, using a brightly lit novel arena. A range of variables can be measured, but most typically locomotion (speed and distance), thigmotaxis (time spent close to the walls), and grooming behaviors are recorded (see review in Prut and Belzung 2003). A variety of modifications have been performed over the years, but always maintaining these basic themes (Prut and Belzung 2003). Anxiety in animals is generally considered to be triggered by two conditions—social isolation and agoraphobia, with the open field test now used in a wide variety of different animals ranging from chickens, pigs, lambs, and rabbits to primates (Forkman et al. 2007). Of the studies using rodents, perhaps the largest and most powerful (in terms of the size of the QTL identified) is the Heterogeneous Stock intercross (Valdar et al. 2006), which utilized >2000 mice and located QTL regions <3 Mb in size.
In humans, anxiety disorders include panic disorders, phobic disorders, generalized anxiety disorder (GAD), obsessive compulsive disorder (OCD), and post-traumatic stress disorder (PTSD) (Smoller et al. 2009). Despite the prevalence of anxiety disorders, there is a lack of knowledge regarding the causal genetic variants. A large genome-wide association study (GWAS) on major depressive disorder (MDD), a disorder that is closely linked with anxiety (Major Depressive Disorder Working Group Of The Psychiatric GWAS Consortium 2013), found no genome-wide significant SNPs, despite a sample size of ∼18,000 subjects. Even more recently, a meta-analysis GWAS study for neuroticism (with a polygenic association with MDD) was performed and found a single significant SNP, which failed to replicate (Genetics of Personality Consortium 2015), while two significant loci were identified in a GWAS using a smaller sample size (9000 cases) but more stringent phenotyping of MDD (Cai et al. 2015).
The relationship between anxiety, bipolar disorder, and schizophrenia is a close and interesting one. Patients with bipolar disorder are found to have far higher incidences of anxiety-related traits (Cosoff and Julian Hafner 1998; Boylan et al. 2004; Harpold et al. 2005), and there is a strong association between the two (often >50% of bipolar patients have diagnosed anxiety disorders; for example, see Faraone et al. 1997). The link between schizophrenia and anxiety-related disorders is more complex, although several studies have shown a correlation between anxiety and both the negative symptoms (Kulhara et al. 1989; Sax et al. 1996; Norman et al. 1998; Huppert et al. 2001) and the positive symptoms (Norman and Malla 1991; Huppert et al. 2001) associated with schizophrenia. This must be taken into consideration with the complex interplay between quality of life, schizophrenia, anxiety, and depression (Huppert et al. 2001; Samsom and Wong 2015). Polygenic risk scores have been shown to possess cross-disorder associations, particularly with regard to adult-onset disorders (Psychiatric Genetics Consortium 2013). Therefore, although large anxiety-based GWASs in humans may not be readily available, large data sets for bipolar disorders (Psychiatric GWAS Consortium Bipolar Disorder Working Group 2011), schizophrenia (Schizophrenia Working Group of The Psychiatric Genomics Consortium 2014), and MDD (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium 2013) are available and may also reveal associations with anxiety behavior.
In this study, we identify a number of QTGs underlying phenotypic differences in anxiety-related open field behavior between wild-derived RJF and domesticated White Leghorn chickens. We expanded an initial wild × domestic F2 intercross up to an eighth-generation advanced intercross line (AIL). This AIL can generate far smaller confidence intervals for mapping than the initial F2 (Darvasi 1998) and was combined with an expression QTL (eQTL) approach, using hypothalamus tissue from the same birds. Hypothalamus tissue was selected for this analysis due to its pivotal role in the hypothalmic-pituitary-adrenal (HPA) axis (integral to stress and anxiety responses), its known effects on anxiety-related behavior (File et al. 2000; McNaughton and Corr 2004; Kallen et al. 2008), and its control of the amygdala (McNaughton and Corr 2004) (also highly involved in the control of anxiety). Correlations between gene expression and behavior in shared QTL/eQTL regions identified 10 putative candidate genes that significantly correlated with anxiety-related open field behavior in chickens.
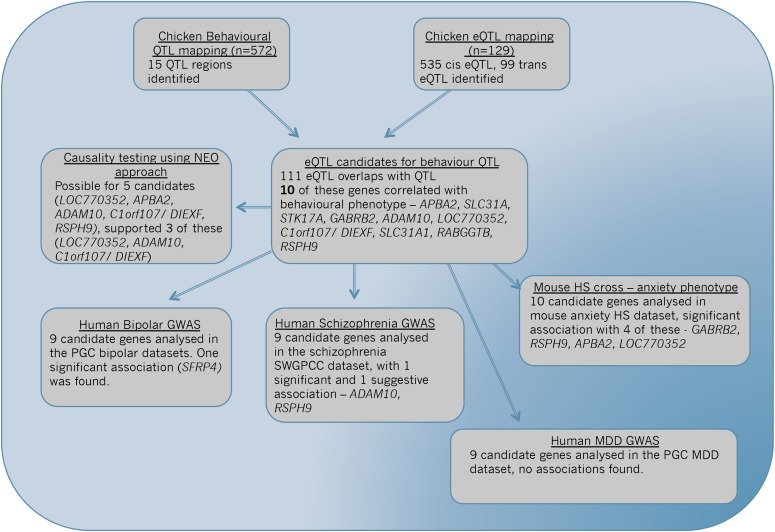
These candidate genes were then further assessed in four published data sets from humans and mice. Orthologous genes in three different human behavior-related GWASs were assayed—a human bipolar disorder data set (Psychiatric GWAS Consortium Bipolar Disorder Working Group 2011), the Psychiatric Genetics Consortium MDD GWAS (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium 2013), and the most recent PGC schizophrenia data set (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). These same genes were also assayed in a large Heterogeneous Stock (HS) mouse cross tested for anxiety behavior, using an open field arena (Valdar et al. 2006). Associations between QTG in chickens and orthologous genes in the human bipolar, MDD, and schizophrenia GWAS data sets were detected for three of the candidate genes, while associations were also observed between four of these candidate genes in the mouse advanced Heterogeneous Stock intercross assayed for open field behavior. A complete summary of all steps taken, the findings from each step, and how they led to further experiments is presented in Figure 1.
Figure 1.
Flow-chart overview of experimental steps taken, plus the findings from each step.
Materials and Methods
Chicken study population and cross design
The intercross population used in this study was an eighth-generation intercross between a line of selected White Leghorn chickens maintained from the 1960s and a population of RJF originally from Thailand (Schutz et al. 2002, 2004). The intercross is based on 1 RJF male and 3 White Leghorn (WL) females. These were expanded into 41 F1 and then 811 F2 progeny and subsequently maintained at a population size of ∼100 birds per generation until the F7 generation. The F2 intercross was previously measured for a variety of behavioral, morphological, and life history traits (Schutz et al. 2002; Kerje et al. 2003; Wright et al. 2008, 2010, 2012). A total of 572 F8 individuals in six batches were generated from 118 families, using 122 F7 individuals (63 females and 59 males), and assayed for behavioral measurements. Average family size was 4.76 ± 3.1 (mean, SD) in the F8. A total of 129 of the 572 F8’s were used in an eQTL experiment, with the hypothalamus/thalamus dissected out at 212 days of age and RNA extracted. For further details on feed and housing see Johnsson et al. (2012). This study was approved by the local Ethical Committee of the Swedish National Board for Laboratory Animals.
Phenotyping
Animals were weighed at hatch, 8 days, 42 days, 112 days, and 212 days. Open field assessment is a standard anxiety measurement, performed in a variety of vertebrates and invertebrates (Prut and Belzung 2003). Trials were performed in a 100 × 80-cm arena at 4 weeks of age. The 60 × 40-cm area in the middle of each arena was considered to be the central zone. Individuals were placed in the corner of the arena in complete darkness, prior to the test starting, with the lights turned on immediately at the commencement of the test. Trials lasted 5 min. Four separate arenas were available, allowing up to four individuals to be tested simultaneously. Measurements were taken using the Ethovision software and continuous video recording (Noldus Information Technology, www.noldus.com). For each trial, total distance moved, velocity, proportion of time spent in the central zone, and frequency (number of times) that the central zone was entered were measured. Each trial was performed twice, with ∼1 week between an individual’s first and second test. Individuals were removed from the arena immediately upon the test finishing to reduce potential habituation, with exposure to the test arena therefore restricted to only the 5 min required for each trial. Correlations between the two trials were found to be significant (total movement Pearson’s correlation coefficient = 0.55, t-value = 13, P < 2.2 × 10−16; velocity Pearson’s correlation coefficient = 0.62, t-value = 19, P < 2.2 × 10−16; time in center correlation coefficient = 0.31, t-value = 8, P = 9.2 × 10−14); see Supporting Information, Figure S1. Additionally, maximum and minimum values were also calculated for distance moved in a further attempt to reduce environmental variation, by finding an individual’s most fearful and least fearful scores. All behavioral phenotypes were correlated with one another (P ≤ 10−15 based on Pearson’s correlation; see Table S1).
RNA isolation and gene expression microarrays
The last three batches of the F8 generation (129 animals, i.e., unselected with respect to behavioral phenotype) were culled at 212 days of age and the hypothalamus was immediately dissected out and snap frozen in liquid nitrogen, prior to storage at −80°. RNA was isolated with Ambion TRI Reagent (Life Technologies), according to the supplier’s protocol. Reverse transcription was performed with the Fermentas Revert Aid Premium First-Strand cDNA Kit (Thermo Fisher Scientific) and oligo-(dT)18 primers, followed by second-strand cDNA synthesis according to the supplier’s protocol. All samples were quality checked on a Bioanalyzer chip (Agilent), and all had a RNA integrity number (RIN) value >8.1.
Gene expression was measured on NimbleGen 12 × 135K Custom Gene Expression Arrays. Array design, hybridization, scanning, and signal preprocessing were performed by Roche NimbleGen Services (Reykjavik, Iceland). The array included all Ensembl (Flicek et al. 2012) and RefSeq (Pruitt et al. 2009) chicken transcripts. In addition to the known transcripts, the array included probe sequences from a chicken brain cDNA library (Boardman et al. 2002). Probes based on the cDNA library were annotated by alignment against the chicken reference genome with Blat (Kent 2002). In total, this yielded an additional 10,686 probe sets. Each transcript was represented by three 60-mer oligonucleotide probes. To avoid SNPs in probe sequences, all known SNP positions derived from the recent resequencing of Red Junglefowl and domestic chickens (Rubin et al. 2010) were masked, so that probes could not be chosen from sequences with known SNPs. Array data were preprocessed with NimbleGen DEVA software (v. 2.6.0.0) and the robust multiarray average (RMA) algorithm (Irizarry et al. 2003). RMA comprises of a background correction, quantile normalization, and summarization of probes to probe set-level expression values with median polish and is the recommended normalization per the manufacturer’s guidelines. Although 36,000 transcripts were tested, there was a high degree of overlap between the EST probes and the known RefSeq genes. The total number of unique probe sets was therefore ∼17,000. All samples were handled in two batches at the Roche Nimblegen central facility in Iceland, with none being discarded. Including the array batch as a covariate did not change the eQTL or trait–expression correlations of the candidate genes. No further corrections for global effects on the expression phenotypes were included after the RMA.
Genotyping and QTL and eQTL mapping
DNA preparation was performed by Agowa (Berlin, Germany), using standard salt extraction, on all 572 F8 individuals. A total of 652 SNP markers were used to generate a map of length ∼9268 cM, with an average marker spacing of ∼16 cM. A full list of marker locations and informativeness can be found in Johnsson et al. (2015). QTL analysis was performed using R/Qtl (Broman et al. 2003) for the standard interval mapping and epistatic analyses. Interval mapping was performed using additive and additive plus dominance models. Map generation was performed on the actual (F8) data set, to account for the map expansion from the F2 to the F8. In this regard the map generation and QTL mapping are precisely like an F2 analysis, only with more recombinations present (hence a larger overall map, with an increase from ∼3000 cM to ∼9300 cM). The increase in map size is approximately equal to that predicted by theory, with a maximum of fourfold increase hypothetically possible for an F8 intercross (Darvasi and Soller 1995). Therefore, given our increased map size, we have on average a threefold increase in resolution compared to that for a standard F2 intercross. This expanded map was also used to generate the significance thresholds, resulting in higher than normal thresholds compared to the F2 (see later in article). In the behavioral QTL analysis batch, sex and arena were always included in the model as fixed effects, while body weight was included as a covariate. To account for a particular QTL varying between the sexes, a sex-interaction effect was added where significant. Digenic epistatic analysis was performed as per the guidelines given in Broman and Sen (2009). A global model that incorporated standard main effects, sex interactions, and epistasis was built up, starting with the most significant loci and working down for each trait.
eQTL mapping was also performed with R/qtl, using RMA preprocessed (Irizarry et al. 2003) expression levels as quantitative phenotypes with sex and batch as additive covariates. Cis-eQTL (defined as QTL that were located close to the target gene they affected) were mapped in an interval from the transcription start positions to the closest flanking markers spanning at least 100 cM (i.e., 50 cM upstream and downstream of the gene, once again using the map generated using the F8 data). A cis-eQTL was called if the LOD score reached above the empirical cis threshold (see below) at any marker in this interval. Note that it is potentially problematic to ascertain at what point a local eQTL is truly cis acting (i.e., potentially on the same strand and acting putatively on a local enhancer to the gene in question), so more accurately our definition should be local rather than cis; however, in eQTL mapping this is a common problem and not one specific to our experiment, and therefore we have opted for the standard terminology used. Similarly, if the eQTL is 50 cM from the gene, it could also be thought of as a more locally acting trans element. The trans-eQTL scan encompassed the whole genome and used a genome-wide empirical significance threshold.
In general an important caveat to mention in terms of estimating the r2 effect of a QTL (i.e., the percentage of variance explained by the QTL) is that this is less effective for smaller sample sizes (leading to an overestimation), with this being known as the Beavis effect (Beavis 1998) or the winner’s curse. This typically starts occurring when the sample size is <n = 1000. The relatively large size of the behavioral QTL study used here (n > 500) reduces this effect. It does, however, pertain more strongly to the eQTL study and results in an overestimation of eQTL effect sizes.
Significance thresholds
Significance thresholds for behavioral QTL analysis were calculated for each trait, using permutation tests (Churchill and Doerge 1994, 1996), with 1000 permutations. As mentioned above, permutations were based on the full F8 map data (∼9000 cM rather than ∼3000 cM). This is important, as the use of the original F2 map would have resulted in far fewer tests being performed and artificially decreased the significance threshold. A suggestive significance level of a genome-wide 20% P-value cutoff threshold was used [principally due to being more conservative than the standard suggestive threshold (Lander and Kruglyak 1995)]. The approximate significance threshold was LOD ∼ 4.4 (5% genome-wide), while the suggestive threshold was ∼3.6 (20% genome-wide). Confidence intervals (C.I.) for each QTL were calculated with a 1.8-LOD drop method (i.e., where the LOD score on either side of the peak decreases by 1.8 LOD); as this threshold gives an accurate 95% confidence interval for an intercross-type population (Manichaikul et al. 2006). The nearest marker to this 1.8-LOD decrease was then used to give the C.I. in megabases. Epistatic interactions were also assessed using permutation thresholds generated using R/qtl, with a 20% suggestive and a 5% significant genome-wide threshold used. In the case of epistatic loci, the approximate average significance thresholds for pairs of loci were as follows (using the guidelines given in Broman and Sen 2009): full model ∼11, full vs. one ∼9, interactive ∼7, additive ∼7, additive vs. one ∼4.
Behavioral QTL significance thresholds were not modified based on the number of different phenotypes measured. In this case the phenotypes were correlated with one another (see Table S1) and were therefore not independent, thereby removing the need to multiple-test correct. It must be noted, however, that despite the P-values of the correlations being highly significant (P < 10−15) some of the pairwise correlations were relatively modest in effect size (most notably between time spent in the central zone and velocity, with a correlation of 33%). This indicates that the traits are also not fully dependent on one another.
eQTL significance thresholds were also generated by permutation, although in this case there were two types of eQTL being mapped—cis (local)-QTL and trans (global)-QTL. In the case of cis-eQTL, only markers surrounding each gene were tested, therefore reducing the multiple-testing correction that is required. To account for both the full genetic map (in the case of trans effects) and the large number of phenotypes (∼36,000) used, these permutations were based not on one phenotype at each time, but by using 100 randomly subsampled probe sets. In the case of cis-eQTL, this procedure was performed in a similar fashion, although only the limited region around each gene locus was used for extracting LOD scores (with 100 probe sets still used simultaneously for each permutation). This generated cis thresholds of LOD 4.0 and trans thresholds of LOD 6.0.
Family structure
Thresholds and analysis for an AIL are potentially problematic, as the family structure can be affected by nonsyntenic association, resulting in false positive results. To avoid this initially, we used a large number of families (n = 118) to generate the total number of individuals, to break down this substructure as much as possible. For instance, if only one offspring were used per family, there would be no family structure and the population would function exactly as recombinant inbred lines (Peirce et al. 2008). A principal components analysis approach was used to control for any residual family structure (Wu et al. 2011). We calculated the first 10 principal components (PCs) and then tested these for significance in each behavioral QTL regression. All significant PCs were then retained in the final model. This approach allowed us to both control for population substructure and test for epistatic interactions, which is impossible using other packages designed for advanced intercross QTL analysis. This was applied only to the behavioral QTL analysis, as the eQTL analysis utilized even smaller family sizes (129 individuals from 44 families), and nonsyntenic linkage (i.e., that regions in linkage disequilibrium would provide false-positive signals) is less of an issue given that only specific loci were tested in the case of the cis-eQTL scan.
Analysis of candidate genes (eQTL genes falling within QTL intervals)
Significant eQTL were overlapped with behavioral QTL, with all significant eQTL genes then being considered candidate genes for the behavioral QTL they overlapped with. To further refine these candidate genes we then modeled the gene expression value on the behavioral trait for the QTL of interest (i.e., if an eQTL overlapped a QTL for open field activity, the eQTL gene expression trait would be correlated with open field activity). For each eQTL overlapping a behavior QTL, a linear model was fitted with the behavior trait as a response variable and the expression traits as predictor, including sex and batch as factors. Weight at 42 days was included for traits where weight was used as a covariate in the QTL analysis (see Table S1). P-values for the regression coefficient were Bonferroni corrected for the number of uncorrelated eQTL in the QTL region. As the eQTL in a given region are often strongly correlated with one another [a typical feature of any genome—genes of similar expression are colocalized (Litvin et al. 2009)], it is required only to correct for the number of these eQTL genes that are independent (i.e., the expression is not significantly correlated). This was especially true in our case, as we had numerous probes that were based on an EST library that frequently overlapped with the actual RefSeq genes (i.e., they were the same genes tested). As we are testing a specific hypothesis (i.e., that a given QTL is caused by the gene expression of an overlapping eQTL), we corrected only for the number of independent eQTL for that particular QTL, rather than for eQTL overlapping other QTL for that trait. The average multiple-testing correction was ∼2 per QTL region. Any eQTL within a QTL C.I. that were significantly correlated with the QTL trait were therefore considered candidate causative genes and were then assessed for causality where possible. One issue with using this approach with this particular data set is that the behavioral QTL were based on up to 572 individuals, whereas the eQTL/expression phenotypes were available only for 129 individuals. Therefore the network edge orienting (NEO) method for causality testing was applied only where the behavioral QTL that a gene was potentially causative to was detectable in the smaller data set (n = 129).
Network edge orientation analysis
Causality analysis was performed using NEO software (Aten et al. 2008) to test whether the expression of correlational candidates was consistent with the transcript having a causal effect on the behavior trait. Single-marker analysis was performed with NEO fitting a causal model (marker → expression trait → behavior) and three other types (reactive, confounded, and collider). The NEO software evaluates the fit of the model with a χ2-test, a higher P-value indicating a better fit of the model. The best-fitting model is chosen based on the ratio of the χ2 P-value to the P-value of the next best model on a logarithmic scale (base 10), called local edge orienting against the next best model (leo.nb) scores. A positive leo.nb score indicates that the causal model fits better than any competing model. Aten et al. (2008) use a single-marker leo.nb score of 1, corresponding to a 10-fold higher P-value of the causal model, as their threshold. They also suggest that users inspect the P-value of the causal model to ensure the fit is good (in this case meaning the model P-value should be nonsignificant if the causal model fits the best). In effect this P-value is the probability of another model fitting the observed data. For each gene, we report the leo.nb score and P-value of the causal model.
Human and mouse GWAS comparisons
A description of the methods for the comparison of candidate genes with human GWAS and mouse QTL data is presented in File S1.
Data availability
Microarray data for the chicken hypothalamus tissue are available at E-MTAB-3154 in ArrayExpress. Full genotype and phenotype data are available on figshare with the following doi: 10.6084/m9.figshare.1265060.
Results
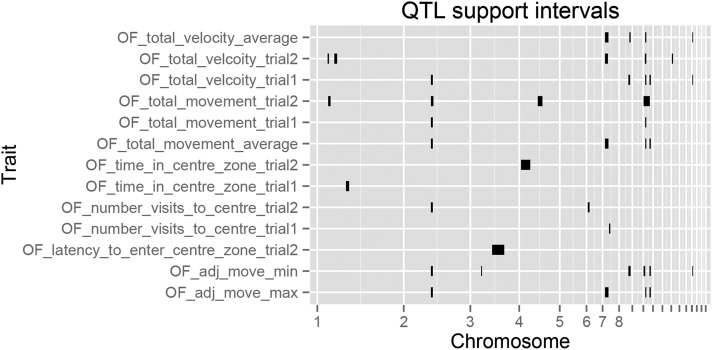
QTL mapping of fearful behavior (n = 572) in the AIL identified a total of 15 distinct QTL regions (comprising 34 QTL) for open field behavior, with the average confidence interval being ∼3 Mb (see Table S2 and Figure 2). Average variance explained (R2) for the behavioral QTL was 5.4%. Expression QTL mapping of hypothalamic gene expression in the AIL detected 535 cis-eQTL and 99 trans-eQTL for 537 genes (Table S3), with effect sizes of these eQTL ranging from 13% to 58% of the variance explained.
Figure 2.
Genomic locations of all detected open field behavioral QTL. Bars indicate 1.8-LOD drop confidence intervals.
Candidate quantitative trait genes
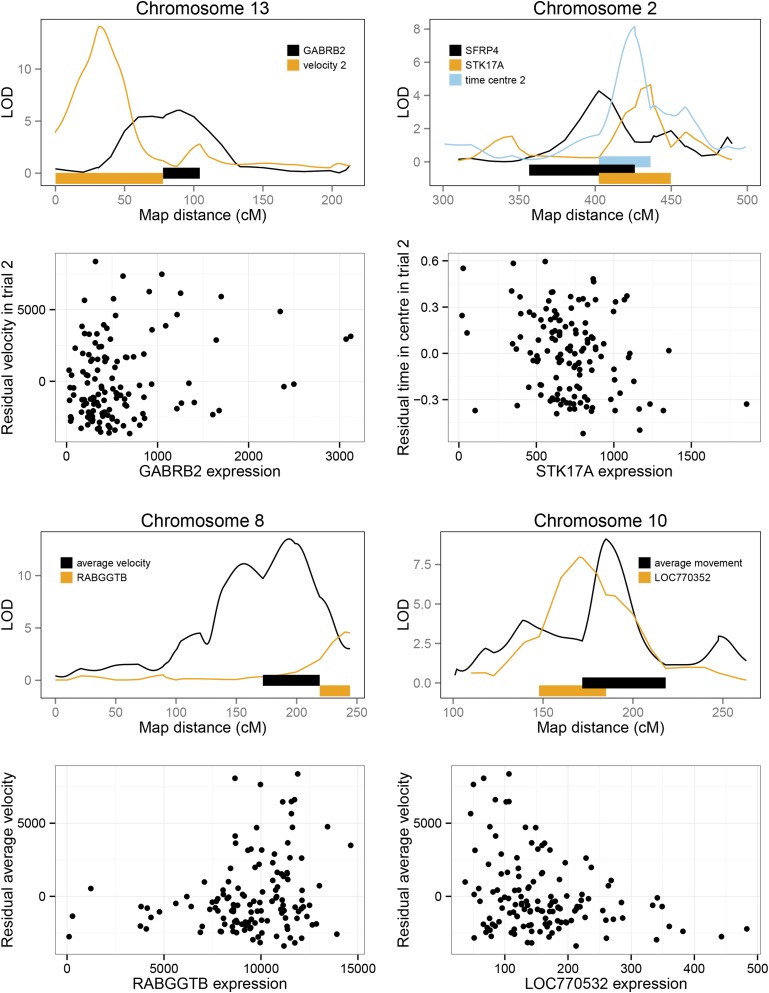
A total of 111 eQTL probe sets were found to overlap behavior QTL, with these all then considered potential candidate genes. A regression between each candidate gene and the respective behavioral trait it overlapped with yielded a total of 10 significant correlational candidate genes, after a Bonferroni correction for the number of uncorrelated eQTL overlapping the behavior QTL region was applied (see Table 1, Figure 3, and Figure S2). These represented seven different QTL regions. Of the significant candidates, 2 genes were strongly significantly correlated (P < 0.001) with their corresponding behavioral trait: GABRB2 and a novel gene Hypothetical protein LOC770352, with a further 4 also with a P < 0.01, including APBA2, SLC31A, STK17A, and C1orf107/DIEXF.
Table 1. Candidate gene expression levels predicting behavior.
| Mouse HS open field LOD scores (additive model) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Trait | QTL C.I. (Mb) | eQTL | PhQTL pos | Gene P-value | NEO leo.nb score | Model P-value | Marker | Latency | Total activity | Time in center | Marker |
| 2 | Time in center zone trial 2 | 45.5–51.9 | SFRP4 | 2@426 | 0.04 | NA | NA | rs2709114 | — | — | — | rs13481716 |
| 2 | Time in center zone trial 2 | 45.5–51.9 | STK17A | 2@426 | 0.004 | NA | NA | rs1181601 | — | — | — | rs6253368 |
| 3 | Minimum total movement | 25.1–27 | RSPH9 | 3@277 | 0.03 | 0.823 | 5.48E-06 | rs3778493 | 4.7 | 7.8 | — | UT_17_45.56 |
| 3 | Minimum total movement | 25.1–27 | C1orf107/DIEXF | 3@277 | 0.003 | 0.368 | 0.804 | rs7549523 | — | — | — | rs3684501 |
| 8 | Total velocity average | 23.9–26.6 | RABGGTB | 8@194.0 | 0.05 | NA | NA | rs7555627 | — | — | — | rs3656403 |
| 10 | Total velocity average | 4.3–6.8 | ADAM10 | 10@101 | 0.03 | 0.44 | 0.396 | rs2242321 | — | — | — | rs13480265 |
| 10 | Total velocity average | 4.3–6.8 | APBA2 | 10@101 | 0.01 | 0.0421 | 0.0719 | rs7680321 | — | 7.7 | — | rs3717846 |
| 10 | Total velocity trial 1 | 14.4–17.4 | LOC770352 | 10@185.0 | 0.002 | 0.943 | 0.687 | rs1480617 | — | — | 4.5 | rs13476065 |
| 10 | Total movement average | 14.4–17.4 | LOC770352 | 10@185.0 | 0.0002 | 1.07 | 0.956 | rs1480617 | — | — | 4.5 | rs13476065 |
| 10 | Minimum total movement | 14.4–17.4 | LOC770352 | 10@182.0 | 0.003 | 0.874 | 0.925 | rs1480617 | — | — | 4.5 | rs13476065 |
| 10 | Total movement trial 2 | 7.6–15.0 | LOC770352 | 10@64.0 | 0.0012 | 0.747 | 0.878 | rs1480617 | — | — | 4.5 | rs13476065 |
| 10 | Maximum total movement | 14.4–17.4 | LOC770352 | 10@185.0 | 0.0003 | 1.13 | 0.985 | rs1480617 | — | — | 4.5 | rs13476065 |
| 10 | Total velocity trial 1 | 4.3–6.8 | APBA2 | 10@99.0 | 0.004 | 0.977 | 0.131 | rs7680321 | — | 7.7 | — | rs3717846 |
| 10 | Total velocity trial 1 | 4.3–6.8 | ADAM10 | 10@99.0 | 0.04 | 0.515 | 0.37 | rs2242321 | — | — | — | rs13480265 |
| 10 | Total movement trial 1 | 4.3–6.8 | LOC770352 | 10@99.0 | 0.008 | 0.815 | 0.852 | rs7680321 | — | — | 4.5 | rs13476065 |
| 10 | Total movement average | 4.3–6.8 | APBA2 | 10@99.0 | 0.02 | −0.966 | 0.0002 | rs7680321 | — | 7.7 | — | rs3717846 |
| 13 | Total velocity trial 2 | 1.5–4.1 | GABRB2 | 13@32.0 | 0.0014 | NA | NA | rs4302629 | 5.1 | 5.1 | — | rs6225242 |
| 17 | Total velocity average | 1–2.5 | SLC31A1 | 17@1 | 0.004 | NA | NA | rs786978 | — | — | — | rs3690581 |
| 17 | Total velocity trial 1 | 1–2.5 | SLC31A1 | 17@1 | 0.003 | NA | NA | rs786978 | — | — | — | rs3690581 |
Chromosome (Chr) and QTL confidence interval are given for the trait that each eQTL significantly correlated with. The phenotypic QTL position (PhQTL pos) is also given in centimorgans. The P-value of the correlation, after Bonferroni correction, is also provided (Gene P-value). The causality modeling scores are also indicated for each gene, as are the WTCCC P-values for each gene region. The association of each gene with the mouse HS cross is also given, with the LOD scores of the gene location for each of the three different open field tests provided.
Figure 3.
Part of LOD curves and scatterplots of trait vs. gene expression for selected candidates GABRB2, RABGGTB, STK17A and SFRP4, and LOC770352. Bars indicate 1.8-LOD intervals, extended to the closest marker.
To attempt to ascertain statistical causation a NEO approach (Aten et al. 2008) was used on 5 of the 10 candidate genes (LOC770352, APBA2, ADAM10, C1orf107, and RSPH9). The remaining 5 genes had an undetectable behavioral QTL in the reduced eQTL sample, preventing this analysis (see Materials and Methods). This approach is designed to identify statistical causation between the phenotype, genotype, and gene expression variables (i.e., if the genotype is the causative locus for a QTL, this should then affect gene expression, which in turn affects the behavioral phenotype). The sole candidate gene LOC770352 for the open field QTL at 14.4–17.4 Mb on chromosome 10 exhibited good statistical evidence for causality as assessed by its local edge orienting score (see Materials and Methods). In the case of the open field QTL at 4.3–7.8 Mb, of the two candidates APBA2 and ADAM10, NEO calculations indicated that ADAM10 was a better candidate (local edge orienting score was low for APBA2 and higher for ADAM10, except for one of the open field variables). NEO also indicated that the gene RSPH9 is unlikely to be causal to the chromosome 3 QTL, whereas there is more support for the gene C1orf107.
As a further assessment of both the 10 candidate genes and the method in general, all significant eQTL were correlated with the behavioral phenotypes and ranked in order. The average rank of these 10 genes was in the top 3% of all eQTL, with 2 of the genes (STK17A and LOC770352) being the top-ranked genes for time in the central zone and maximum total movement, respectively (see Table 2). The total number of eQTL correlated for each trait for which a significant candidate was obtained is given in Table S4.
Table 2. Candidate genes trait correlations and ranking vs. all eQTL.
| Gene | Trait | Overall rank (of 535 eQTL) | Trait–gene correlation P-value |
|---|---|---|---|
| ADAM10 | Average velocity | 52 | 0.027 |
| ADAM10 | Velocity trial 1 | 35 | 0.018 |
| APBA2 | Average total movement | 34 | 0.024 |
| APBA2 | Average velocity | 27 | 0.010 |
| APBA2 | Velocity trial 1 | 11 | 0.002 |
| C1orf107/DIEXF | Minimum total movement | 7 | 0.003 |
| GABRB2 | Velocity trial 2 | 7 | 0.006 |
| LOC770352 | Total movement trial 1 | 11 | 0.004 |
| LOC770352 | Total movement trial 2 | 2 | 0.001 |
| LOC770352 | Maximum total movement | 1 | <0.001 |
| LOC770352 | Minimum total movement | 4 | 0.001 |
| LOC770352 | Average total movement | 2 | <0.001 |
| LOC770352 | Velocity trial 1 | 13 | 0.002 |
| RABGGTB | Average velocity | 23 | 0.008 |
| RSPH9 | Minimum total movement | 39 | 0.033 |
| SFRP4 | Time in center trial 2 | 24 | 0.018 |
| SLC31A1 | Average velocity | 17 | 0.004 |
| SLC31A1 | Velocity trial 1 | 15 | 0.003 |
| STK17A | Time in center trial 2 | 1 | 0.002 |
Associations with MDD, bipolar disorder, and schizophrenia
To test for the presence of associations with human bipolar disorder, 9 of the 10 candidate genes (on closer examination LOC770352 did not have a suitably conserved ortholog and was excluded) were examined in the PGC bipolar data set (n ∼ 7500 cases) (Psychiatric GWAS Consortium Bipolar Disorder Working Group 2011). Of the genes tested, there was one significant association with SFRP4 (P = 0.0001).
The PGC study for MDD is perhaps the closest human GWAS study to the anxiety phenotype measured here in chickens (Levinson 2006). However, this study identified no genome-wide significant SNPs in humans, indicating that the phenotype is less repeatable, has a different genetic architecture (smaller-effect loci), or is harder to accurately phenotype. It is also a more common disorder than the other behavioral disorders used here, with such disorders typically requiring a larger sample size than their less common counterparts. No associations were found with any of the genes tested.
Although farther from a standard anxiety phenotype, schizophrenia also has similar symptoms, making analysis of nine of the candidate genes also potentially relevant in a schizophrenia GWAS. The largest data set available has been collected by the PGC once again (n ∼ 36,000 cases) (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). Using this data set, a significant association was found between ADAM10 and schizophrenia (P = 1 × 10−5), while a suggestive association was found with RSPH9 (P = 9 × 10−5); see Table 3.
Table 3. Association of the nine genes tested in the GWASs.
| Bipolar: | Schizophrenia: | Major depressive disorder: | ||||
|---|---|---|---|---|---|---|
| PGCBD 2011 study | SWGPCCa 2014 | PGCMDD | ||||
| Gene | P-value | Marker | P-value | Marker | P-value | Marker |
| SFRP4 | 0.0001* | rs7807315 | NS | — | NS | — |
| STK17A | NS | — | NS | — | NS | — |
| RSPH9 | NS | — | 0.00009# | rs72859036 | NS | — |
| DIEXF | NS | — | NS | — | NS | — |
| RABGGTB | NS | — | NS | — | NS | — |
| ADAM10 | NS | — | 0.00001* | rs6494035 | NS | — |
| APBA2 | NS | — | NS | — | NS | — |
| GABRB2 | NS | — | NS | — | NS | — |
| SLC31A1 | NS | — | NS | — | NS | — |
Significant (*) and suggestive (#) P-values are given for each gene, along with the marker showing the highest association in the gene interval. In the case of RSPH9 and ADAM10 the distance of the marker from the gene is also given.
Schizophrenia Working Group of the Psychiatric Genomics Consortium.
Associations with mouse models of anxiety
One of the largest studies performed using a mouse model to study anxiety, using an open field assay, utilized a Heterogeneous Stock cross to obtain far smaller confidence intervals than a standard F2 intercross (average QTL C.I. <3 Mb in size) (Valdar et al. 2006). Testing for an association between the nearest SNPs to each of the 10 candidate genes under investigation and three open field variables (latency to move, total activity, and time spent in the center of the arena) revealed a total of four significant loci (see Table 2). Of the significant loci, several were replicated for the same behavioral measurement in both chickens and mice. RSPH9 showed a significant correlation with total movement in chickens and for total activity in mice (LOD = 7.8). APBA2 and GABRB2 both showed a correlation with velocity in chickens and with total activity in mice (APBA2 LOD = 7.7, GABRB2 LOD = 5.1).
Discussion
In this study we use a combination of QTL and eQTL mapping in a novel anxiety model, chickens, and identify multiple candidate gene variants affecting anxiety behavior. We also present evidence that some of these gene polymorphisms may have some translational effects, although care must be taken not to overstress these results. This approach is obviously costly in terms of the number of individuals required for the eQTL analysis and to date has to our knowledge been used only to identify QTG for startle-induced locomotion behavior (among other ecological phenotypes) in Drosophila (Ayroles et al. 2009), exercise behavior in mice (Kelly et al. 2012), and tameness in rats (Heyne et al. 2014), but has enormous potential. Here we use this technique to identify gene variants affecting anxiety for the first time in chickens. Chickens have many advantages, both genetic and phenotypic, making them attractive as a model for anxiety-related disorders. When this is coupled with the in-depth knowledge of the neural development of chickens (a classical model of such development), this represents a powerful resource.
Of the 10 candidate genes, 6 have been previously identified as having some bearing on behavior (mainly anxiety and stress related) or neuronal development, while 3 have no previous evidence (SFRP4, C1orf107, and RSPH9) and 1 is a novel gene (LOC770352). Such an enrichment gives us a great deal of confidence about the genes that have been identified, as well as providing far greater evidence for their effects on behavior, in many cases, than was available previously. The 6 genes with previous implications [SLC31A (Jones et al. 2008; Lagus et al. 2010; Goerlich et al. 2012), ADAM10 (Postina et al. 2004; Schmitt et al. 2006; Bell et al. 2008; Jorissen et al. 2010), GABRB2 (Moriarty 1995; Marín et al. 1997; Salvatierra and Arce 2001), APBP2 (Kirov et al. 2008), and STK17A and RABBGTB (Kõks et al. 2004)] have a variety of effects in neural tissue or on behavior. ADAM10 is required for embryonic brain development, (Jorissen et al. 2010), protects against amyloid plaque formation (Postina et al. 2004), increases synaptogenesis (Bell et al. 2008), and also causes behavioral differences in learning and memory (Schmitt et al. 2006). Similarly, APBA2 has been implicated in schizophrenia by one study (Kirov et al. 2008), although this gene was not identified in the more recent PGC GWASs. SLC31A and RABGGTB have both been shown to have some links with anxiety behavior and memory. SLC31A (a copper transporter) has been linked with depression in rats (Lagus et al. 2010) and chickens exposed to social isolation and restraint stress (Goerlich et al. 2012), and genetic correlations suggest a link between copper and anxiety behaviors in BXD mice (Jones et al. 2008). RABGGTB has been linked with fear responsiveness in rats (Kõks et al. 2004). Although little has been characterized of STK17A [a serine/threonine kinase and a part of the death-associated protein kinase group (Kögel et al. 2001)], barring a link with p53 in cancer (Mao et al. 2011), there are several examples of other serine/threonine kinases affecting behavior, specifically learning and anxiety in Drosophila (Choi et al. 1991) and mice (Silva et al. 1992) (Hodge et al. 2002), with links to bipolar disorder (Klein and Melton 1996) and depression (Gould et al. 2004; O’Brien et al. 2004). In a parallel fashion, GABRB2 has been found to have prior associations with schizophrenia (Lo et al. 2007; Pun et al. 2011) and bipolar disorder in German and Chinese cohorts, while the closely related GABRB1 has associations with alcohol dependence (Parsian and Zhang 1999).
A point of note is how potentially translational the gene polymorphisms that can influence quantitative variation in behavior can be and the applicability between different model organisms and humans. It is striking how an open field assay in chickens and mice can show associations in the same genes for even the same aspects of the assay (activity in the case of RSPH9, APBA2, and GABRB2). Assessing the candidate genes in human data sets also gave some potential indications of translational effects in these gene polymorphisms. The data sets analyzed indicated putative effects for ADAM10 and RSPH9 with regard to schizophrenia and SFRP4 with regard to bipolar disorder. Care must be taken in interpreting these results from human GWASs, however. It can be problematic to control for such variables as gene density and also recombination rate, the size of haplotype blocks, and so forth, when calculating suitable thresholds for significance for these selected-region scans. Similarly, the exact phenotype used must be chosen with care, with more stringent phenotyping criteria for the human disorder potentially leading to a more relevant trait comparison. Although the associations found here would be subthreshold values in a full GWAS analysis, for the targeted approach used here they offer indications that some of the genes may also play a role in human quantitative variation and are worthy of further study to ascertain whether this is the case. Such an approach also offers a method of exploratory analysis to gain additional information from such large GWASs. Due to the issue of massive multiple-testing correction, it has been conjectured that many true significant associations are buried in the “noise” of such necessarily stringent thresholds. For example, the “missing” heritability that such GWASs fail to find has potentially been identified by considering the joint effect of all SNPs in linear mixed models [an excellent example of this is with human height (Yang et al. 2010), with this technique also used for psychiatric disorders (Cross-Disorder Group of the Psychiatric Genomics Consortium 2013)], in essence decreasing the significance threshold for inclusion into the genetic model. Assessing candidate genes supported by evidence from model organisms in such data sets offers a method of maximizing the potential from such studies
One potential issue with the use of chickens as model organisms for human behavior is the relatively compact nature of the chicken genome. It is possible that given the reduced genome size chickens are missing key regulatory elements that may have evolved in the noncoding DNA found in humans, which potentially harbor variants that affect the fine temporo-spatial regulation of the expression of such genes and may thus mask cross-species correspondences between risk loci. This is obviously true for all model organisms, however. Similarly, it is less likely the exact mutation affecting a gene is replicated between species, but the gene itself may still be functioning in a similar fashion. In this way, cross-species modeling is more likely to find candidate genes than candidate mutations.
In conclusion, we have used a combination of an advanced intercross, QTL, and eQTL analysis in a chicken model to identify 10 putative QTGs affecting anxiety behavior. These genes were then tested in published data sets using mice and humans and significant associations were identified. By using multiple forms of validation, this will hopefully provide greater weight of evidence for the effects of a particular gene. Similarly, in many cases not all of the different validation tests will positively associate with a given gene. The final form of validation for all of these genes would ideally be transgenic verification. This study represents the first of its kind for anxiety-related behavior in chickens and establishes them as a strong model for the genetic dissection of anxiety behavior and of personality in general.
Ethics statement
This study was approved by the Regional Committee for Ethical Approval of Animal Experiments (Jordbruksverket DNR 122-10). Birds were killed by cervical dislocation and decapitation, as per the guidelines of the permit.
Acknowledgments
The authors thank Leif Andersson for help with the design and implementation of this study. This research was carried out within the framework of the Swedish Centre of Excellence in Animal Welfare Science and the Linköping University Neuro-network. SNP genotyping was performed by the Uppsala SNP&SEQ Platform. This project was supported by grants from the Swedish Research Council, the Swedish Research Council for Environment (to D.W.), Agricultural Sciences and Spatial Planning (to D.W.), and the European Research Council (advanced research grant GENEWELL 322206) (to P.J.).
Footnotes
Communicating editor: N. R. Wray
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179010/-/DC1.
Literature Cited
- Anholt R. R. H., Mackay T. F. C., 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat. Rev. Genet. 5: 838–849. [DOI] [PubMed] [Google Scholar]
- Archer J., 1973. Tests for emotionality in rats and mice: a review. Anim. Behav. 21: 205–235. [DOI] [PubMed] [Google Scholar]
- Aten J., Fuller T., Lusis A., Horvath S., 2008. Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst. Biol. 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W. D., 1998. QTL analyses: power, precision and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by Paterson A. H. CRC Press, Boston. [Google Scholar]
- Bell K. F. S., Zheng L., Fahrenholz F., Cuello A. C., 2008. ADAM-10 over-expression increases cortical synaptogenesis. Neurobiol. Aging 29: 554–565. [DOI] [PubMed] [Google Scholar]
- Belzung, C., 1999 Measuring rodent exploratory behavior. Techniques Behav. Neural Sci. 13: 738–752.
- Boardman P. E., Sanz-Ezquerro J., Overton I. M., Burt D. W., Bosch E., et al. , 2002. A comprehensive collection of chicken cDNAs. Curr. Biol. 12: 1965–1969. [DOI] [PubMed] [Google Scholar]
- Boylan K. R., Bieling P. J., Marriott M., Begin H., Young L. T., et al. , 2004. Impact of comorbid anxiety disorders on outcome in a cohort of patients with bipolar disorder. J. Clin. Psychiatry 65: 1106–1113. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen S., 2009. A Guide to QTL Mapping with R/qtl. Springer-Verlag, New York. [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Buitenhuis A., Rodenburg T., Siwek M., Cornelissen S., Nieuwland M., et al. , 2004. Identification of QTLs involved in open-field behavior in young and adult laying hens. Behav. Genet. 34: 325–333. [DOI] [PubMed] [Google Scholar]
- Burmeister M., McInnis M. G., Zöllner S., 2008. Psychiatric genetics: progress amid controversy. Nat. Rev. Genet. 9: 527–540. [DOI] [PubMed] [Google Scholar]
- Chiavegatto S., Izidio G. S., Mendes-Lana A., Aneas I., Freitas T. A., et al. , 2008. Expression of [alpha]-synuclein is increased in the hippocampus of rats with high levels of innate anxiety. Mol. Psychiatry 14: 894–905. [DOI] [PubMed] [Google Scholar]
- Choi K. W., Smith R. F., Buratowski R. M., Quinn W. G., 1991. Deficient protein kinase C activity in turnip, a Drosophila learning mutant. J. Biol. Chem. 266: 15999–16006. [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE Consortium , 2015. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosoff S. J., Julian Hafner R., 1998. The prevalence of comorbid anxiety in schizophrenia, schizoaffective disorder and bipolar disorder. Aust. N. Z. J. Psychiatry 32: 67–72. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium , 2013. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A., 1998. Experimental strategies for the genetic dissection of complex traits in animal models. Nat. Genet. 18: 19–24. [DOI] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic-mapping. Genetics 141: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge R. W., Churchill G. A., 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone S. V., Biederman J., Wozniak J., Mundy E., Mennin D., et al. , 1997. Is comorbidity with ADHD a marker for juvenile-onset mania? J. Am. Acad. Child Adolesc. Psychiatry 36: 1046–1055. [DOI] [PubMed] [Google Scholar]
- File S. E., Kenny P. J., Cheeta S., 2000. The role of the dorsal hippocampal serotonergic and cholinergic systems in the modulation of anxiety. Pharmacol. Biochem. Behav. 66: 65–72. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick M. J., Ben-Shahar Y., Smid H. M., Vet L. E. M., Robinson G. E., et al. , 2005. Candidate genes for behavioural ecology. Trends Ecol. Evol. 20: 96–104. [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Barrell D., Beal K., Brent S., et al. , 2012. Ensembl 2012. Nucleic Acids Res. 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., 2003. Analysis of quantitative trait loci that influence animal behaviour. J. Neurobiol. 54: 46–77. [DOI] [PubMed] [Google Scholar]
- Forkman B., Boissy A., Meunier-Salaün M.-C., Canali E., Jones R., 2007. A critical review of fear tests used on cattle, pigs, sheep, poultry and horses. Physiol. Behav. 92: 340–374. [DOI] [PubMed] [Google Scholar]
- Genetics of Personality Consortium , 2015. MEta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry 72: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerlich V. C., Nätt D., Elfwing M., Macdonald B., Jensen P., 2012. Transgenerational effects of early experience on behavioral, hormonal and gene expression responses to acute stress in the precocial chicken. Horm. Behav. 61: 711–718. [DOI] [PubMed] [Google Scholar]
- Gould T. D., Einat H., Bhat R., Manji H. K., 2004. AR-A 014418, a selective GSK-3 inhibitor, produces antidepressant-like effects in the forced swim test. Int. J. Neuropsychopharmacol. 7: 387–390. [DOI] [PubMed] [Google Scholar]
- Gyetvai B., Simonyi A., Oros M., Saito M., Smiley J., et al. , 2011. mGluR7 genetics and alcohol: intersection yields clues for addiction. Neurochem. Res. 36: 1087–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. S., 1934. Emotional behavior in the rat. I. Defecation and urination as measures of individual differences in emotionality. J. Comp. Psychol. 18: 385. [Google Scholar]
- Harpold T. L., Wozniak J., Kwon A., Gilbert J., Wood J., et al. , 2005. Examining the association between pediatric bipolar disorder and anxiety disorders in psychiatrically referred children and adolescents. J. Affect. Disord. 88: 19–26. [DOI] [PubMed] [Google Scholar]
- Heyne H. O., Lautenschläger S., Nelson R., Besnier F., Rotival M., et al. , 2014. Genetic influences on brain gene expression in rats selected for tameness and aggression. Genetics 198: 1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C. W., Raber J., McMahon T., Walter H., Sanchez-Perez A. M., et al. , 2002. Decreased anxiety-like behavior, reduced stress hormones, and neurosteroid supersensitivity in mice lacking protein kinase Cε. J. Clin. Invest. 110: 1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert J. D., Weiss K. A., Lim R., Pratt S., Smith T. E., 2001. Quality of life in schizophrenia: contributions of anxiety and depression. Schizophr. Res. 51: 171–180. [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., et al. , 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson M., Gustafson I., Rubin C.-J., Sahlqvist A.-S., Jonsson K. B., et al. , 2012. A sexual ornament in chickens is affected by pleiotropic alleles at HAO1 and BMP2, selected during domestication. PLoS Genet. 8: e1002914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson M., Jonsson K. B., Andersson L., Jensen P., Wright D., 2015. Genetic regulation of bone metabolism in the chicken: similarities and differences to mammalian systems. PLoS Genet. 11: e1005250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L. C., Beard J. L., Jones B. C., 2008. Genetic analysis reveals polygenic influences on iron, copper, and zinc in mouse hippocampus with neurobiological implications. Hippocampus 18: 398–410. [DOI] [PubMed] [Google Scholar]
- Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., et al. , 2010. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 30: 4833–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen V. L., Tulen J. H. M., Utens E. M. W. J., Treffers P. D. A., De Jong F. H., et al. , 2008. Associations between HPA axis functioning and level of anxiety in children and adolescents with an anxiety disorder. Depress. Anxiety 25: 131–141. [DOI] [PubMed] [Google Scholar]
- Kaluev, A. V., and J. M. Cachat, 2012 Zebrafish Models in Neurobehavioral Research. Humana Press–Springer, New York. [Google Scholar]
- Kas M., Fernandes C., Schalkwyk L., Collier D., 2007. Genetics of behavioural domains across the neuropsychiatric spectrum; of mice and men. Mol. Psychiatry 12: 324–330. [DOI] [PubMed] [Google Scholar]
- Kelly S. A., Nehrenberg D. L., Hua K., Garland T., Pomp D., 2012. Functional genomic architecture of predisposition to voluntary exercise in mice: expression QTL in the brain. Genetics 191: 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S., Carlborg O., Jacobsson L., Schutz K., Hartmann C., et al. , 2003. The twofold difference in adult size between the red junglefowl and White Leghorn chickens is largely explained by a limited number of QTLs. Anim. Genet. 34: 264–274. [DOI] [PubMed] [Google Scholar]
- Kim S., Zhang S., Choi K. H., Reister R., Do C., et al. , 2009. An E3 ubiquitin ligase, really interesting new gene (RING) finger 41, is a candidate gene for anxiety-like behavior and β-carboline-induced seizures. Biol. Psychiatry 65: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G., Gumus D., Chen W., Norton N., Georgieva L., et al. , 2008. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 17: 458–465. [DOI] [PubMed] [Google Scholar]
- Klein, P. S., and D. A. Melton, 1996 A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93: 8455–8459. [DOI] [PMC free article] [PubMed]
- Kögel D., Prehn J. H. M., Scheidtmann K. H., 2001. The DAP kinase family of pro-apoptotic proteins: novel players in the apoptotic game. BioEssays 23: 352–358. [DOI] [PubMed] [Google Scholar]
- Kõks S., Luuk H., Nelovkov A., Areda T., Vasar E., 2004. A screen for genes induced in the amygdaloid area during cat odor exposure. Genes Brain Behav. 3: 80–89. [DOI] [PubMed] [Google Scholar]
- Kulhara P., Avasthi A., Chadda R., Chandiramani K., Mattoo S. K., et al. , 1989. Negative and depressive symptoms in schizophrenia. Br. J. Psychiatry 154: 207–211. [DOI] [PubMed] [Google Scholar]
- Lagus M., Gass N., Saharinen J., Saarela J., Porkka-Heiskanen T., et al. , 2010. Gene expression patterns in a rodent model for depression. Eur. J. Neurosci. 31: 1465–1473. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Kruglyak L., 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- Levinson D. F., 2006. The genetics of depression: a review. Biol. Psychiatry 60: 84–92. [DOI] [PubMed] [Google Scholar]
- Litvin O., Causton H. C., Chen B.-J., Pe’er D., 2009. Modularity and interactions in the genetics of gene expression. Proc. Natl. Acad. Sci. USA 106: 6441–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.-S., Harano M., Gawlik M., Yu Z., Chen J., et al. , 2007. GABRB2 association with schizophrenia: commonalities and differences between ethnic groups and clinical subtypes. Biol. Psychiatry 61: 653–660. [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., 2004. The genetic architecture of quantitative traits: lessons from Drosophila. Curr. Opin. Genet. Dev. 14: 253–257. [DOI] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium , 2013. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Dupuis J., Sen S., Broman K. W., 2006. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics 174: 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P., Hever M. P., Niemaszyk L. M., Haghkerdar J. M., Yanco E. G., et al. , 2011. Serine/threonine kinase 17A is a novel p53 target gene and modulator of cisplatin toxicity and reactive oxygen species in testicular cancer cells. J. Biol. Chem. 286: 19381–19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín R. H., Martijena I. D., Arce A., 1997. Effect of diazepam and a β-carboline on open-field and T-maze behaviors in 2-day-old chicks. Pharmacol. Biochem. Behav. 58: 915–921. [DOI] [PubMed] [Google Scholar]
- McGue M., Bouchard T. J., Jr, 1998. Genetic and environmental influences on human behavioral differences. Annu. Rev. Neurosci. 21: 1–24. [DOI] [PubMed] [Google Scholar]
- McNaughton N., Corr P. J., 2004. A two-dimensional neuropsychology of defense: fear/anxiety and defensive distance. Neurosci. Biobehav. Rev. 28: 285–305. [DOI] [PubMed] [Google Scholar]
- Moriarty D. D., 1995. Anxiogenic effects of a β-carboline on tonic immobility and open field behavior in chickens (Gallus gallus). Pharmacol. Biochem. Behav. 51: 795–798. [DOI] [PubMed] [Google Scholar]
- Murray C. J., Lopez A. D., 1996. Evidence-based health policy—lessons from the Global Burden of Disease study. Science 274: 740–743. [DOI] [PubMed] [Google Scholar]
- Norman R. M. G., Malla A. K., 1991. Dysphoric mood and symptomatology in schizophrenia. Psychol. Med. 21: 897–903. [DOI] [PubMed] [Google Scholar]
- Norman R. M. G., Malla A. K., Cortese L., Diaz F., 1998. Aspects of dysphoria and symptoms of schizophrenia. Psychol. Med. 28: 1433–1441. [DOI] [PubMed] [Google Scholar]
- O’Brien W. T., Harper A. D., Jové F., Woodgett J. R., Maretto S., et al. , 2004. Glycogen synthase kinase-3β haploinsufficiency mimics the behavioral and molecular effects of lithium. J. Neurosci. 24: 6791–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian A., Zhang Z. H., 1999. Human chromosomes 11p15 and 4p12 and alcohol dependence: possible association with the GABRB1 gene. Am. J. Med. Genet. 88: 533–538. [DOI] [PubMed] [Google Scholar]
- Peirce, J. L., K. W. Broman, L. Lu, E. J. Chesler, G. Zhou et al., 2008 Genome reshuffling for advanced intercross permutation (GRAIP): simulation and permutation for advanced intercross population analysis. PLoS ONE 3: e1977. [DOI] [PMC free article] [PubMed]
- Postina R., Schroeder A., Dewachter I., Bohl J., Schmitt U., et al. , 2004. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 113: 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price E. O., 1984. Behavioral aspects of animal domestication. Q. Rev. Biol. 59: 1–32. [Google Scholar]
- Pruitt K. D., Tatusova T., Klimke W., Maglott D. R., 2009. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 37: D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prut L., Belzung C., 2003. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur. J. Pharmacol. 463: 3–33. [DOI] [PubMed] [Google Scholar]
- Psychiatric Genetics Consortium , 2013. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group , 2011. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pun F. W., Zhao C., Lo W. S., Ng S. K., Tsang S. Y., et al. , 2011. Imprinting in the schizophrenia candidate gene GABRB2 encoding GABAA receptor [beta]2 subunit. Mol. Psychiatry 16: 557–568. [DOI] [PubMed] [Google Scholar]
- Ramos A., Mormède P., 1997. Stress and emotionality: a multidimensional and genetic approach. Neurosci. Biobehav. Rev. 22: 33–57. [DOI] [PubMed] [Google Scholar]
- Robinson G. E., Grozinger C. M., Whitfield C. W., 2005. Sociogenomics: social life in molecular terms. Nat. Rev. Genet. 6: 257–270. [DOI] [PubMed] [Google Scholar]
- Robinson G. E., Fernald R. D., Clayton D. F., 2008. Genes and social behavior. Science 322: 896–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C.-J., Zody M. C., Eriksson J., Meadows J. R. S., Sherwood E., et al. , 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464: 587–591. [DOI] [PubMed] [Google Scholar]
- Salvatierra N. A., Arce A., 2001. Day-old chicks categorised on latency to peck, exhibit a stable fear pattern until 15 days of age. Appl. Anim. Behav. Sci. 73: 103–116. [DOI] [PubMed] [Google Scholar]
- Samsom J. N., Wong A. H. C., 2015. Schizophrenia and depression co-morbidity: what we have learned from animal models. Front. Psychiatry 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K. W., Strakowski S. M., Keck P. E., Jr, Upadhyaya V. H., West S. A., et al. , 1996. Relationships among negative, positive, and depressive symptoms in schizophrenia and psychotic depression. Br. J. Psychiatry 168: 68–71. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium , 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt U., Hiemke C., Fahrenholz F., Schroeder A., 2006. Over-expression of two different forms of the α-secretase ADAM10 affects learning and memory in mice. Behav. Brain Res. 175: 278–284. [DOI] [PubMed] [Google Scholar]
- Schutz K., Kerje S., Carlborg O., Jacobsson L., Andersson L., et al. , 2002. QTL analysis of a red junglefowl x White Leghorn intercross reveals trade-off in resource allocation between behavior and production traits. Behav. Genet. 32: 423–433. [DOI] [PubMed] [Google Scholar]
- Schütz K. E., Forkman B., Jensen P., 2001. Domestication effects on foraging strategy, social behaviour and different fear responses: a comparison between the red junglefowl (Gallus gallus) and a modern layer strain. Appl. Anim. Behav. Sci. 74: 1–14. [Google Scholar]
- Schutz K. E., Kerje S., Jacobsson L., Forkman B., Carlborg O., et al. , 2004. Major growth QTLs in fowl are related to fearful behavior: possible genetic links between fear responses and production traits in a red junglefowl x white leghorn intercross. Behav. Genet. 34: 121–130. [DOI] [PubMed] [Google Scholar]
- Sih A., Bell A., Johnson J. C., 2004. Behavioral syndromes: an ecological and evolutionary overview. Trends Ecol. Evol. 19: 372–378. [DOI] [PubMed] [Google Scholar]
- Silva A., Paylor R., Wehner J., Tonegawa S., 1992. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science 257: 206–211. [DOI] [PubMed] [Google Scholar]
- Smoller J. W., Block S. R., Young M. M., 2009. Genetics of anxiety disorders: the complex road from DSM to DNA. Depress. Anxiety 26: 965–975. [DOI] [PubMed] [Google Scholar]
- Tomida S., Mamiya T., Sakamaki H., Miura M., Aosaki T., et al. , 2009. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 41: 688–695. [DOI] [PubMed] [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., et al. , 2006. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887. [DOI] [PubMed] [Google Scholar]
- Vos, T., R. M. Barber, B. Bell, A. Bertozzi-Villa, S. Biryukov et al., 2015 Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386: 743–800. [DOI] [PMC free article] [PubMed]
- Wang X., Mozhui K., Li Z., Mulligan M. K., Ingels J. F., et al. , 2012. A promoter polymorphism in the Per3 gene is associated with alcohol and stress response. Transl. Psychiatry 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren A., Gunnarsson U., Andersson L., Jensen P., 2009. Domestication-related genetic effects on social behavior in chickens—effects of genotype at a major growth quantitative trait locus. Poult. Sci. 88: 1162–1166. [DOI] [PubMed] [Google Scholar]
- Wirén A., Wright D., Jensen P., 2013. Domestication‐related variation in social preferences in chickens is affected by genotype on a growth QTL. Genes Brain Behav. 12: 330–337. [DOI] [PubMed] [Google Scholar]
- Wright D., Kerje S., Brändström H., Schütz K., Kindmark A., et al. , 2008. The genetic architecture of a female sexual ornament. Evolution 62: 86–98. [DOI] [PubMed] [Google Scholar]
- Wright D., Rubin C. J., Martinez Barrio A., Schütz K., Kerje S., et al. , 2010. The genetic architecture of domestication in the chicken: effects of pleiotropy and linkage. Mol. Ecol. 19: 5140–5156. [DOI] [PubMed] [Google Scholar]
- Wright D., Rubin C., Schutz K., Kerje S., Kindmark A., et al. , 2012. Onset of sexual maturity in female chickens is genetically linked to loci associated with fecundity and a sexual ornament. Reprod. Domest. Anim. 47: 31–36. [DOI] [PubMed] [Google Scholar]
- Wu C., DeWan A., Hoh J., Wang Z., 2011. A comparison of association methods correcting for population stratification in case–control studies. Ann. Hum. Genet. 75: 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Willis-Owen S. A. G., Fullerton J., Meesaq A., Deacon R. M., et al. , 2004. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat. Genet. 36: 1197–1202. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Microarray data for the chicken hypothalamus tissue are available at E-MTAB-3154 in ArrayExpress. Full genotype and phenotype data are available on figshare with the following doi: 10.6084/m9.figshare.1265060.