Abstract
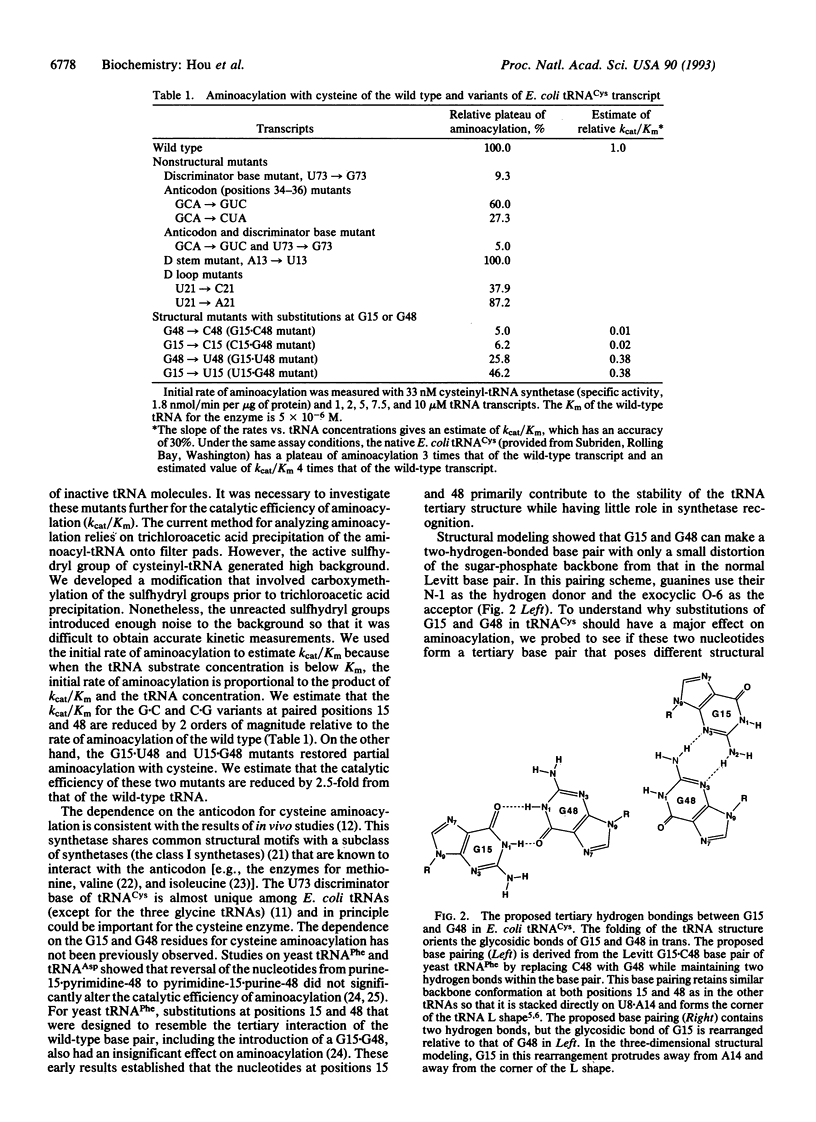
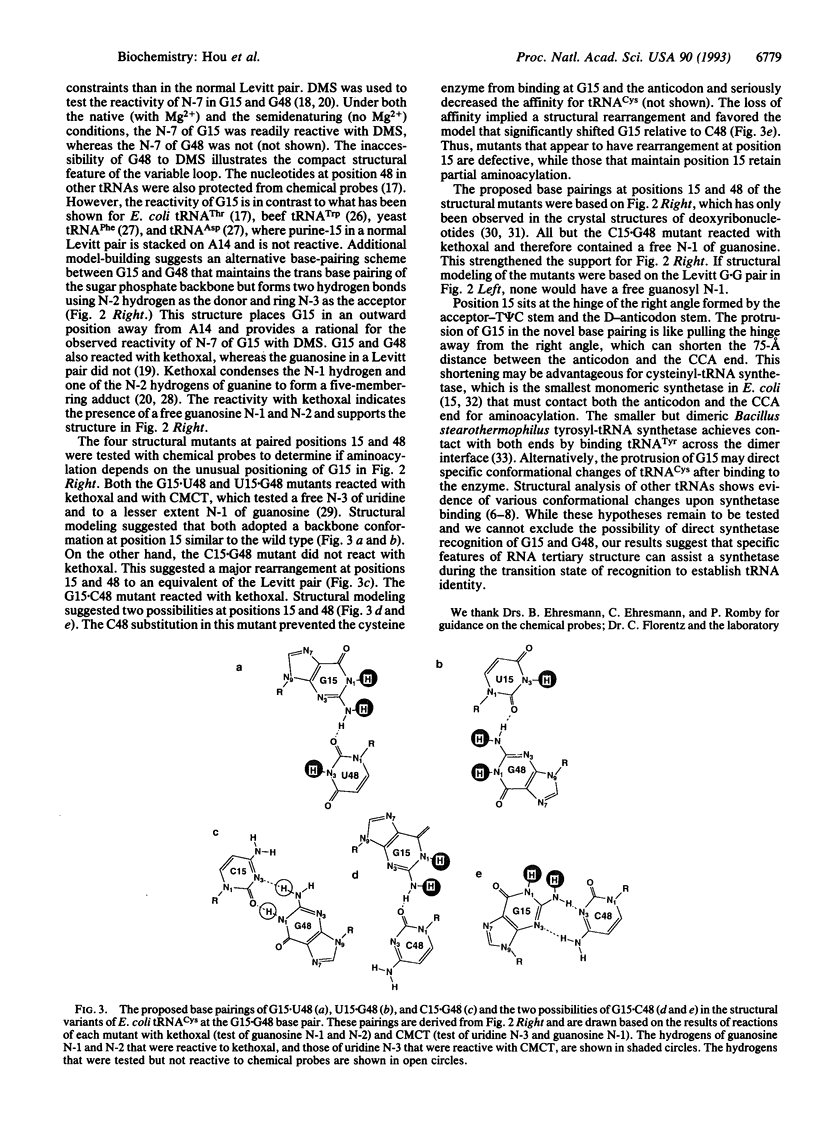
The nucleotides in a tRNA that specifically interact with the cognate aminoacyl-tRNA synthetase have been found largely located in the helical stems, the anticodon, or the discriminator base, where they vary from one tRNA to another. The conserved and semiconserved nucleotides that are responsible for the tRNA tertiary structure have been shown to have little role in synthetase recognition. Here we report that aminoacylation of Escherichia coli tRNA(Cys) depends on the anticodon, the discriminator base, and a tertiary interaction between the semiconserved nucleotides at positions 15 and 48. While all other tRNAs contain a purine at position 15 and a complementary pyrimidine at position 48 that establish the tertiary interaction known as the Levitt pair, E. coli tRNA(Cys) has guanosine -15 and -48. Replacement of guanosine -15 or -48 with cytidine virtually eliminates aminoacylation. Structural analyses with chemical probes suggest that guanosine -15 and -48 interact through hydrogen bonds between the exocyclic N-2 and ring N-3 to stabilize the joining of the two long helical stems of the tRNA. This tertiary interaction is different from the traditional base pairing scheme in the Levitt pair, where hydrogen bonds would form between N-1 and O-6. Our results provide evidence for a role of RNA tertiary structure in synthetase recognition.
Full text
PDF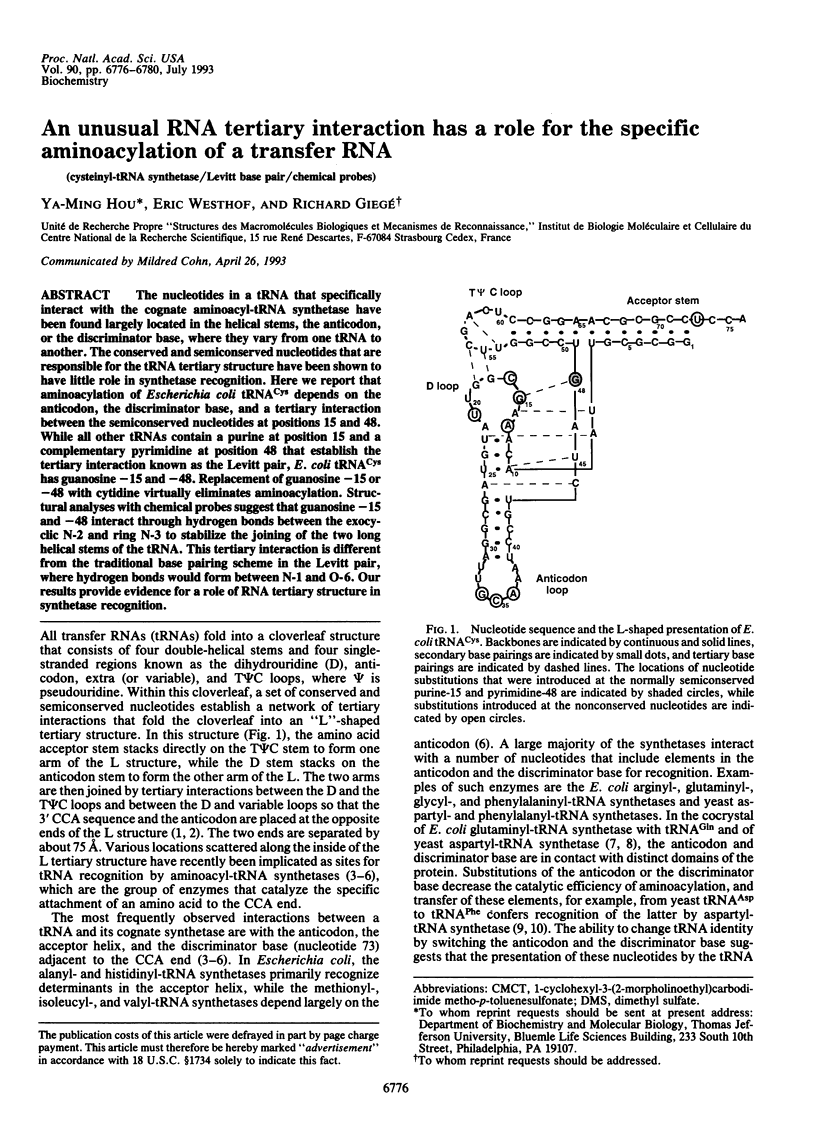


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carter P., Bedouelle H., Winter G. Construction of heterodimer tyrosyl-tRNA synthetase shows tRNATyr interacts with both subunits. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1189–1192. doi: 10.1073/pnas.83.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse W. B., Egert E., Kennard O., Sala G. B., Salisbury S. A., Viswamitra M. A. Self base pairing in a complementary deoxydinucleoside monophosphate duplex: crystal and molecular structure of deoxycytidylyl-(3'-5')-deoxyguanosine. Biochemistry. 1983 Apr 12;22(8):1833–1839. doi: 10.1021/bi00277a014. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Cysteinyl-tRNA synthetase: determination of the last E. coli aminoacyl-tRNA synthetase primary structure. Nucleic Acids Res. 1991 Jan 25;19(2):265–269. doi: 10.1093/nar/19.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garret M., Labouesse B., Litvak S., Romby P., Ebel J. P., Giegé R. Tertiary structure of animal tRNATrp in solution and interaction of tRNATrp with tryptophanyl-tRNA synthetase. Eur J Biochem. 1984 Jan 2;138(1):67–75. doi: 10.1111/j.1432-1033.1984.tb07882.x. [DOI] [PubMed] [Google Scholar]
- Giegé R., Florentz C., Garcia A., Grosjean H., Perret V., Puglisi J., Théobald-Dietrich A., Ebel J. P. Exploring the aminoacylation function of transfer RNA by macromolecular engineering approaches. Involvement of conformational features in the charging process of yeast tRNA(Asp). Biochimie. 1990 Jun-Jul;72(6-7):453–461. doi: 10.1016/0300-9084(90)90069-s. [DOI] [PubMed] [Google Scholar]
- Giegé R., Puglisi J. D., Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Shiba K., Mottes C., Schimmel P. Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):976–980. doi: 10.1073/pnas.88.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Pallanck L., Li S., Schulman L. H. The anticodon and discriminator base are major determinants of cysteine tRNA identity in vivo. J Biol Chem. 1992 Apr 15;267(11):7221–7223. [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Dumas P., Ebel J. P., Giegé R. Comparison of the tertiary structure of yeast tRNA(Asp) and tRNA(Phe) in solution. Chemical modification study of the bases. J Mol Biol. 1987 May 5;195(1):193–204. doi: 10.1016/0022-2836(87)90336-6. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry. 1990 Mar 13;29(10):2523–2532. doi: 10.1021/bi00462a014. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Shapiro R., Hachmann J. The reaction of guanine derivatives with 1,2-dicarbonyl compounds. Biochemistry. 1966 Sep;5(9):2799–2807. doi: 10.1021/bi00873a004. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald A., Springer M., Grunberg-Manago M., Ebel J. P., Giege R. Tertiary structure of Escherichia coli tRNA(3Thr) in solution and interaction of this tRNA with the cognate threonyl-tRNA synthetase. Eur J Biochem. 1988 Aug 15;175(3):511–524. doi: 10.1111/j.1432-1033.1988.tb14223.x. [DOI] [PubMed] [Google Scholar]
- Van Stolk B. J., Noller H. F. Chemical probing of conformation in large RNA molecules. Analysis of 16 S ribosomal RNA using diethylpyrocarbonate. J Mol Biol. 1984 Nov 25;180(1):151–177. doi: 10.1016/0022-2836(84)90435-2. [DOI] [PubMed] [Google Scholar]
- Wakao H., Romby P., Westhof E., Laalami S., Grunberg-Manago M., Ebel J. P., Ehresmann C., Ehresmann B. The solution structure of the Escherichia coli initiator tRNA and its interactions with initiation factor 2 and the ribosomal 30 S subunit. J Biol Chem. 1989 Dec 5;264(34):20363–20371. [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]