Abstract
Early detection and reliable diagnostics are keys to effectively design cancer therapies with better prognoses. Simultaneous detection of panels of biomarker proteins holds great promise as a general tool for reliable cancer diagnostics. A major challenge in designing such a panel is to decide upon a coherent group of biomarkers which have higher specificity for a given type of cancer. The second big challenge is to develop test devices to measure these biomarkers quantitatively with high sensitivity and specificity, such that there are no interferences from the complex serum or tissue matrices. Lastly, integrating all these tests into a technology that doesn’t require exclusive training to operate, and can be used at point-of-care (POC) is another potential bottleneck in futuristic cancer diagnostics. In this article, we review electrochemistry-based tools and technologies developed and/or used in our laboratories to construct low-cost microfluidic protein arrays for highly sensitive detection of the panel of cancer-specific biomarkers with high specificity and at the same time have the potential to be translated into a POC.
1. Introduction
The U.S. National Institutes of Health (NIH) defines biomarkers as “molecules that can be objectively measured and evaluated as indicators of normal or disease processes and pharmacologic responses to therapeutics”.1 Levels of these molecules are representative of changes occurring in the basic cell regulatory functions and cellular physiology, such as cell division or contact inhibition, and may vary from their normal levels in case of diseases and disorders.2 Cancer is one of the best examples of impaired cell functioning where cells lose their basic regulation and become dysplastic and neoplastic.3 This condition has no immediate effects on the body and can stay undetected until the late stages of the disease. Thus, cancer is sometimes referred to as the ‘silent killer’. Therefore, early detection of neoplasia is the key to effectively treat cancer.
Currently, most cancers are diagnosed by quantification of the biomarkers, by the analysis of cellular packaging and morphology in tissue biopsies, and by imaging such as mammograms and colonoscopy.4,5 As biopsies are highly invasive and may miss tumor tissue, while imaging is restricted to detect only tumors, biomarker quantification is a preferred future clinical diagnostics approach. Given the current scenario, finding accurate and highly sensitive biomarkers is of utmost importance that can enable us to define cancers at the earliest stages.4 Currently, there are only 24 biomarkers approved by the US Food and Drug Administration (FDA) associated with different types of cancers (Table 1) that include proteins, genetic hotspots, and most recently the glycans.6 The restricted number of approved biomarker is attributed to the high heterogeneity within cancer cells, even of the same origin.7 In addition, for several of these biomarkers the individual prognostic and diagnostic value is critically low which is the biggest challenge in developing highly efficient cancer detection system, e.g. prediction success is ~70% for PSA, which is one of the better single biomarkers.8 Therefore, validated protein biomarkers associated with cancer development and progression must have excellent clinical specificity, which is the ability of the assay to rule out a condition when it is absent, and clinical sensitivity, which is the ability of the assay to detect the condition when it is present. High clinical specificity and sensitivity are important to avoid false positives and false negatives, which is crucial to avoid misdiagnosis, and needs to be high, preferably >90%.9,10 During the validation step low specificity of the selected biomarkers impacts their advancement to the next stage.
Table 1.
| Biomarker | Class | Source | Cancer | Clinical use |
|---|---|---|---|---|
| Prostate | ||||
| PSA (total) | P | Serum | Prostate | S, M |
| PSA (complex) | P | Serum | Prostate | S, M |
| PSA (free %) | P | Serum | Prostate | Benign hyperplasia vs cancer diagnosis |
| Breast | ||||
| HER2 | P | Serum | Breast | M |
| CA15-3 | GP | Serum | Breast | M |
| CA27–29 | GP | Serum | Breast | M |
| OPR | P | Tumor | Breast | Selection of hormonal therapy |
| HER2/NEU | P | Tumor | Breast | Prognosis and selection of therapy |
| HER2/NEU | DNA | Tumor | Breast | --ditto-- |
| Bladder | ||||
| FDP | P | Urine | Bladder | M |
| BTA | P | Urine | Bladder | M |
| HMW CEA and mucin | P | Urine | Bladder | M |
| NMP | P | Urine | Bladder | S, M |
| Chromosome 3, 7, 9p21, 17 | DNA | Urine | Bladder | S, M |
| Testicular | ||||
| α-fetoprotein | GP | Serum | Testicular | St |
| HCG-β | GP | Serum | Testicular | St |
| Colon | ||||
| CEA | P | Serum | Colon | M |
| EGFR | P | Colon | Colon | Selection of therapy |
| Others | ||||
| CA19-9 | C | Serum | Pancreatic | M |
| CA125 | GP | Serum | Ovarian | M |
| Thyroglobulin | P | Serum | Thyroid | M |
| KIT | P | Tumor | GIST | Diagnosis and selection of therapy |
HMW: High Molecular Weight; P: Protein; GP: Glycoprotein; C: Carbohydrate; S: Screening; M: Monitoring; St: Staging
BTA- Bladder tumor-associated antigen; CA- Cancer antigen; CEA- Carcinoemryonic antigen; EGFR- Epidermal growth factor receptor; FDP- Fibrin degradation protein; HCG- Human chorionic gonadotropin; HER- Human epidermal growth factor receptor; NMP- Nuclear matrix protein; OPR- Oestrogen and progesterone receptor; PSA-Prostate-specific antigen
Clinical reference limits: HE4 <3.5ug/mL; CEA non-smoker <3ng/mL, smoker <5ng/mL; AFP <6ng/mL; PSA upper limit <2ng/mL (<40years); HER2 <15 ng/mL
Increasing the predictive value of a single biomarker is a bottleneck for statistical and biochemical reasons particularly when biomarkers lack specificity to a particular disease. For example, PSA levels can elevate in several benign prostate diseases as well as in prostate cancer,11 introducing diagnostic ambiguity. Therefore, the key to the successful detection of cancers at early stages lies in the parallel measurements of groups of biomarkers.5 Measurement of the levels of four to ten biomarkers is likely to provide more statistically relevant information of prognostics and higher diagnostics value. However, inclusion of a specific set of biomarkers in the panel will be crucial.12,13 For example, Levels of PSA in normal subjects range between 0.5 – 2 ng/mL and 4 – 10 ng/mL for cancer patients while for IL-6 normal levels are <7 pg/mL and for cancer patients up to 5 ng/mL, respectively. Given this situation, we need to develop approaches such that these biomarkers can be placed together on a single detection assay.14 The multiplexed assays must detect an analyte of the cancer panel whose concentration significantly differs from the others without affecting the detection accuracy of the other, particularly cut-off values for distinguishing cancer from healthy subjects. It is often useful to devise a global parameter reflecting the combined values of all the proteins in the panel, and define a diagnostic cut-off value based on this measured value.15
Other critical factors that will drive the clinical applications of biomarker panels include low cost, and easy to perform automated assays. Reliable and accurate detection of cancer biomarkers remains limiting factor, and can be attributed to the insufficient sensitivity of many of the assays used. In addition, several of the current commercially available immunoassay methods lack the needed dynamic range to detect proteins of interest that are often expressed, as discussed above, at levels in the low pg/mL range in serum, and in some cases even below the detection limit.16 Therefore, immunoassay design becomes crucial in such cases for obtaining a balance between detection specificity and sensitivity. The use of new recombinant antibodies for different cancer biomarkers may allow for highly specific and sensitive detection but still needs to be tested in multiplexed detection formats.
When packaging such complex multiplexed cancer immunoassays for point-of-care (POC) applications, on-chip reagent storage, automation, low cost, accuracy, and achieving desired clinical selectivity and sensitivity are the other important challenges, and must be addressed according to the Clinical Laboratory Improvement Amendments of 1988 (CLIA).17 Commercially developed tests allow for minimizing these challenges by following strict norms and regulations set forth by guiding authorities such as FDA.
There are numerous commercial diagnostic tests for cancer but most are dedicated to genetic testing while the rest are microtitre plate-based immunoassays. These commercial assays thus can only be performed in laboratory settings. Recently, microfluidics-based devices have significantly impacted in-vitro diagnostics by enabling tests that can be used at the POC. FDA has approved 23 companion diagnostics (CDx) tests for cancer screening and staging but only three of these are for detecting protein biomarkers. Ventana Medical Systems, Inc. received approval for anaplastic lymphoma kinase protein detection while Leica Biosystems and Dako Denmark A/S have got their HER2 protein detection tools approved. All these three CDx tools are mainly for immunohistochemical applications. Pandora CDx has developed a CD-based centrifugal diagnostics system for POC detection of breast cancer. This test costs as low as $2 per person for complete screening using molecular detection of protein and nucleic acid biomarkers. OPKO Health, Inc. has recently launched microfluidics-based POC with fluorescence detection for PSA panel that includes total and free PSA with testosterone. HalioDx has launched Immunoscore™ companion diagnostics that pools in on-chip immunohistochemistry and clinical scoring. There are several other tools that have been under translational research and have tremendous potential to be the components of future diagnostics platforms. We discuss these modern tools for immunoassay development in the next section of this review.
2. Tools for building immunoassays
2.1 Analytical consideration and tool designing
Detection limit and dynamic range of the devised test along with their biomarker specificity and sensitivity are critical analytical considerations that must be addressed while designing diagnostic tools.18 Several attempts have been made to develop highly sensitive microplate-based test that can be performed quicker with wider dynamic ranges and high test recoveries than conventional tests.19,20 However, these tools are still in nascent stages of development and the time caveat associated to the bulk-matrix testing will still be there.21 In addition, volumes as large as 100µL are required in standard operational procedure.
Conversely, numerous attempts are being made to address the bulk-matrix challenge by employing microfluidic tools. Examples of such tools are fluidic-integrated microarrays22,23 and microplate-based tools24, bead-based fluidic systems and fluidic nanoarrays25. These tools are low volume, moderate sensitivity that mainly represents biomarker levels in several clinical conditions including cancer. However, the most significant drawback with these tools is the involvement of lithography process that makes them laborious to develop and thus to mass produce.
We have taken cognizance of these restrictions and have addressed each of them specifically in our tools. Our tools are based on electrochemical detection thus allow lower fg/mL level sensitivity, broader ng-fg/mL dynamic ranges, which in our case suits to the bioassay requirements, attributed to better signal to noise ratio26 obtained by such type of systems. We have integrated fluidic components thus addressing the diffusion limitedness and reducing the sample volumes down to 2–5µL. Assays on these platforms can be performed quicker (~10–15 min) that make these as ideal candidates for POC applications.
2.2 Printed electrodes
Electrochemical tools address most of the CLIA recommendations and can be packed into small, cheap devices that could be designed for POC. The major challenges include automation and meeting the immunoassay requirements for sensitive and specific cancer diagnostics. Immunoassay performance of amperometric immunoarrays is highly dependent on the electrode properties as the signal response is generated near to sensor-electrode surface. Strategies usually involve attachment of antibodies to the sensors, which often employ nanostructure-coated surfaces.27 Electrode type, material, dimensions, and surface properties are crucial components that must be given detailed consideration prior to designing the detection system.26 These electrode properties are mainly governed by the method of electrode fabrication; choice of material used and design-related restrictions are imposed by the chosen fabrication method. For example, chemical vapor deposition (CVD),28,29 can deposit most of the materials on surface in desired patterns and shapes given an appropriate mask is employed; other popular and equally effective fabrication methods are photolithography,30,31 screen-printing,32,33 stencil printing,34 and inkjet printing35,36.
Some of these techniques such as CDV and photolithography are more costly and time consuming than others, but may produce better quality electrodes.27 Screen printing technology is widely used for the production of low-cost thin film electronics, especially for fabricating disposable electrodes used to develop electrochemical immunoassays.32,33 Shi et al demonstrated ultrasensitive detection of IL-6 and matrix metallopeptidase-9 (MMP-9) with the fabricated electrically heated screen-printed carbon electrodes (SPEs).32 Our team has developed highly sensitive amperometric immunoassays for detecting a series of oral and prostate cancer-specific protein biomarkers by employing commercially available screen printed Kanichi carbon arrays (Figure 1A).15,37–39 These SPEs are cheap due to the manufacturing procedures employed, e.g each array from Kanichi costs ~$5. In addition, these SPEs allow further customization of the electrode surface with 5 nm Au-glutathione (Au-GSH) nanoparticles to increase the surface area and to provide attachment of significantly higher number of antibodies. Shi and colleagues further modified their SPEs with graphene nanoribbons (GNRs) to immobilize capture antibodies and for amplifying electrochemical signals.32 They have achieved detection limits in fg/mL range (100 fg/mL for IL-6 and 5 fg/mL for MMP-6) by developing assays on polymer-nanocomposite deposited electrodes. Our group obtained detection limits in the 5–40 fg/mL range using the Kanichi arrays modified with 5 nm glutathione-decorated gold nanoparticles (GSH-AuNPs)40 on a 0.3–0.5 nm under-layer of cationic poly(diallydimethylammonium) ions (PDDA).38
Figure 1.
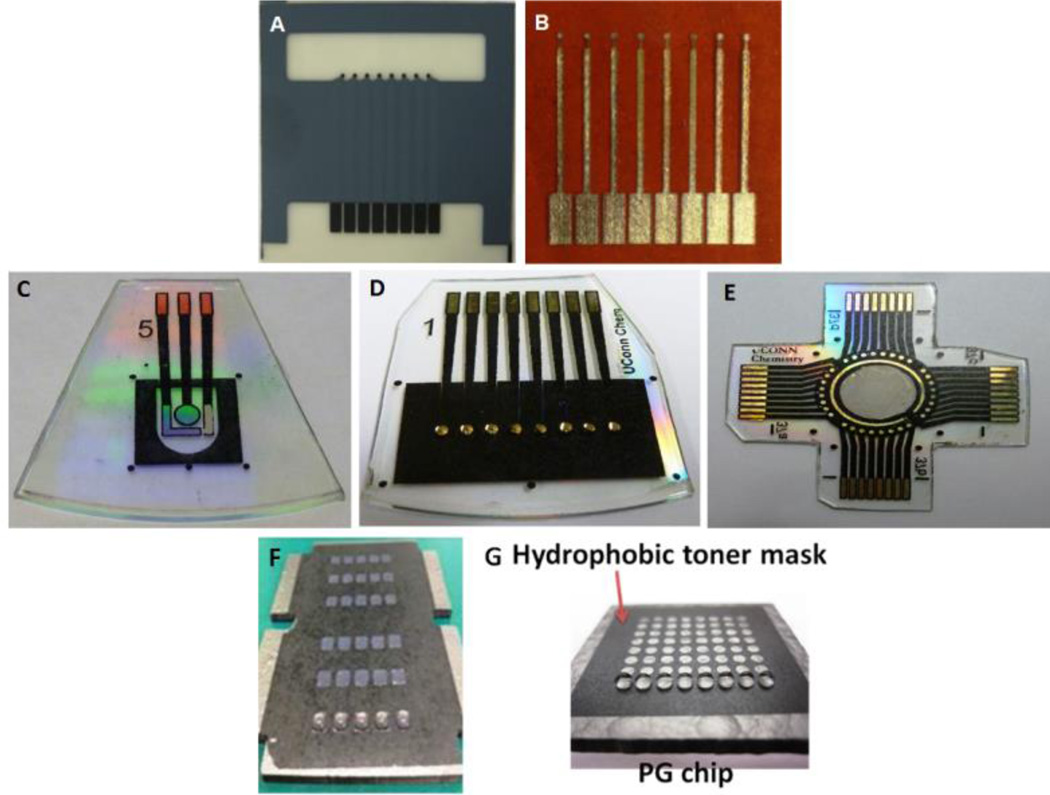
Electrode arrays for electrochemical protein determinations: A) screen printed carbon electrode arrays (Kanichi Research Ltd); B) in-house inkjet-printed gold nanoparticle arrays. C-E) three types of gold arrays fabricated using the print/heat/peel methods: (C) single sensor chip with on-chip reference and counter electrodes; (B) an 8-sensor array; (C) a 32-sensor array with on chip reference and counter electrodes. (F) a 30-microwell and (C) a 64-microwell patterned pyrolytic graphite (PG) chip for electrochemiluminescence-based detection with 2 µL and 1.5 µL droplets of aqueous solution as shown, respectively.
An alternative is to fabricate AuNP arrays by inkjet printing (Figure 1B) on flexible, heat resistant polyimide Kapton plastic sheets to reduce the overall cost of the sensor to$0.2.35,41 Au-NP ink was prepared with 4 nm dodecane thiol-protected AuNPs in toluene and was printed in multiple copies with a Dimatrix ink-jet printer. Printed electrode arrays were annealed to create a continuous conducting layer followed by insulating the leads with poly(amic) acid, a Kapton precursor, and heating it to polymerize. Electrodes prepared with this approach have reproducible surface areas with RSD <3%. These electrode arrays were used to develop immunoassays for detection of a panel of cancer biomarkers.39 This non-contact inkjet printing to produce AuNPs arrays demonstrated an elegant, cheap and simple technique for production of electrochemical sensor array.35,41
2.3 Chemically etched arrays
We also fabricated gold arrays by using a simple print-heat-peel method, such that electrodes are surrounded by tiny hydrophobic nanowells. This approach transfers computer generated patterns onto gold CD-Rs followed by selective chemical etching.42,43 Instead of inkjet printing gold nanoparticles, we printed laser jet patterns of desired shape and size under computer control onto glossy paper, and transferred them on to a gold CD-R via heat transfer. Later, the gold around the transferred patterns was etched out to generate a pattern of electrodes. These electrodes (Fig 1 C–E) were highly reproducible with ~2% RSD and have a working area of 0.4 mm2, as measured electrochemically.
This is a cheap and rapid method for fabricating nL-volume microwells for amperometry and electrochemiluminescence (ECL)-based cancer detection. This fabrication approach can create a desired number of microwells of preferred dimensions. A layer of printed patterns of wells with diameter of 0.8 mm and a thickness of 6–14 µm on hydrophobic sheet was transferred over the sensing surface via heat-transfer. These hydrophobic microwells around the sensor elements have nL-volumes, but can hold up to 1 µL drops of aqueous reagents (Fig. 1F, G) due to the high contact angle exhibited by the well edge. These microwell gold arrays were tested by detecting interleukin (IL)-6 in diluted serum with a detection limit of 10 fg/mL. We are currently extending this fabrication technique to create 32-sensors arrays (Figure 1E).
The microwell-patterning technique is quite general, and we have adapted it to other substrates including PG chips for automated multiplexed detection of four biomarker proteins.44 On a 30-well array (Fig 1F), single-wall carbon nanotube forests were grown in patterned microwell bottoms to facilitate the development of a highly sensitive surface for ECL detection. A sensor chamber incorporating this chip was used to perform fully automated, 30 minute electrochemical immunoassays to detect PSA, prostate-specific membrane antigen (PSMA), IL-6, and platelet factor 4 (PF4) in undiluted serum with detection limits of 10–100 fg/mL.44 The automated system for reagent addition was controlled by a microprocessor as described in a later section. A 64-microwell PG detection chip (Fig. 1G) was adapted to detection of DNA damage caused by reactive metabolites.45
These low cost, simple sensor fabrication approaches hold tremendous potential to help bring protein-based cancer testing to POC. However, the challenges associated with the protein diagnostics, such as non-specificity and poor signal-to-noise ratio (SNR), needs to be addressed to achieve highly sensitive protein detection. In the next section we discuss potential strategies to overcome these restrictions.
2.4 Nano/microparticles for signal enhancement
Significantly increasing the number of biorecognition elements on the sensors is crucial for sensitively detecting protein-based cancer biomarkers. In order to achieve high Signal-to-noise ratios (SNR), multiple labels can be used for enhancing the assay-specific signal by many fold. Both these aims can be accomplished by conjugating the target antibody/label to nano/microparticles. Each of these particles can accommodate a huge number of biomolecules, which is significantly higher than the area of a corresponding flat surface employed in typical immunoassay systems.5,27,46 In such applications, magnetic particles offer many advantages over non-magnetic particles in terms of the ease of manipulation for labelling, antibody attachment, and purification of bead bioconjugates with inexpensive magnets.27,47 Many magnetic particles are commercially available with a range of sizes and surface chemistry functionalities.48
We have extensively explored 1 µm magnetic beads with surface functionalities, such as tosyl, carboxylate, and biotin, for ultrasensitive detection of cancer biomarkers in microfluidic immunoarrays. Practically, these magnetic particles can be loaded with many thousands of antibodies and horseradish peroxidase (HRP) labels. By using 1 µm magnetic beads having up to 500,000 HRPs and 100,000 antibodies to capture analyte proteins, we have achieved ultralow detection limits down to 5 fg/mL (attomolar) levels for electrochemical detection of cancer biomarker proteins in serum.49
We have also employed Ru-(bpy)32+, (RuBPY)-doped silica nanoparticles (Ru-SiNP) as labels for ECL-based immunoarrays. These ~100 nm diameter nanoparticles were prepared in water-in-oil microemulsions in the presence of water soluble RuBPY to give an estimated half-million RuBPY ions per particle.50 The Ru-SiNPs were decorated with analyte-specific detection antibodies, and were used to detect PSA, PSMA, PF4, and IL6 by using ECL-based detection method.44 We have obtained 10–100 fg/mL detection limits for all the four biomarkers with 30 min assay time. To further automate Ru-SiNP-based detection method, we employed PDMS-based microfluidic system that has three 90 µL channels conformally placed over a PG block encased in a PMMA holder. Each channel was incorporated with an Ag/AgCl reference and a Pt counter electrode running along the length of the channel while PG served as working electrode. We have achieved zeptomolar detection limits for IL-6 and PSA.51,52
2.5 Microfluidic prototyping
2.5.1 Precision blade cutting
In addition to the development of platforms, overall time, sample volume, and automation are other important factors to be addressed for POC applications. In accordance with the CLIA regulations, integration of microfluidics with the analytical system is crucial for reducing the overall immunoassay times and sample volumes, and for introducing pump-less reagent delivery. Designing microfluidic by lithography is a mature area53, but lithography-based prototyping can be costly and time consuming due to the requirement of mask designing and master-mold development. New tools for enabling faster microfluidic prototyping are sought that are also easy to use even for new entrants. Simple machined metal molds for patterning soft polymers, such as polydimethylsiloxane (PDMS), can also be employed for developing modular microfluidic components.54 However, unlike soft lithography, achieving sub-hundred micron resolution is a challenge. In addition, machining a series of prototypic designs during device optimization is laborious and time-consuming. We recently tested precision blade cutting of cheap 800 µM thick silicone gaskets in desired patterns for the microfluidic protoyping of ECL-based array (SI Fig 1).54 This technique is very simple, even for the inexperienced, and tools such as desktop craft cutters (SI Fig 1a), are cheap with operation costs ranging in only few cents per prototype. However, cut resolution is a trade-off in this method where the best resolutions do not exceed 800 µm. However, this is sufficient resolution for many practical bioanalytical applications. There are other more advanced precision cutting methods, such as waterjet cutting, wire electrical discharge machining (EDM), laser-mediated, plasma-based, and milling but they are costly and require operational training. Milling and wire EDM allows a precision cut with 2.5 µm resolution. However, the major restriction is that only fluidics can be created while other device components need to be assembled separately. These restrictions can be addressed with 3D printing methods that in principle can print complete devices including fluidic components.55
2.5.2 3D Printing
3D printing, or additive manufacturing, provide revolutionary new opportunities for bioanalysis and biotechnology.56 High resolution desktop 3D printers are cheap (currently €2000–4500) and enable one-step rapid prototyping of complete 3D structures, whereas popular conventional techniques like lithography require numerous steps to design, visualize and optimize a 3D object.57 Several researchers have employed either 3D printed parts as molds for microfluidic designs or have printed complete microfluidic devices. We recently employed an inexpensive desktop fused filament fabrication (FFF) 3D printer Makerbot Replicator 2X to develop a nanoparticle (NP) synthesizer as well as a simple microfluidic sensor to detect hydrogen peroxide.58 These devices were designed to have smooth bottom surfaces with semi-transparent channels to visualize colored solutions. We made NP-synthesis devices with poly(ethyleneterephthalate) (PET) having Y-shaped mixing channel leading to a serpentine channel to facilitate mixing to make Prussian blue nanoparticles (PBNPs). These PBNP’s were then deposited onto gold sensor electrodes for amperometric detection of hydrogen peroxide (H2O2) inside another 3D printed microfluidic device having threaded electrode fittings equipped with working, reference, counter electrodes. We achieved detection limits of 100 nM with good linear responses for 0.1 to 20 µM peroxide. These 3D printed fluidic devices were reproducible with channel widths as small as ~250 µm and were reusable (SI Fig 2). This entire 3D printed device has a manufacturing cost of ~ €0.5.
Filament-based 3D prints were not transparent and can be used only for microfluidic prototyping of non-optical applications. Recently, we started using commercial desktop SLA printer Form1+ 3D printer (Formlabs) and methacrylate-based clear resin. These prints were optically clear and can be used for ECL light detection. With these tools at our disposal, our aim is to develop automated, multiplexed, and high-sensitivity immunoassays for simultaneous detection of virtually any small panel of cancer biomarker proteins.
3. Automation for Multiplexed Biomarker Detection
3.1 Automated Electrochemical Detection System
Low cost automation is a key to realize the routine clinical or POC biomarker panel-based cancer diagnostics in the future. This will require integration and co-ordination of micropumps, mixers, and valves with the reagent storage and delivery, and sensor arrays. Use of the integrated microfluidic systems facilitates automation and decreases the overall assay time.
We have integrated these features into a partly automated modular microfluidic device featuring a capture and a detection chamber made from PDMS encased in polymethylmethacrylate plastic (Fig 2). The sensor array was arranged in the detection chamber such that the sensing surface lies within the microfluidic channel hosting platinum (Pt) wire as counter and silver/silver chloride (Ag/AgCl) wire as reference electrode. (Fig 2C). Initially, we tested this semi-automatic detection device with prepared antibody and enzyme-label reagents out of the device ‘off-line capture’.37 Later, we included capture and detection steps in the microfluidic chambers.38 Off-line capture was used to measure protein biomarkers including IL-6, IL-8, vesicular endothelial growth factor (VEGF), and VEGF-C for clinical diagnosis of oral cancer14 at ultra-low detection limits (5–50 fg mL−1) in 50 min duration. Results of these immunoassays were strongly correlated with standard ELISA measurements of the same patient samples.38 We were also able to trade sensitivity for shorter assay times to decreased the total assay duration ~8 min.41 Using on-line capture system, we performed all the immunoassay steps in the microfluidic chip (Fig 2B) for a multiplexed detection of IL6, IL-1β, tumor necrosis factor (TNF) α, and C-reactive protein (CRP) associated with oral cancer mucositis, a serious therapy side effect.
Figure 2.
(A) Illustrated representations of the modular microfluidic system used for amperometric immunoassays. The device has a capture chamber (B) for capturing analytes on magnetic beads and washing upstream of a detection chamber (C) for housing the Ab1-decorated sensor array. ‘D’ depicts the current v/s time response curve for various concentrations of IL-6, while ‘E’ shows the immunoassay parameters as concentration of IL-6-specific current-response (Reprinted from ref. 30, Copyright 2014, with permission from Elsevier.
We detected these proteins as low as 10–40 fg/mL in 30 min assays with a good correlation to ELISA detection.39 For a four protein multiplexed assay, total cost of the assay reagents and sample for our microfluidic system is~$6 while to set-up the whole instrumentation the total cost ranges between €8000 and €25,000. The total cost is much lower than the cheapest automated multiplexed immunoassay instruments, providing a low-cost alternative that could be set up in virtually any biomedical laboratory.
3.2 Automated ECL system
3.2.1 Electronic Reagent Delivery
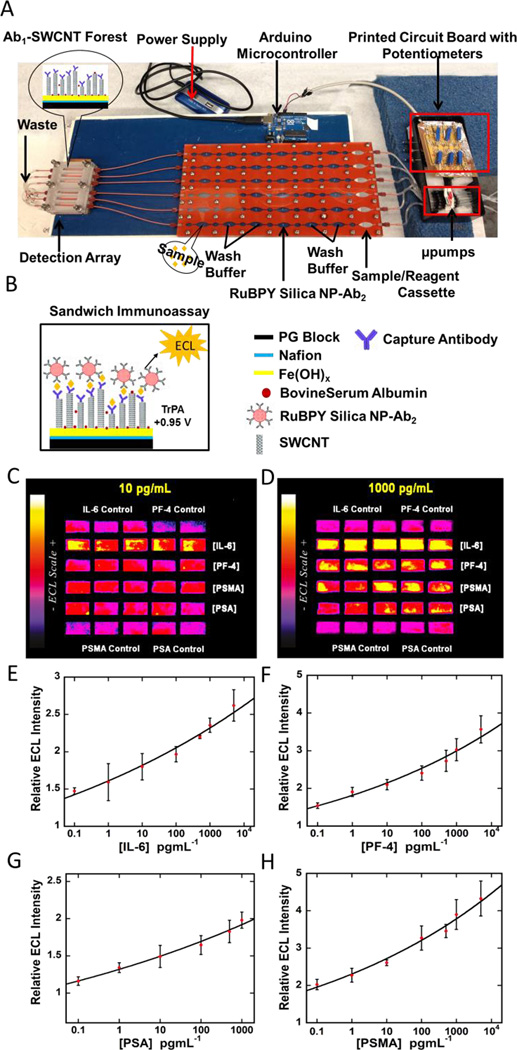
We addressed automating sample and reagent delivery for Ru-SiNP-based ECL detection. We integrated precision cut silicon microfluidic channels (SI Fig 1) sandwiched between PMMA cover panels with a programmed on-board Arduino microcontroller for sample and reagent delivery (Fig 3).44 This microcontroller-programmed micropump system has six pumps mounted on a printed circuit board along with six potentiometers to adjust the flow rates for reagent delivery. The controller maintains an initial flow rate of 155 µL/min and then enables pump switching to either deliver sample and reagents or stop flow for incubations. We used this integrated system to simultaneously detect four prostate cancer biomarkers in 30 min with a good dynamic range and low fg/mL detection limits. We tested this system with patient samples and found good correlations with the ELISA assays for each biomarker separately. The systems is very easy to setup and is reusable, and can be adapted to more proteins as required. In addition, the overall cost is less than $500 not including the CCD camera for ECL measurements.
Figure 3.
(A) Microprocessor-controlled automated microfluidic immunoarray featuring 30 microwell SWCNT modified detection array fed with sample/immunoreagents from a reagent cassette (red) using inexpensive micropumps. The entire assay takes 36 min. Immunoassay steps (B) are automated and controlled by a microprocessor.. C-D shows ECL immunoassay results for four cancer biomarkers, viz. IL-6, PF4, PSMA, PSA, at specific analyte concentrations. E-H are immunoassay calibration curves for IL-6, PF4, PSA, and PSMA. Adapted from ref 36, Copyright 2015. American Chemical Society.
While such integrated systems hold tremendous commercial viability, difficulties are associated with building and housing these components into a single automated machine for POC. We are currently exploring 3D printing to develop a more fully integrated assay device for automated sample/reagent delivery and detection.
3.2.2 Gravity-driven Reagent Delivery
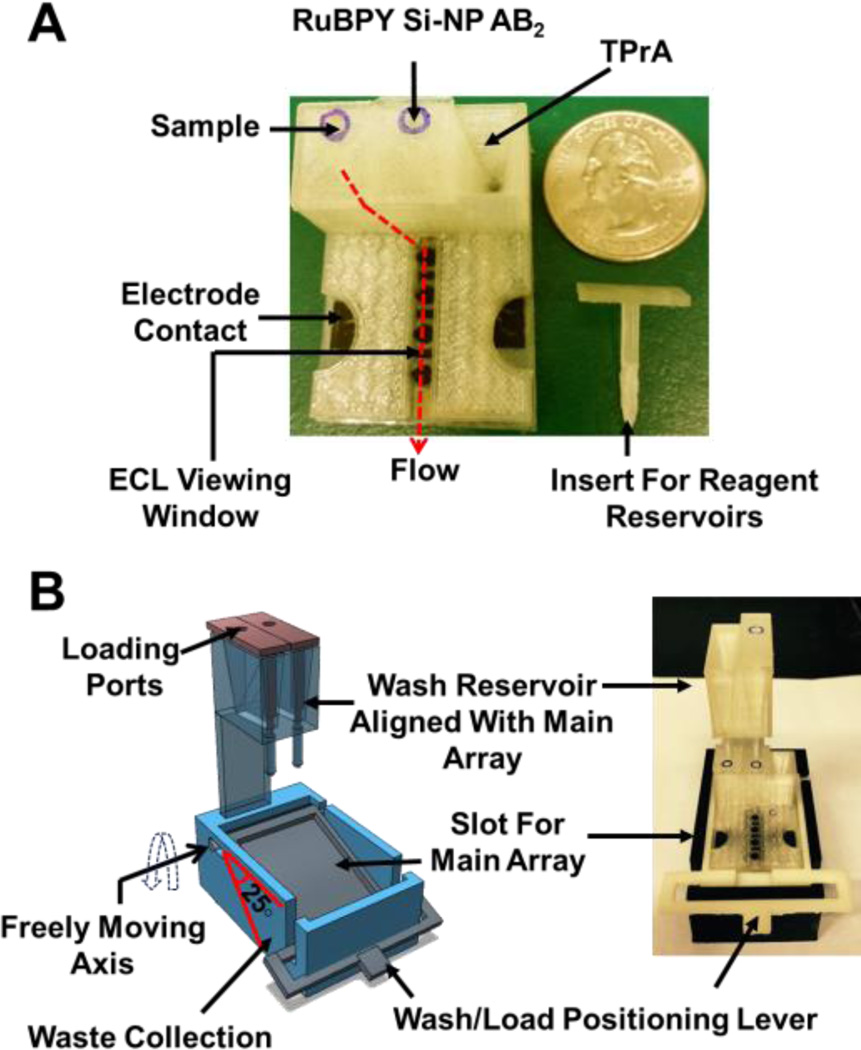
Our first venture into 3D-printed devices was a gravity-operated reservoir module developed using a Fused Deposition Modeling (FDM) desktop 3D printer MakerBot Replicator 2X and polylactic acid polymer (PLA).59 Sample and reagent reservoirs, with a capacity of ~150 µL, are integrated with the main array (Fig 4). These reservoirs (sample, wash, and Ru-SiNP reagents) facilitate the complete immunoassay using gravity flow. We also replaced the potentiostat with a small inexpensive supercapacitor that helped to package the detection system in a housing measuring a few cm3.
Figure 4.
3D-printed immunoarray (A) Main unit showing sample and 2 reagent reservoirs equipped with inserts along with flow path for reagents to reach microfluidic channel. (B) Wash reservoir module (B Left) 3D model showing freely moving lever to change between wash and load position along with wash reservoirs aligned with main unit, (B Right) assembled immunoarray with main unit and wash module. Reprinted from ref. 59, Copyright 2015, with permission from Elsevier.
This system was tested by detecting PSA, PSMA, and PF4 in serum with detection limits of 300, 535, 420 fg/mL, respectively. The overall fabrication cost for this immunoarray was approximately less than €30, included €10 for supercapacitor and €12 for solar panel, ~€1 for 3D printed parts along with 4-sensor array, and ~€0.5 for immunoassay reagents. For detection, the device used CCD camera (€20,000) but for a functional POC unit, it might be possible to use a much less expensive camera.
4. Cancer detection at POC – Research update
Efforts have been made by other researchers to achieve commercial grade POC detection systems. While there has been good progress, lack of sensitivity is still an issue in most of published reports. The need of sensitivity is to ensure effective correlation studies in panels of several biomarkers where slight change in one biomarker can lead to a significant change in the receiver-operator functions (ROC)-based analyses. Although, clinical reference values, as described in Mayo clinic’s biomarker listing, are in ng/mL range for healthy subjects but these can fluctuate in either direction depending on the nature and state of cancer, treatment, and recurrence. Tools that can only detect biomarkers in the over-expression scenario will become obsolete. Therefore, we devised tools that can be used to detect broader dynamic ranges of biomarkers with higher sensitivity. A comparative research update has been summarized in Table 2.
Table 2.
Comparative summary of tools for cancer diagnosis
| Tool | Principle | Analytical Parameters | |||||
|---|---|---|---|---|---|---|---|
| Number of Samples/Matrix |
Biomarkers tested |
Sample (µL) |
LOD/Clinic al Value (pg/mL) |
Assay time (min) |
Cost/test (USD) |
||
| µChip60 | µFluidics; Colorimetric IA | 1/Urine | HE4 for ovarian cancer | 100 | 19000 | 300 | Not mentioned |
| LOC61 | µFluidics; Fluorescence IA | 1/Serum | CEA, PSA, AFP | 1000 | <5000 | ~30 | Not mentioned |
| LOC62 | µFluidic plug-n-play PDMS system; Colorimetric IA |
1/Whole blood | CEA, CYFRA21-1 | <3 | <60 | ~60 | Not mentioned |
| LOC63 | µFluidics; IHC | 1/Tissues | ER, HER2, PR, Ki-67 | Not mentioned |
|||
| LOC23 | µFluidics; AlphaLISA | Sequential 8 samples /Cells |
Cytokine secretion | ~10 | ~10 | 45/~6 per sample |
Not mentioned |
| LOC44 | µFluidics; ECL | 1/Serum | PSA, PSMA, PF4 | <10 | <0.5 | 35 | ~0.5 |
| µChip59 | µFluidics; ECL | 1/Serum | IL-6, PSA, PSMA, Pf-4 | <10 | <0.1 | 36 | ~ 3.0 |
| µChip38 | µFluidics; Amperometry |
1/ Serum | IL-6, IL-8, TNF-α IL-1β,CRP |
5 | <0.007, <0.01 |
30 | ~ 5.0 |
| µChip37 | µFluidics; Amperometry |
1/Serum | IL-6, IL-8 |
5 | <0.08 <5 |
45 8 |
~ 5.0 |
| µChip43 | µFluidics; DPV on Paper | 1/Serum | PSA | 10 | <5 | 15 | ~ 1.5 |
µChip-microchip; µFluidics-microfluidics; LOC-Lab-on-a-Chip; IA-Immunoassay; IHC-Immunohistochemistry; ECL-Electrochemiluminescence; DPV-Differential Pulse Voltametry; HE4-Human epididymis protein 4; CEA-Carcinoembryonic antigen; AFP-Alphafetoprotein; HER- Human epidermal growth factor receptor; PSA-Prostate-specific antigen; ER-Estrogen receptor; PR-Progesterone receptor; Ki67-Proliferation marker; CYFRA-Cytokeratin fragment; IL-Interleukin; PSMA-Prostate-specific membrane antigen; CRP-C-reactive protein; Pf4- Platelet factor
Demirci and group have developed a simple and inexpensive microchip ELISA-based detection module that can be coupled with a portable detection system such as a cell phone or CCD camera.60 Their system has a simple microfluidic design with a capillary reagent delivery system. Incubations were performed without mixing. They employed this device for quantification of HE4 protein, an ovarian cancer biomarker, in urine. The specificity and sensitivity using either detection module was 90% and 89.5%, respectively, for the single protein. CCD-coupled device also has similar receiver-operator functions under same analytical conditions. Therefore, with further automation this device has good commercial promise to be used at POC. However, lack of multiplexing is currently a problem with this tool.
Hu et al. developed a portable lab-on-a-chip (LOC) microfluidic device for multiplexed detection of CEA, PSA, and AFP using on-chip fluorescence.61 This device was made up of several serially connected specially-treated glass capillaries. They have coupled a homemade handheld analyzer and an automatic pump with this capillary system. They obtained detection limits in a range of 1–5 ng/mL with wide dynamic ranges for all the analyzed biomarkers in serum. They claim that their new LOC device eliminates the requirement of expensive micro-fabrication, and offers inexpensive and disposable, but replaceable tube-type “microchannels” for multiplexed detection and can potentially be used at POC.
Zhang and colleagues developed a simple, power-free microfluidic system incorporated with a place-and-play PDMS pump and nano-ELISA for simple detection of CEA and CYFRA21-1 as model cancer biomarkers in microliter volumes of whole blood. This system was used to automatically extract blood plasma from less than 3 µL of whole blood and performed a multiplex sample-to-answer assay (nano-ELISA technique) without the use of external power or extra components. This device has a top layer of glass immobilized with antibody strips, a middle PDMS layer having microfluidic channels, and a bottom glass support layer. The chip has six independent microchannels, and each divided in three operational segments: a sedimentation area, a reaction area, and a pumping/waste area. For analysis, each channel intakes approximately a drop of whole blood loaded via a self-priming, degassing-driven flow technique. PDMS slab at the outlet pulls the liquid further into the channel via degassing mechanism.62 The sedimentation area is a trench like structure which is deeper and wider than other areas to slow down the flow rate and allows blood cells to sediment under gravity. All the reagents including plasma, wash buffers, and detection antibody-gold-silver complex were pumped to the reaction site at different times with the PDMS pump. They have achieved low detection limits in pg/mL range with CEA at a concentration of 50 pg/mL and CYFRA21-1 at a concentration of 60 pg/mL in less than 60 min.62
Kim et al developed a microfluidics-integrated device for the quantitative, reproducible, fast, and inexpensive multiplexed detection of breast cancer biomarkers using immunohistochemistry. They have incorporated a fluidic control system with this device for efficient reagent delivery.
They have employed this device at POC for detecting four biomarkers, estrogen receptor (ER), human epidermal growth factor receptor 2 (HER2), progesterone receptor (PR) and Ki-67 on breast cancer cells and human breast cancer tissues. They have obtained semi-quantitative analysis with this device by comparing results against Western blot. Also, they found strong agreement within their results and conventional whole-section analysis (lowest Kendall's coefficient of concordance, 0.90).63
Recently, Fu and colleagues have developed a microfluidics-based system for POC detection of cancer by integrating commercial no-wash, homogeneous AlphaLisa-based immunoassay technology. Their system enables a multiplexed detection of up to eight samples, given one sample at a time with lower detection limits of ~10 pg/mL. The total analysis time for all the eight samples was 45 min. The major advantage of this chip is that it allows the immunophenotyping of the cell samples to examine cytokine secretion behavior.23
Sia and colleagues developed a smartphone accessory that serves as an integrated system replicating mechanical, optical, and electronic functions of a laboratory-based ELISA.64 This device is a plug-n-play type where power is obtained from the smartphone/tablet. This device has pre-loaded reagents and detection zones and was hosted in an injection-molded casing. This assay uses a drop of whole blood to detect HIV and Syphilis within 15 min. They have validated the results from this device with gold-standard immunoassays for 96 subjects in Rwanda. Given the ease of handling at POC this device can also be employed for detecting cancer biomarkers and holds commercial potential.
5. Outlook for the future
A realizable hope to decrease mortality from cancer and improve therapeutic outcome for patients may be offered by earliest possible detection coupled with new targeted drug delivery therapies featuring personalized biomarker-based monitoring.5,65,66 There are several commercially available cancer diagnostics systems with moderate sensitivity, but these can be used only in central lab facilities. Realizing the current needs of early detection as well as timely diagnostics during therapy, user-friendly tools and methods are required that can easily be performed by minimally trained clinic employees. Minimally invasive sampling for the analysis is also important for patient comfort. The next generation CDx and POC development will be governed by the demands of target users in resource-limiting settings. In addition to the manufacturing, stringent FDA and CLIA approvals pose major restriction for rapid development of diagnostics for POC.
Developments in biotechnology and biosensor technology have significantly improved the outlook associated with the biorecognition elements, immunoassay methods, and sensitivity of the detection platforms. However, advancement and optimization on these fronts should not significantly raise the overall cost in design and development. On the manufacturing front, rapid prototyping of essential POC components, such as valves, mixers, micropumps, and microfluidic chips, allow minimizing such costs. In addition, use of inexpensive materials, small reagent volumes, and mass scale industrial production will significantly bring down the total costs. Therefore, integration of biological and engineering components and achieving desired automation without drastically increasing the manufacturing cost should lead to cost effective POC device in the future.
There are research groups dedicated toward developing products for POC applications, but a majority of their research is never translated to the clinic. We must question why this is the case if progress is to be made. Smart choices must be made in selecting detection platforms and methods of detection for CDx as this will influence integration with other components. Ability to detect false positives vs false negative,67 detection limits, and thorough clinical validation are also important issues. Multiplexity will be essential to achieve the ultimate goal of widespread protein-based cancer diagnostics because it is now a fact that cancer can only be reliably diagnosed with panels of several biomarkers. In order to address these issues, researchers in academic settings must start thinking on the lines of development in accordance with the guidelines of FDA and CLIA. This will increase the translational output of their research in form of commercial grade products.
Supplementary Material
Acknowledgements
This work was supported financially by grants No. EB016707 and EB014586 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
References
- 1.Karley D, Gupta D, Tiwari A. World J. Oncol. 2011;2:151–157. doi: 10.4021/wjon352w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strimbu K, Tavel JA. Curr. Opin. HIV AIDS. 2010;5:463–466. doi: 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Kalia M. Metab. - Clin. Exp. 2015;64:S16–S21. doi: 10.1016/j.metabol.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 5.Rusling JF, Kumar CV, Gutkind JS, Patel V. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Füzéry AK, Levin J, Chan MM, Chan DW. Clin. Proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magee JA, Piskounova E, Morrison SJ. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokoll LJ, Sanda MG, Feng Z, Kagan J, Mizrahi IA, Broyles DL, Partin AW, Srivastava S, Thompson IM, Wei JT, Zhang Z, Chan DW. Cancer Epidemiol. Biomarkers Prev. 2010;19:1193–1200. doi: 10.1158/1055-9965.EPI-10-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moncada V, Srivastava S. Biomark. Med. 2008;2:181–195. doi: 10.2217/17520363.2.2.181. [DOI] [PubMed] [Google Scholar]
- 10.Henry NL, Hayes DF. Mol. Oncol. 2012;6:140–146. doi: 10.1016/j.molonc.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson IM. J. Nutr. 2006;136:2704S–2704S. doi: 10.1093/jn/136.10.2704S. [DOI] [PubMed] [Google Scholar]
- 12.Zhu CS, Pinsky PF, Cramer DW, Ransohoff DF, Hartge P, Pfeiffer RM, Urban N, Mor G, Bast RC, Moore LE, Lokshin AE, McIntosh MW, Skates SJ, Vitonis A, Zhang Z, Ward DC, Symanowski JT, Lomakin A, Fung ET, Sluss PM, Scholler N, Lu KH, Marrangoni AM, Patriotis C, Srivastava S, Buys SS, Berg CD. Cancer Prev. Res. (Phila. Pa.) 2011;4:375–383. doi: 10.1158/1940-6207.CAPR-10-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung KYC, Tabor B, Buckley MJ, Priebe IK, Purins L, Pompeia C, Brierley GV, Lockett T, Gibbs P, Tie J, McMurrick P, Moore J, Ruszkiewicz A, Nice E, Adams TE, Burgess A, Cosgrove LJ. PLoS ONE. 2015;10:e0120425. doi: 10.1371/journal.pone.0120425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusling JF. Bioanalysis. 2010;2:847–850. doi: 10.4155/bio.10.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, Abraham MT, Dey DK, Gutkind JS, Rusling JF. Anal. Chem. 2012;84:6249–6255. doi: 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang H-J, Yang M-J, Yang Y-H, Hou M-F, Hsueh E-J, Lin S-R. Oncol. Rep. 2009;22:1119–1127. doi: 10.3892/or_00000544. [DOI] [PubMed] [Google Scholar]
- 17.C. for Medicare, M. S. 7500 S. B. Baltimore and M. Usa. 2015 [Google Scholar]
- 18.Chin CD, Linder V, Sia SK. Lab. Chip. 2012;12:2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 19.Dixit CK, Vashist SK, MacCraith BD, O’Kennedy R. Nat. Protoc. 2011;6:439–445. doi: 10.1038/nprot.2011.304. [DOI] [PubMed] [Google Scholar]
- 20.Dixit CK, Vashist SK, O’Neill FT, O’Reilly B, MacCraith BD, O’Kennedy R. Anal. Chem. 2010;82:7049–7052. doi: 10.1021/ac101339q. [DOI] [PubMed] [Google Scholar]
- 21.Vashist SK, O’Sullivan SA, O’Neill FT, Holthofer H, O’Reilly B, Dixit CK. WIPO Pat. WO2010044083. 2010;24 [Google Scholar]
- 22.Dixit CK, Aguirre GR. Microarrays. 2014;3:180–202. doi: 10.3390/microarrays3030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tak Z, For Yu, Guan H, Ki Cheung M, McHugh WM, Cornell TT, Shanley TP, Kurabayashi K, Fu J. Sci. Rep. 2015;5:11339. doi: 10.1038/srep11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kai J, Puntambekar A, Santiago N, Lee SH, Sehy DW, Moore V, Han J, Ahn CH. Lab. Chip. 2012;12:4257–4262. doi: 10.1039/c2lc40585g. [DOI] [PubMed] [Google Scholar]
- 25.Dixit CK, Kaushik A. Biochem. Biophys. Res. Commun. 2012;419:316–320. doi: 10.1016/j.bbrc.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Grieshaber D, MacKenzie R, Vörös J, Reimhult E. Sensors. 2008;8:1400–1458. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusling JF, Bishop GW, Doan NM, Papadimitrakopoulos F. J. Mater. Chem. B. 2013;2:12–30. doi: 10.1039/C3TB21323D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metters JP, Banks CE. Vacuum. 2012;86:507–519. [Google Scholar]
- 29.Chen Z, Ren W, Gao L, Liu B, Pei S, Cheng H-M. Nat. Mater. 2011;10:424–428. doi: 10.1038/nmat3001. [DOI] [PubMed] [Google Scholar]
- 30.Rauf S, Shiddiky MJA, Asthana A, Dimitrov K. Sens. Actuators B Chem. 2012;173:491–496. [Google Scholar]
- 31.Rauf S, Shiddiky MJA, Trau M. Sens. Actuators B Chem. 2013;185:543–547. [Google Scholar]
- 32.Shi J-J, He T-T, Jiang F, Abdel-Halim ES, Zhu J-J. Biosens. Bioelectron. 2014;55:51–56. doi: 10.1016/j.bios.2013.11.056. [DOI] [PubMed] [Google Scholar]
- 33.Fakunle ES, Fritsch I. Anal. Bioanal. Chem. 2010;398:2605–2615. doi: 10.1007/s00216-010-4098-5. [DOI] [PubMed] [Google Scholar]
- 34.Siegel AC, Phillips ST, Dickey MD, Lu N, Suo Z, Whitesides GM. Adv. Funct. Mater. 2010;20:28–35. [Google Scholar]
- 35.Jensen GC, Krause CE, Sotzing GA, Rusling JF. Phys. Chem. Chem. Phys. 2011;13:4888–4894. doi: 10.1039/c0cp01755h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ihalainen P, Majumdar H, Viitala T, Törngren B, Närjeoja T, Määttänen A, Sarfraz J, Härmä H, Yliperttula M, Österbacka R, Peltonen J. Biosensors. 2012;3:1–17. doi: 10.3390/bios3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Biosens. Bioelectron. 2011;26:4477–4483. doi: 10.1016/j.bios.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otieno BA, Krause CE, Latus A, Chikkaveeraiah BV, Faria RC, Rusling JF. Biosens. Bioelectron. 2014;53:268–274. doi: 10.1016/j.bios.2013.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krause CE, Otieno BA, Bishop GW, Phadke G, Choquette L, Lalla RV, Peterson DE, Rusling JF. Anal. Bioanal. Chem. 2015;407:7239–7243. doi: 10.1007/s00216-015-8873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krause CE, Otieno BA, Latus A, Faria RC, Patel V, Gutkind JS, Rusling JF. ChemistryOpen. 2013;2:141–145. doi: 10.1002/open.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang CK, Vaze A, Rusling JF. Lab. Chip. 2011;12:281–286. doi: 10.1039/c1lc20833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang CK, Vaze A, Rusling JF. Anal. Methods. 2014;6:8878–8881. doi: 10.1039/C4AY01962H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadimisetty K, Malla S, Sardesai NP, Joshi AA, Faria RC, Lee NH, Rusling JF. Anal. Chem. 2015;87:4472–4478. doi: 10.1021/acs.analchem.5b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wasalathanthri DP, Malla S, Bist I, Tang CK, Faria RC, Rusling JF. Lab. Chip. 2013;13:4554–4562. doi: 10.1039/c3lc50698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rusling JF. Chem. Rec. 2012;12:164–176. doi: 10.1002/tcr.201100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mani V, Chikkaveeraiah BV, Rusling JF. Expert Opin. Med. Diagn. 2011;5:381–391. doi: 10.1517/17530059.2011.607161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tekin HC, Gijs MAM. Lab. Chip. 2013;13:4711–4739. doi: 10.1039/c3lc50477h. [DOI] [PubMed] [Google Scholar]
- 49.Munge BS, Coffey AL, Doucette JM, Somba BK, Malhotra R, Patel V, Gutkind JS, Rusling JF. Angew. Chem. Int. Ed. 2011;50:7915–7918. doi: 10.1002/anie.201102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sardesai N, Pan S, Rusling J. Chem. Commun. 2009:4968–4970. doi: 10.1039/b909220j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sardesai NP, Kadimisetty K, Faria R, Rusling JF. Anal. Bioanal. Chem. 2013;405:3831–3838. doi: 10.1007/s00216-012-6656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mani V, Kadimisetty K, Malla S, Joshi AA, Rusling JF. Environ. Sci. Technol. 2013;47:1937–1944. doi: 10.1021/es304426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonald JC, Whitesides GM. Acc. Chem. Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 54.Chikkaveeraiah BV, Liu H, Mani V, Papadimitrakopoulos F, Rusling JF. Electrochem. Commun. 2009;11:819–822. doi: 10.1016/j.elecom.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leigh SJ, Bradley RJ, Purssell CP, Billson DR, Hutchins DA. PLoS ONE. 2012;7:e49365. doi: 10.1371/journal.pone.0049365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gross BC, Erkal JL, Lockwood SY, Chen C, Spence DM. Anal. Chem. 2014;86:3240–3253. doi: 10.1021/ac403397r. [DOI] [PubMed] [Google Scholar]
- 57.Shallan AI, Smejkal P, Corban M, Guijt RM, Breadmore MC. Anal. Chem. 2014;86:3124–3130. doi: 10.1021/ac4041857. [DOI] [PubMed] [Google Scholar]
- 58.Bishop GW, Satterwhite JE, Bhakta S, Kadimisetty K, Gillette KM, Chen E, Rusling JF. Anal. Chem. 2015;87:5437–5443. doi: 10.1021/acs.analchem.5b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadimisetty K, Mosa IM, Malla S, Satterwhite-Warden JE, Kuhns TM, Faria RC, Lee NH, Rusling JF. Biosens. Bioelectron. 2016;77:188–193. doi: 10.1016/j.bios.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Zhao X, Khimji I, Akbas R, Qiu W, Edwards D, Cramer DW, Ye B, Demirci U. Lab. Chip. 2011;11:3411–3418. doi: 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu W, Lu Z, Liu Y, Chen T, Zhou X, Li CM. Lab. Chip. 2013;13:1797–1802. doi: 10.1039/c3lc41382a. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H, Li G, Liao L, Mao H, Jin Q, Zhao J. Biomicrofluidics. 2013;7 doi: 10.1063/1.4807803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim MS, Kwon S, Park J-K. Microfluidic Diagnostics: Methods and Protocols. Vol. 949. LLC: Springer Science+Business Media; 2013. pp. 349–364. [Google Scholar]
- 64.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. Sci. Transl. Med. 2015;7:273re1–273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 65.Weinstein IB, Joe AK. Nat. Clin. Pract. Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 66.Le Tourneau C, Faivre S, Siu LL. Eur. J. Cancer. 2007;43:2457–2466. doi: 10.1016/j.ejca.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 67.Ao L, Yan H, Zheng T, Wang H, Tong M, Guan Q, Li X, Cai H, Li M, Guo Z. Sci. Rep. 2015;5:11895. doi: 10.1038/srep11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.