ABSTRACT
Avian leukosis virus (ALV) induces B-cell lymphoma and other neoplasms in chickens by integrating within or near cancer genes and perturbing their expression. Four genes—MYC, MYB, Mir-155, and TERT—have previously been identified as common integration sites in these virus-induced lymphomas and are thought to play a causal role in tumorigenesis. In this study, we employ high-throughput sequencing to identify additional genes driving tumorigenesis in ALV-induced B-cell lymphomas. In addition to the four genes implicated previously, we identify other genes as common integration sites, including TNFRSF1A, MEF2C, CTDSPL, TAB2, RUNX1, MLL5, CXorf57, and BACH2. We also analyze the genome-wide ALV integration landscape in vivo and find increased frequency of ALV integration near transcriptional start sites and within transcripts. Previous work has shown ALV prefers a weak consensus sequence for integration in cultured human cells. We confirm this consensus sequence for ALV integration in vivo in the chicken genome.
IMPORTANCE
Avian leukosis virus induces B-cell lymphomas in chickens. Earlier studies showed that ALV can induce tumors through insertional mutagenesis, and several genes have been implicated in the development of these tumors. In this study, we use high-throughput sequencing to reveal the genome-wide ALV integration landscape in ALV-induced B-cell lymphomas. We find elevated levels of ALV integration near transcription start sites and use common integration site analysis to greatly expand the number of genes implicated in the development of these tumors. Interestingly, we identify several genes targeted by viral insertions that have not been previously shown to be involved in cancer.
INTRODUCTION
Avian leukosis virus (ALV) is a simple retrovirus that infects chickens and some other avian species (1). Like all retroviruses, ALV reverse transcribes its RNA genome in the cytoplasm, and then the proviral DNA enters the nucleus, where it integrates into the genomic DNA of the host cell. Several studies have shown ALV integration occurs in a quasi-random fashion in human and chicken cells grown in culture, with only slight preference for active transcription units (2–4). In addition, a weak consensus sequence for ALV integration was observed (5, 6).
Infection of chicken embryos or young chicks with ALV has been shown to induce metastatic B-cell lymphoma and occasionally other types of neoplasms. The latency of these tumors can vary between 1.5 and 6 months and is dependent on the strain of ALV injected and the age of the bird at the time of infection. The lymphomas typically begin in the bursa (an avian organ in which B cells mature) and then metastasize to distant organs such as the liver, kidney, and spleen (7).
Unlike the closely related Rous sarcoma virus (RSV), ALV does not carry a transforming oncogene. Instead, ALV induces tumors by insertional mutagenesis (8, 9). ALV is a potent insertional mutagen because the provirus contains strong promoter and enhancer sequences in its viral long terminal repeats (LTRs). This means that when ALV integrates into the genome, it can perturb the expression of genes in the vicinity of the proviral integration site. Hence, if the virus integrates near a cancer gene, the ALV-induced misexpression of that gene may contribute to the transformation of the cell and potentially tumorigenesis. Depending on where ALV integrates and its relationship to the nearby genes, the virus can have other effects as well. For example, the virus could potentially reduce or eliminate the expression of a gene, it could induce expression of a truncated gene product (10), or it could potentially perturb splicing or polyadenylation of a host transcript (9).
Much previous work has been done to identify genes that drive ALV-induced oncogenesis by locating clusters of proviral integration in these tumors. MYC was the first gene shown to be affected by ALV integrations in long-latency B-cell lymphomas (8, 9). These birds were infected 2 to 7 days after hatching and developed tumors by 4 to 6 months of age. Later c-bic was shown to be a common integration site, and c-bic integrations often occurred in the same tumors as MYC integrations (11). It turns out the c-bic gene is not protein coding but instead is the precursor for an oncogenic microRNA that was later given the name Mir-155 (12). Later work showed that infection of 10-day embryos with a different strain of ALV, strain EU-8, resulted in short-latency tumors harboring integrations at the MYB locus (13). Recent work studying ALV subgroup J has shown that MYC, TERT, and ZIC1 are targets of integration in ALV-J-induced myeloid leukosis, and MET is a common target in ALV-J-induced hemangiomas (14, 15).
Both the viral strain and the time of infection are important in determining how quickly tumors develop and what genes are affected. EU-8, the strain that first caused a high incidence of rapid-onset B-cell lymphomas, is a recombinant strain of ALV that contains parts of ALV strain UR2AV and ring-necked pheasant virus (13). Importantly, only embryonic EU-8 infections produced rapid-onset B-cell lymphomas. Infection of birds early with a different virus (UR2AV) produced mainly long-latency MYC tumors, as was the case if birds were infected with EU-8 after hatching.
Follow-up studies showed that EU-8 is able to rapidly induce tumors because it contains a 42-nucleotide deletion that disrupts the viral negative regulator of splicing (NRS) (16). This NRS disruption reduces the efficiency of polyadenylation, increases the rate of viral readthrough, and increases the efficiency of splicing to downstream genes—factors that are thought to enable the virus to induce tumors rapidly (16–19). Later, several modifications were made to ALV strain LR-9, a strain incapable of inducing rapid-onset B-cell tumors, and these changes were able to mimic the NRS deficiency of EU-8. These LR-9 mutant strains, LR9-Δ42, LR9-U916A, and LR9-G919A, were able to rapidly induce B-cell tumors (18, 20).
In this study, we generated rapid-onset B-cell lymphomas by infecting 5- and 10-day embryos with either ALV-A viral strain LR-9, LR9-Δ42, LR9-U916A, or LR9-G919A (see Table S1 in the supplemental material). A subset of these tumors were analyzed previously by lower-throughput methods (18, 20, 21). Some tumors were shown to harbor MYB integrations via locus-specific nested PCR, and inverse PCR identified TERT as common integration site in some tumors (see Table S1). Southern blot analysis showed several tumors appeared to be clonal or oligoclonal for TERT integrations, while others were clonal for MYB (21). In this study, we use high-throughput sequencing to identify proviral integration sites. High-throughput sequencing enables a more complete characterization of the integration landscape in these tumors and the genes that are perturbed by ALV integration.
RESULTS
We sequenced 37 tissue samples from 27 different birds (see Table S1 in the supplemental material) and obtained approximately 2.39 million reads originating from viral integrations in tumor and non-tumor tissues. These reads mapped to 32,050 unique viral integration sites. Among these unique integration sites, we identified 43,000 unique sonication breakpoints. The average number of breakpoints per integration was 1.342, with the vast majority of integrations (86.8%) showing only a single sonication breakpoint and therefore no evidence of clonal expansion.
Increased clonality in metastatic tumors versus bursal tumors.
The bursa is believed to act as the primary organ of transformation in cases of ALV-induced B-cell lymphoma. Laboratory-infected chickens typically develop multiple primary neoplastic follicles in the bursa, some of which may eventually form primary tumors. Secondary tumors are also commonly found in the liver, spleen, kidneys, and some other organs. These tumors are believed to arise when a single cell within the bursa acquires a combination of integrations and possibly other mutations that enable the cell to proliferate and then metastasize to a distant organ. Once at the distant location, the progenitor cell is thought to clonally expand and form a tumor, which typically presents as a nodular or diffuse tumor in the distant organ (7).
The extent to which the progenitor cell has clonally expanded can be measured by determining the number of different sonication breakpoints observed for an integration (22, 23). Sonication breakpoints are generated during library preparation by the shearing of genomic DNA followed by ligation of adapters onto the sheared ends. When an integration occurs in a cell that later divides by clonal expansion, multiple sonication breakpoints can potentially be observed for that integration. In this way, it is possible to obtain a metric of relative clonal expansion for each integration in a given sample.
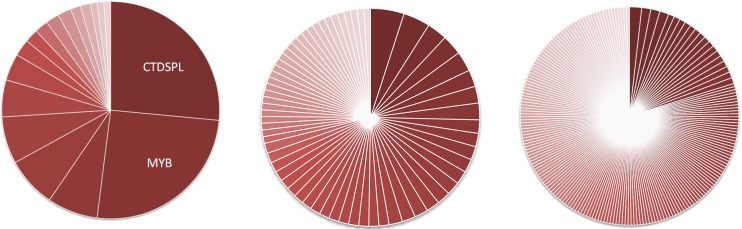
Consistent with the clonal expansion hypothesis, we observed that metastatic tumors often contained one or more integrations that have a high number of breakpoints, whereas bursal tumors only occasionally exhibited highly expanded integrations. This can be visualized via a pie chart, where the pie represents a tumor, each slice represents a specific integration, and the size of the slice corresponds to the number of sonication breakpoints observed for that integration. Pie charts for a typical metastatic liver tumor, bursa with neoplastic follicles, and liver exhibiting no tumor are shown in Fig. 1. The liver tumor contains several integrations that show a high level of clonal expansion. The bursa contains many different neoplastic follicles, each with a unique complement of integrations and all with low levels of clonal expansion. Lastly, a chart for a non-tumor liver is shown for comparison, which as expected, exhibits almost no clonally expanded integrations.
FIG 1 .
Metastatic tumors contain integrations within clonally expanded cells. Each pie represents a specific tissue that underwent high-throughput integration site sequencing. Each slice represents a unique integration, and the size of each slice corresponds to the number of sonication breakpoints observed for that integration. The integrations that exhibit the greatest clonal expansion (i.e., the most breakpoints) are shown. A total of 200 breakpoints are shown for each sample. (Left) C3-B256 metastatic liver tumor exhibits extensive clonal expansion. (Middle) D1-G157 bursa with neoplastic follicles contains some integrations in moderately expanded clones. (Right) D4-G163 non-tumor liver exhibits very few integrations in expanded clones.
Common integration sites.
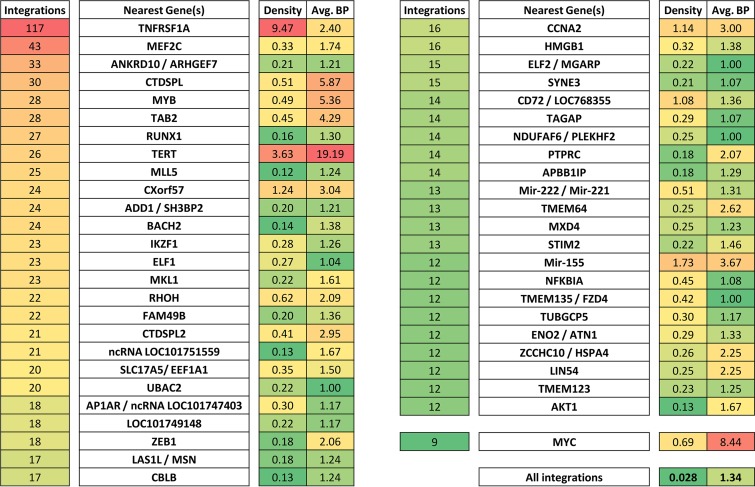
A total of 37 tissues, including 13 primary neoplasms and 17 metastatic tumors, were sequenced. Analysis of the resulting integrations identified a diverse array of genes as targets of ALV integration. A list of the top 48 targets of integration is shown in Fig. 2. All of these common integration sites exhibited at least 12 unique integrations within a single 50-kb sliding window. Several of the most targeted genes have been identified in previous ALV insertional mutagenesis screens. For example, the first gene identified as a common integration site in long-latency ALV-induced lymphomas was MYC in 1981 (8). Although MYC is not among the top 50 common targets of integration, we did identify nine unique integrations into the MYC gene. In addition, the MYC cluster was among the most clonally expanded clusters in our study, with 8.44 breakpoints per integration, second only to TERT (Fig. 2). MYB, first seen as a common integration site in rapid-onset lymphomas in 1988 (13), is tied for the fifth-most-targeted gene, with 28 unique integrations. Likewise, Mir-155 was first seen as an ALV common integration site in 1989 (11), and we observe it in our tumors as well with 12 unique integrations, making it tied for the 40th-most-common target of integration.
FIG 2 .
Common sites of ALV proviral integration. The top 48 common integration sites are shown. Integration clusters were defined as any 50-kb region that harbors 12 or more unique ALV integrations. If an integration cluster was within or near a gene, all integrations within that gene and ±10 kb from the gene transcript were also included. “Density” represents the number of integrations per kilobase in a given cluster. The average number of sonication breakpoints per integration is shown for each gene. A higher number of breakpoints indicates increased clonal expansion of the cells carrying that integration. MYC did not penetrate the 12-integration threshold but is shown for comparison.
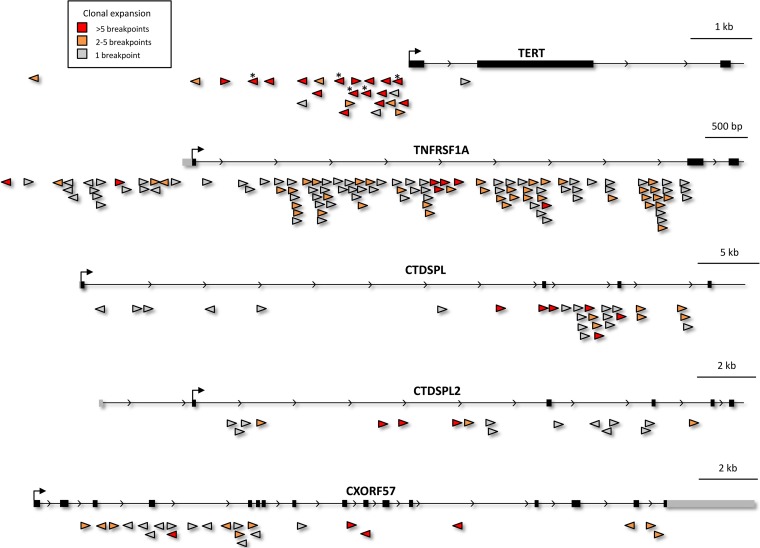
TERT had the most clonally expanded integrations identified in our study, with an average of 19.19 breakpoints per integration. This is consistent with earlier work analyzing a subset of the same tumors that identified 5 clonal or oligoclonal integrations upstream of the TERT transcription start site by inverse PCR (21). The position and orientation of each of these previously characterized integrations was successfully verified by high-throughput sequencing. In addition, 20 integrations upstream of or within the TERT promoter were identified that had not been seen previously (Fig. 3). Like the integrations identified earlier, most of the novel TERT integrations (16/20) were in the opposite orientation of the TERT gene, and all but one occurred in birds infected at embryonic day 10 (see Table S2 in the supplemental material).
FIG 3 .
Selected common integration sites. Integration clusters for TERT, TNFRSF1a, CTDSPL, CTDSPL2, and CXorf57 are shown. The orientation of each integrated provirus is indicated by the direction of the triangle, and the tip of the triangle corresponds to the exact location of integration. The extent of clonal expansion is indicated by the color of the integration marker—integrations with 1 breakpoint are gray, those with 2 to 5 breakpoints are orange, and those with greater than 5 breakpoints are red. TERT integrations marked with an asterisk (*) are the same integrations identified previously via inverse PCR (21).
Although MYC, MYB, Mir-155, and TERT have been seen in previous ALV insertional mutagenesis screens, most of the top targets of integration that we identified have not been identified in similar lower-throughput studies conducted previously. One such gene is TNFRSF1a; it was the most frequent target of integration that we observed, with a total of 117 unique viral integrations at this locus. TNFRSF1a is a member of the tumor necrosis factor (TNF) receptor superfamily and is one of the major receptors for tumor necrosis factor alpha (TNF-α). TNFRSF1a can activate NF-κB and has known roles mediating apoptosis and regulating inflammation and cell proliferation (24). The vast majority of the integrations (82.9%) are within TNFRSF1a intron 1, and most are in the same orientation as the gene (92.3%) (Fig. 3; see Table S2 in the supplemental material). The location and orientation of these integrations suggest that the virus is promoting the transcription of a TNFRSF1a transcript lacking exon 1. Exon 1 encodes part of the protein’s extracellular domain, which is crucial for the binding to its ligand TNF-α (25). Although this is a frequent target for ALV integration, it was only identified in two highly expanded clones (>10 breakpoints) and was almost always restricted to the bursa (113/117, 96.6% bursa [see Table S2]). These results suggest that ALV may be inducing a truncated receptor that is unable to bind TNF-α and mediate apoptosis. The fact that this integration is rarely found outside the bursa suggests that this truncated gene product does not contribute to metastasis of the neoplasm to distant organs.
MEF2C was the second-most-targeted gene for ALV integrations, with a total of 43 unique integrations within 10 kb of this gene. MEF2C belongs to a family of transcription factors that have been shown to be important regulators of apoptosis, proliferation, survival, differentiation, and cancer (26). MEF2C has been observed as a common integration site in other retroviral insertional mutagenesis screens conducted in mice. This work has observed integrations most often within introns 1 and 2 and in the same orientation as the gene (27–30). We observe a similar pattern of MEF2C integrations, with 21 of the 43 MEF2C integrations occurring in intron 1 or 2, although we observed no preference for integration in the same orientation as the gene (see Table S2 in the supplemental material).
Two related phosphatase genes, CTDSPL (also known as RBSP3 or HYA22) and CTDSPL2, were also common integration sites, with 30 and 21 unique integrations, respectively. Both genes belong to a gene family of RNA polymerase II C-terminal domain phosphatases and contain a conserved Dullard-like phosphatase domain (31). CTDSPL is a known tumor suppressor that can dephosphorylate RB1 and affect cell cycle progression (32). It is downregulated in primary non-small-cell lung cancer and has been shown to promote proliferation by modulating pRB/E2F1 in acute myeloid leukemia (33, 34). CTDSPL2 is less studied and has not been linked to cancer. Recent work has shown that CTDSPL2 directly interacts with and dephosphorylates SMAD 1/5/8, which negatively regulates bone morphogenetic protein (BMP) signaling (35). We observed a strong cluster of integrations for both genes. Integrations were clustered within intron 2 in CTDSPL and within introns 2 and 3 for CTDSPL2. A strong preference for integration in the forward orientation was observed for both genes (Fig. 3). This pattern suggests the virus may be producing a truncated protein product in both cases. The relatively high number of breakpoints—5.87 on average for CTDSPL and 2.95 for CTDSPL2—indicates that the cells harboring these integrations experienced a moderate level of clonal expansion. Interestingly, liver tumors from 2 different birds accounted for 16/30 of the CTDSPL integrations and 17/21 of the CTDSPL2 integrations (see Fig. S1 in the supplemental material). This suggests that these genes may cooperate in ALV-induced lymphomagenesis.
CXorf57 was the 10th-most-frequently targeted common integration site and is among the most enigmatic genes that we identified. CXorf57 is conserved in humans but has never been characterized and hence has no known function. CXorf57 encodes a protein that has a conserved putative replication factor A protein 1 domain. Genes with this domain that have been characterized have been shown to be involved in recognition of DNA damage for nucleotide excision repair (31, 36). CXorf57 contains 24 unique integrations that are spaced throughout the gene and in no preferred orientation (Fig. 3). This integration pattern indicates that these proviral integrations may be disrupting the normal transcription of this gene, suggesting that it could be a novel tumor suppressor. Interestingly, a strong preference for integration in B-cell lymphomas in the liver was observed (18 of 24 integrations [see Table S2 in the supplemental material]).
Functional annotation enrichment analysis of ALV common integration sites.
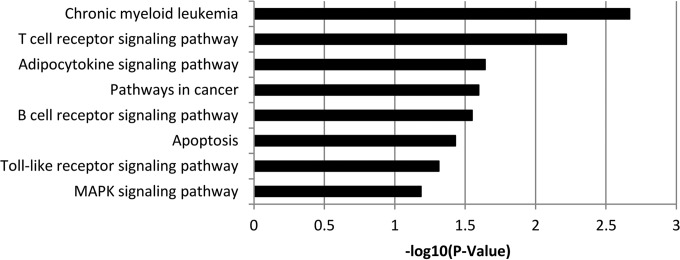
To determine whether these 48 major common integration sites (Fig. 2) are enriched for genes of specific functions or involved in specific pathways, we conducted gene annotation enrichment analysis with DAVID (37). We identify six enriched KEGG pathways and processes, most of which are related to cancer or are pathways active in immune cells (Fig. 4). Gene Ontology (GO) term analysis revealed strong enrichment (P < 0.005) for a number of different gene ontologies (see Fig. S2 in the supplemental material). The most significant enrichment was seen for regulators of transcription (both positive and negative). Additionally, strong enrichment was observed for several types of positive regulators of metabolic and biosynthetic processes, as well as several antiapoptotic functional terms.
FIG 4 .
KEGG pathway analysis. KEGG pathways enriched among the top 48 common integration sites are shown. MAPK, mitogen-activated protein kinase.
ALV integration has a weak palindromic consensus sequence in vivo.
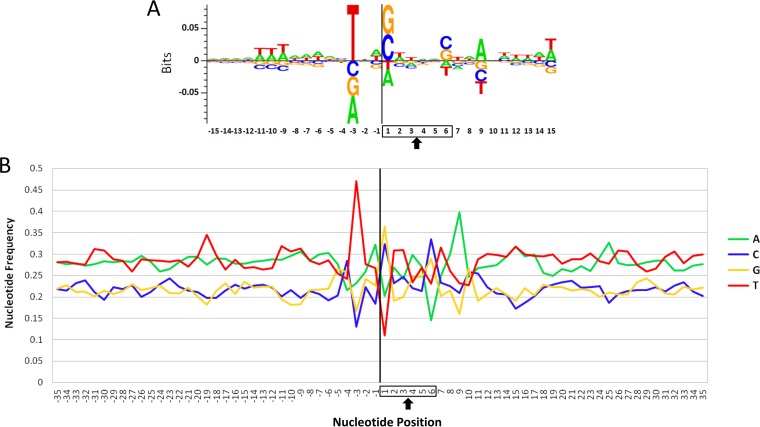
It was shown in earlier work that ALV integration has a weak palindromic consensus sequence when integrating into human DNA (5, 6). These analyses were performed in human cells in culture that had been engineered to express the TVA receptor, enabling them to be infected with ALV. To determine whether ALV exhibits a similar preference in its canonical host in vivo, we performed a similar analysis of our full data set of integrations in chicken. We observed very similar results to those seen in human cell culture (Fig. 5). For example, a strong preference for a T −3 nucleotides from the viral integration site was observed. In addition, strong preferences for G/C at position 1 and A at position 9 were also observed. Notably, the nucleotide frequencies that we observe are nearly exactly what were seen in cultured human cells. For example, we calculated the frequency of T at position −3 to be 47%, which is exactly the same frequency reported in human cells (5). The preference for G/C at position 1 was 68.8% in our study and 71% in human cells, and the preference for A at position 9 was 39.8% in our study and 43% in human cells. These results show that the consensus sequence observed in human cells infected with ALV is the same as that seen in vivo in the virus’ natural host.
FIG 5 .
Consensus target integration site. (A) Sequence logo displaying the consensus sequence surrounding ALV integration sites in this study. The vertical black line represents the viral integration site, and the 6 nucleotides of sequence duplicated during viral integration are boxed. The arrow indicates the axis of symmetry. (B) Base frequencies in the chicken genome at ALV integration sites are shown.
Interestingly, as with previous studies (5, 6), we observed that the ALV consensus sequence is slightly asymmetric. This contrasts with other retroviruses such as HIV and murine leukemia virus (MLV) that have perfectly symmetric consensus sequences (5, 6). Although it has been shown that ALV integrase typically generates 6-base duplications, there are indications that 5-base duplications are possible under certain circumstances (38, 39). If a 5-base duplication is generated by ALV integrase at sufficient frequency, this could reduce the nucleotide preferences that we observe to the right of the duplication (Fig. 5) but not to its left, which could explain the asymmetry that we observe.
ALV prefers integration near promoters and within genes in vivo.
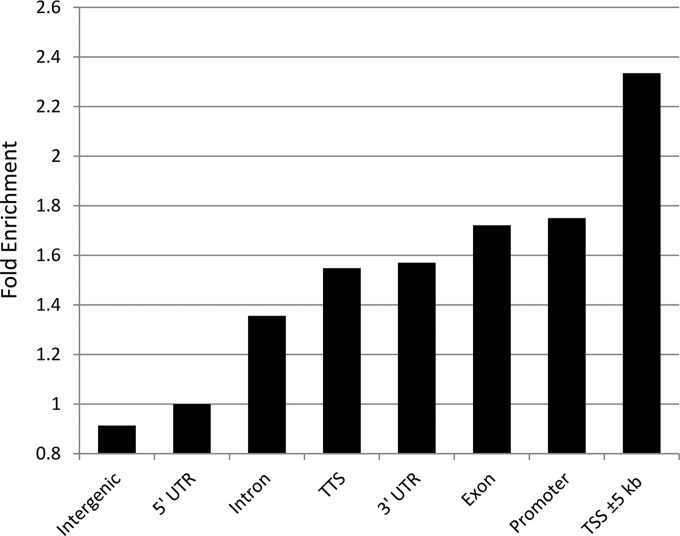
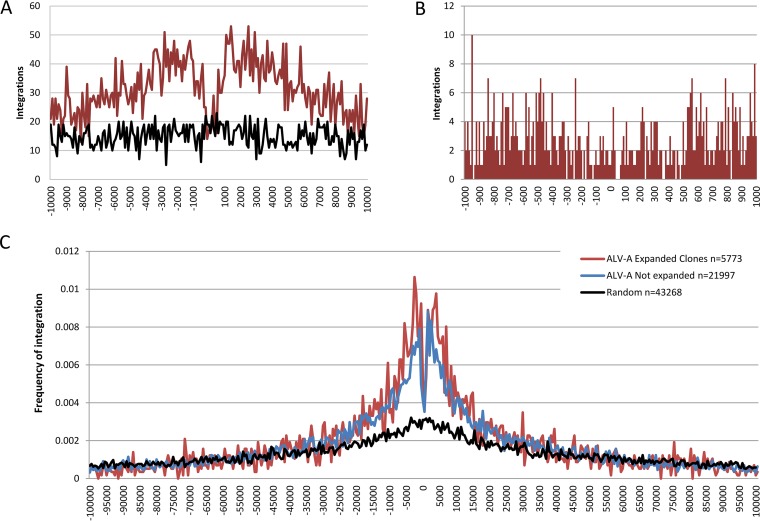
To determine whether ALV prefers integration near certain features in vivo, we employed the HOMER software suite (40). A total of 27,770 unique ALV integrations and an equal number of random, computer-generated integrations were annotated with the nearest genomic feature. This analysis revealed a preference for integration near transcription start sites (TSSs) (Fig. 6). To better understand the pattern of integrations surrounding TSSs, we plotted all integrations with respect to the nearest TSS (Fig. 7). We observed enrichment for ALV integration extending 30 kb on either side of the TSS. In addition, we observed a sharp drop in integration frequency in the immediate vicinity of the TSS (Fig. 7B). This pattern is similar to that seen in studies of murine leukemia virus (MLV) and is believed to be due to the occupancy of this area by basal transcriptional machinery such as transcription factor IID (TFIID) (41).
FIG 6 .
Preference for integration near genomic features. Enrichment for integration near genomic features was calculated with HOMER (40). Fold enrichment was calculated by comparing ALV integrations to a randomly generated integration data set. Promoters are defined as the region from −1 kb to +100 bp from transcription start sites, while transcription termination sites (TTS) are defined as the region from −100 bp to +1 kb flanking the transcription termination site.
FIG 7 .
ALV integrations mapped with respect to transcription start sites. (A) Integrations within 10 kb of transcription start sites are shown placed into 100-bp bins. The red line represents ALV-A integrations, and the black line represents randomly simulated integrations. A preference for integration flanking TSSs is observed. (B) Integrations within 1 kb of TSSs are shown in 10-bp bins. A striking lack of integrations was observed in the immediate vicinity of TSSs. (C) Integration frequency was calculated for expanded clones (red), nonexpanded clones (blue), and randomly generated integrations (black), and integrations are presented in 500-bp bins. Integration frequency is the fraction of total integrations that fall into each 500-bp bin. Integrations near the TSS are shown to be slightly more likely to result in clonal expansion.
Earlier work on ALV integration in cell culture has shown that the virus has a slight preference for integration near transcribed elements, but a preference for integration centered on transcription start sites was not seen in these earlier studies (2–4). There are several ways to explain this inconsistency with earlier reports. First, this pattern may be explained by the fact that we sequenced integrations that occurred in vivo. Hence, many of the integrations have been subject to selection, especially those found in clonally expanded cells. To determine the extent to which integrations in clonally expanded cells are affecting observed enrichment for integrations near TSSs, integrations that show evidence of clonal expansion were analyzed separately from those for which only a single sonication breakpoint was observed. This analysis shows that even integrations that show no evidence of clonal expansion show enrichment for integration near TSSs (Fig. 7C). It is possible that selection is still at work in the cases of integrations that are not clonally expanded: if, for example, the gene near the integration promotes cell survival but not proliferation.
This analysis also revealed preference for integration near other genomic features as well (Fig. 6). Integration near promoters (−1 kb to +100 bp from transcription start sites) was the most enriched compared to the control, with a 1.75-fold increase. Other features for which enrichment was observed include exons (1.72-fold), 3′ untranslated regions (3′ UTRs) (1.57-fold), transcription termination sites (−100 bp to +1 kb, 1.55-fold), and introns (1.36-fold). 5′ UTRs exhibited no increase in ALV integration versus the control, while intergenic regions were less likely to harbor ALV integrations than random (0.91-fold).
DISCUSSION
In this study, we characterized the integration of proviruses in ALV-A-induced B-cell lymphomas with high-throughput sequencing. This method allows for a much more detailed analysis of integration sites than was possible in earlier studies of these types of neoplasms.
We observed that promoters and TSSs are the most preferred sites of ALV integration in vivo (Fig. 6 and 7). This preference had not been seen in previous studies of ALV integration. Analyses of other retroviruses such as HIV and murine leukemia virus (MLV) have shown that MLV but not HIV prefers integration near TSSs and CpG islands (41, 42). MLV’s integration site preference is mediated by the binding of bromodomain and extraterminal domain (BET) proteins to the MLV integrase, although a slight preference for TSSs and CpG islands persists in the absence of this interaction (43–45). MLV is also known to prefer integration within 2.5 kb of TSSs, and a strong decrease in MLV integration frequency has been shown within 100 bp of TSSs (41).
The pattern of ALV integration that we report is very similar to MLV but not identical. For example, while we observed a strong preference for integration on both sides of TSSs and a sharp drop-off within 100 bp of TSSs (Fig. 7), we did not observe a narrow peak of increased integration frequency ±2.5 kb from the TSS. Instead, we saw a broader peak of elevated integration frequency that stretches as far as 30 kb on either side of the TSS (Fig. 7C). Also, we observed a weaker preference for ALV integration in the immediate vicinity of TSSs than has been seen for MLV. Previous work calculated a 4.7-fold increase in the frequency of MLV integrations within 5 kb of the TSS, although recent work has shown this can vary by cell type (42, 46). In contrast, we observed only a 2.3-fold increase for ALV over that range (Fig. 6). Because our experiments were conducted in vivo, where cells are subject to selection and clonal expansion, the preference for ALV integration that we observe may be partially due to these additional variables. This may explain why a preference for integration centered on TSSs was not observed in earlier studies in cell culture.
To date, only four genes had been shown to be common integration sites in ALV-A-induced B-cell lymphoma: MYC, MYB, Mir-155, and TERT. Here we identify all four of these genes as common integration sites, as well as a host of new genes that had not been previously implicated in ALV-induced lymphomagenesis.
Three reports had been published previously that partially characterize 8 of the 28 tumors that we analyzed in this study (see Table S1 in the supplemental material). Two of these publications utilized nested PCR to map proviral integrations at the MYB promoter and showed some tumors contained one or more integrations into the MYB locus (18, 20). A third report used inverse PCR to map proviral integrations (which is not biased to a specific locus) and showed multiple integrations in the TERT promoter in the opposite orientation (21).
By reanalyzing these tumors, we were able to verify many of the integrations seen in previous studies. First, with regard to TERT, we verified by deep sequencing all 5 TERT promoter integrations that were described previously and identified an additional 21 integrations at the TERT locus in both newly analyzed and reanalyzed tumors. Previous work also showed that these integrations were clonal or oligoclonal by Southern blotting, meaning that the integrations were present in a large fraction of cells in the tumor (21). Deep-sequencing results confirm this finding; all of these integrations exhibited extensive clonal expansion by breakpoint analysis. Overall, TERT was the eighth-most frequent target of integration, with 26 unique integration sites identified by deep sequencing. Although it was not the most frequent target of integration, TERT integrations were often highly expanded, with an average of 19.19 sonication breakpoints observed per integration, which may explain why it was identified so readily by inverse PCR in previous work. The extensive expansion of clones containing TERT integrations is consistent with the hypothesis that TERT activation is an early event in tumorigenesis.
MYB was the fifth-most-targeted gene, with 28 unique integrations. Only one of these integrations was described in previous work (see A2-R588, liver, in Table S1 in the supplemental material), suggesting that many of the MYB integrations identified in earlier work were not clonal and were possibly only present in a small number of cells (18, 20).
Historically, MYC and Mir-155 were often seen in ALV-induced B-cell lymphomas. Both genes were prominent integration clusters in this study (Fig. 2). As for Mir-155, we identified 12 unique Mir-155 integrations. Earlier studies have shown that Mir-155 integrations are often seen in metastatic tumors, which led to the hypothesis that Mir-155 is a late event in ALV tumor induction and may play a role in metastasis (8). Eleven of the 12 Mir-155 integrations we observed occurred in metastatic liver tumors, with only one seen in the primary bursa in our study (see Table S2 in the supplemental material), which is consistent with this hypothesis.
In this study, we identified only 9 integrations in the MYC locus (Fig. 2). MYC was the first gene ever identified as a common integration site in ALV-induced lymphomas, and MYC integrations have since been seen in many studies of these neoplasms (8, 9, 47). The time of infection is thought to be an important factor in the development of MYC-associated tumors, with later infections (especially after hatching) more likely to induce tumors with MYC integrations. In contrast, we infected birds much earlier, at embryonic day 5 or day 10. Interestingly, all 9 of the MYC integrations occurred in birds that were infected at day 10, while no MYC integrations were observed at day 5 (see Table S2 in the supplemental material). This supports the idea that the early timing of injections may explain why we see fewer MYC integrations than in earlier work.
Interestingly, the most frequent target of integration was TNFRSF1a. This gene codes for a receptor for tumor necrosis factor alpha (TNF-α). TNFRSF1a can activate NF-kappaB and has known roles mediating apoptosis and regulating inflammation and cell proliferation (24). Although TNFRSF1a harbored 117 unique integrations, it was only highly clonally expanded (>10 breakpoints) in two cases. This lack of highly expanded clones may explain why this gene was not identified in previous experiments mapping ALV integration sites. The vast majority of the integrations occurred in the first intron of the gene and in the same orientation as the gene, and integrations were almost exclusively found in bursal tissues and not in metastatic tumors (see Table S2 in the supplemental material). These data suggest that the integration may be producing a truncated protein product and that this product does not contribute to metastasis or proliferation but gives the cell a survival advantage in the bursa.
Although we identified many clusters of integration that appear to be driving ALV-induced lymphomagenesis, it is important to note that integration clusters do not necessarily have to arise by selection postintegration. It is possible, for example, that some clusters could form due to preferential ALV integrase targeting in the chicken genome, although this has not previously been seen. Clearly, in some cases, selection appears to be driving clustering. For example, when bias for integration in a specific orientation or location within a gene is observed, selection is likely involved.
While ALV-A induces lymphoid neoplasms, ALV-J is known to induce myeloid neoplasms and hemangiomas. We recently reported integrations in ALV-J-induced hemangiomas, and interestingly we see very little overlap between the common integration sites in ALV-A-induced lymphoid tumors and ALV-J-induced hemangiomas. The only gene that appears to be shared as a common integration site between the two studies is ELF1, which was the second-most-frequently targeted gene in ALV-J hemangiomas and the 13th-most-frequent target of integration in ALV-A lymphoid tumors. The striking lack of overlap between these data sets is likely due to the biological differences between the types of cells affected and the genes involved in inducing lymphomas versus hemangiomas.
Recent work characterizing HIV integrations identified BACH2 and MKL2 as common integration sites in individuals on suppressive combination antiretroviral therapy (cART) (48, 49). We identify BACH2 but not MKL2 as a common integration site in this study. In one earlier study, BACH2 integrations showed a strong preference for integration in the forward orientation (15/15 integrations), and 6 of 15 integrations were found in expanded clones. In ALV-induced lymphomas, we see a weaker preference for integration in the forward orientation (17/24 [70.8%]), with 5 of 24 present in clonally expanded cells. Although MKL2 was not a common integration site in our study, we did identify the related gene MKL1 as a common integration site. Both MKL1 and -2 are coactivators of the transcription factor serum response factor (SRF), which regulates genes involved in many biological processes, including cell growth and migration (50).
In conclusion, this study greatly expands the number of genes known to be common integration sites in ALV-induced B-cell lymphoma. As one might expect, many of the genes we identified have well-characterized roles in cancer and related processes. These genes include RUNX1, Mir-221, Mir-222, IKZF1, CCNA2, ZEB1, CBLB, and HMGB1, as well as many others. In addition to canonical cancer genes, we identified a number of genes as common integration sites that are conserved in humans but have never been linked to cancer. These include CXorf57, CTDSPL2, TMEM135, ZCCHC10, FAM49B, and MGARP. In fact, three of these six genes, CXorf57, ZCCHC10, and FAM49B, have never undergone any characterization and have no known functions. We think these genes as well as others that we identify in this study are interesting targets for further research.
MATERIALS AND METHODS
Tumor induction.
Five- and 10-day-old chicken embryos were injected with either ALV-LR9, ALV-ΔLR9, ALV-G919A, or ALV-U916A. The chickens injected at 5 days were SPAFAS embryos (Charles River) and were injected via the yolk sac route. The chickens injected at 10 days were inbred SC White Leghorn line embryos (Hy-Line International, Dallas Center, IA), and viruses were injected into the chorioallantoic veins as described previously (18). A total of 10 birds were infected on embryonic day 5, and 15 birds were infected on day 10. Chickens were observed daily and were euthanized when apparently ill or at 12 weeks (for the day-5-injected cohort) or 10 weeks (for the day-10-injected cohort). IACUC approval was obtained. A total of 37 tissues were selected for characterization by high-throughput sequencing (see Table S1 in the supplemental material). Two uninfected tissues and several non-tumor tissues from infected birds were sequenced to serve as controls (see Table S1). Additional birds were infected, but not all birds were analyzed in this study.
DNA extraction and deep sequencing.
DNA was isolated, and sequencing libraries were prepared as described previously (15). Briefly, 5 µg of purified genomic DNA was sonicated with a Bioruptor UCD-200. End repair, A-tailing, and adapter ligation were performed as described by Gillet et al. (22) (adapter short arm, P-GATCGGAAGAGCAAAAAAAAAAAAAAAA, and adapter long arm, CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T, where “X’s” denote the barcode sequence, “P” denotes phosphorylation, and “*” denotes a phosphorothioate bond). Nested PCR was performed to enrich the library for proviral junctions. The first PCR was 23 cycles and employed an ALV-A-specific primer (CGCGAGGAGCGTAAGAAATTTCAGG) between the 3′ LTR and env and a primer (CAAGCAGAAGACGGCATACGAGAT) within the adapter that was attached by ligation in the previous step. In the second round of PCR, a primer (AATGATACGGCGACCACCGAGATCTACACTCGACGACTACGAGCACATGCATGAAG) at the 3′ end of the LTR was used. This primer ended 12 nucleotides short of the junction between virus and genomic DNA. This primer was paired with an adapter-specific primer on the opposite side of the fragment, which overlapped the adaptor’s bar code sequence (CAAGCAGAAGACGGCATACGAGATXXXXXX). Libraries were quantified by quantitative PCR (qPCR) and then underwent single-end 75- or 100-bp multiplexed sequencing on the Illumina Hi-Seq 2000. A custom sequencing primer (ACGACTACGAGCACATGCATGAAGCAGAAGG) was used which hybridized near the end of the viral 3′ LTR, 5 nucleotides short of the proviral/genomic DNA junction. The resulting reads could be validated as genuine integrations by verifying that they began with the last 5 nucleotides of the proviral DNA, CTTCA. The last two nucleotides of the unintegrated proviral DNA, TT, are cleaved by ALV integrase upon integration, so the lack of these 2 nucleotides in the read acted as further validation of a true viral integration.
Sequence analysis.
Reads were first filtered with a custom Python script to remove sequences that did not begin with the last 5 nucleotides of viral DNA, CTTCA. Files were then uploaded to Galaxy (51–53), which was used to perform some downstream analyses. In Galaxy, the quality scores were first converted to Sanger format with FastQ Groomer v1.0.4 (54). Adapters were trimmed using the Galaxy Clip tool v1.0.1. This tool also removed reads containing an N and reads less than 20 nucleotides in length after adapter removal. The remaining reads were mapped with Bowtie (55), using the Gallus gallus 4.0 genome (November 2011). A total of 100,000 random mapped reads were selected from each sample to be used for further analysis. If less than 100,000 reads were present for a sample, all available reads were used.
A custom Perl pipeline was developed to analyze the aligned reads’ output from Bowtie. Briefly, reads containing sequencing errors were filtered, and read counts and sonication breakpoints were quantified. Integrations found in multiple samples were assigned to the sample with the highest number of breakpoints. Files were annotated with refseq features, and the orientation and distance to the nearest gene were calculated for each integration. Integrations into repetitive regions were then manually removed from the data set. In all, 32,050 unique ALV integrations were obtained. Integration clusters were identified via a sliding window approach. If 12 or more integrations were observed within a 50-kb window, they were considered a cluster of viral integration. If the cluster was located in or near a gene, all additional integrations in that gene were also counted, as were any integrations within 10 kb upstream or downstream of that gene. If the cluster encompassed two genes, both genes were recorded and any integrations between the two genes and within 10 kb of either end were included in the cluster. The source code for this pipeline is available upon request.
Consensus sequence, feature, and Gene Ontology analysis.
Reads were mapped with Bowtie (55). Only reads that mapped uniquely to the genome were kept, and any reads that mapped equally well to two locations were discarded. This step filtered out reads that originate from repetitive elements. Mapped reads from all samples were then combined into a single file and analyzed with HOMER (40). HOMER calculates the nucleotide composition and enriched features at each integration locus. A random integration control data set was generated with Bedtools Random (56). The genomic DNA sequences corresponding to the genomic coordinates obtained from Bedtools Random were extracted from the Gallus gallus 4 genome using the Galaxy tool Extract Genomic DNA (51–53). Control sequences were mapped with Bowtie and analyzed with HOMER using the same conditions as above. A consensus Logo plot was constructed with Seq2Logo (57). Gene Ontology analysis for the top 48 clusters of integration was conducted with DAVID (37, 58).
SUPPLEMENTAL MATERIAL
Two liver tumors exhibited multiple unique integrations within CTDSPL and CTDSPL2. The tumors G-158-L and B-256-L are each represented by a bar chart. Each bar represents a unique viral integration, and the height of the bar corresponds to the number of sonication breakpoints observed for that integration (which is a measure of clonal expansion). Multiple high-breakpoint CTDSPL (red) and CTDSPL2 (blue) integrations can be seen in these tumors. In addition, 5 CTDSPL and 8 CTDSPL2 one- or two-breakpoint integrations were observed for G-158-L, and 1 each CTDSPL and CTDSPL2 single-breakpoint integration was observed for B-256-L (not shown). Download
Gene Ontology analysis. Enriched Gene Ontology terms identified by DAVID for the top 48 common integration sites are shown. Download
Birds infected and neoplasms observed. T, B-cell lymphoma; NF, neoplastic follicles; I, inflammation; H, hemangioma; X, tissue collected and no neoplasm observed. Shaded boxes represent samples that underwent high-throughput sequencing. A superscript 1 indicates MYB integration identified by nested PCR. R794 bursa MYB rearrangement was confirmed by Southern blotting (20). A superscript 2 indicates TERT integration observed via inverse PCR and confirmed via Southern blotting (21).
Expanded statistics for common integration sites. For each common integration site, the number of integrations is shown, followed by the nearest gene. If the integration cluster encompassed two genes, they are both listed. Integration density is the number of integrations observed per kilobase for the area of the integration cluster. “Avg. BP” is the mean number of breakpoints observed per integration in the cluster. “Tissue” refers to the tissue that harbored the integration. “Injection” refers to the age of the chicken embryo when it was infected with ALV: 5- or 10-day embryos were used in this experiment. “Virus” refers to the strain of virus that produced each integration (see Materials and Methods for details). “Orientation” refers to the orientation of the provirus with respect to the nearest gene. If the integration cluster encompasses two genes and they are in opposite orientations, the orientation with respect to each gene in the cluster is shown.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01CA124596, R01CA48746, and T32GM007231.
We thank Paul Neiman, Sandra Bowers, Miguel Ruano, Erin Bernburg, Amy Anderson, Grace Isaacs, Milos Markis, and Grace Lagasse for help with chickens.
Footnotes
Citation Justice JF, IV, Morgan RW, Beemon KL. 2015. Common viral integration sites identified in avian leukosis virus-induced B-cell lymphomas. mBio 6(6):e01863-15. doi:10.1128/mBio.01863-15.
REFERENCES
- 1.Justice J IV, Beemon KL. 2013. Avian retroviral replication. Curr Opin Virol 3:664–669. doi: 10.1016/j.coviro.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell RS, Beitzel BF, Schroder ARW, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J Virol 78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. 2005. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J Virol 79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. 2005. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol 79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holman AG, Coffin JM. 2005. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci U S A 102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadly AM, Nair V. 2008. Leukosis/sarcoma group, p 514–568. In Saif YM, Fadly AM, Glisson JR, McDougald LK (ed), Diseases of poultry, 12th ed. Blackwell Publishing, Hoboken, NJ. [Google Scholar]
- 8.Hayward WS, Neel BG, Astrin SM. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475–480. [DOI] [PubMed] [Google Scholar]
- 9.Payne GS, Bishop JM, Varmus HE. 1982. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 10.Jiang W, Kanter MR, Dunkel I, Ramsay RG, Beemon KL, Hayward WS. 1997. Minimal truncation of the c-myb gene product in rapid-onset B-cell lymphoma. J Virol 71:6526–6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clurman BE, Hayward WS. 1989. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol 9:2657–2664. doi: 10.1128/MCB.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam W, Ben-Yehuda D, Hayward WS. 1997. Bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol 17:1490–1502. doi: 10.1128/MCB.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanter MR, Smith RE, Hayward WS. 1988. Rapid induction of B-cell lymphomas: insertional activation of c-myb by avian leukosis virus. J Virol 62:1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Liu X, Yang Z, Xu C, Liu D, Qin J, Dai M, Hao J, Feng M, Huang X, Tan L, Cao W, Liao M. 2014. The MYC, TERT, and ZIC1 genes are common targets of viral integration and transcriptional deregulation in avian leukosis virus subgroup J-induced myeloid leukosis. J Virol 88:3182–3191. doi: 10.1128/JVI.02995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Justice J IV, Malhotra S, Ruano M, Li Y, Zavala G, Lee N, Morgan R, Beemon K. 2015. The MET gene is a common integration target in avian leukosis virus subgroup J-induced chicken hemangiomas. J Virol 89:4712–4719. doi: 10.1128/JVI.03225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith MR, Smith RE, Dunkel I, Hou V, Beemon KL, Hayward WS. 1997. Genetic determinant of rapid-onset B-cell lymphoma by avian leukosis virus. J Virol 71:6534–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Sullivan CT, Polony TS, Paca RE, Beemon KL. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses src mRNA splicing and promotes polyadenylation. Virology 302:405–412. doi: 10.1006/viro.2002.1616. [DOI] [PubMed] [Google Scholar]
- 18.Polony TS, Bowers SJ, Neiman PE, Beemon KL. 2003. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J Virol 77:9378–9387. doi: 10.1128/JVI.77.17.9378-9387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilusz JE, Beemon KL. 2006. The negative regulator of splicing element of Rous sarcoma virus promotes polyadenylation. J Virol 80:9634–9640. doi: 10.1128/JVI.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neiman PE, Grbiç JJ, Polony TS, Kimmel R, Bowers SJ, Delrow J, Beemon KL. 2003. Functional genomic analysis reveals distinct neoplastic phenotypes associated with c-myb mutation in the bursa of Fabricius. Oncogene 22:1073–1086. doi: 10.1038/sj.onc.1206070. [DOI] [PubMed] [Google Scholar]
- 21.Yang F, Xian RR, Li Y, Polony TS, Beemon KL. 2007. Telomerase reverse transcriptase expression elevated by avian leukosis virus integration in B cell lymphomas. Proc Natl Acad Sci U S A 104:18952–18957. doi: 10.1073/pnas.0709173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillet NA, Malani N, Melamed A, Gormley N, Carter R, Bentley D, Berry C, Bushman FD, Taylor GP, Bangham CRM. 2011. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood 117:3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CRM, Bushman FD. 2012. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics 28:755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wertz IE. 2014. TNFR1-activated NF-κB signal transduction: regulation by the ubiquitin/proteasome system. Curr Opin Chem Biol 23:71–77. doi: 10.1016/j.cbpa.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Balkwill F. 2006. TNF-alpha in promotion and progression of cancer. Cancer Metastasis Rev 25:409–416. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Zhu B, Davie J. 2015. Alternative splicing of MEF2C pre-mRNA controls its activity in normal myogenesis and promotes tumorigenicity in rhabdomyosarcoma cells. J Biol Chem 290:310–324. doi: 10.1074/jbc.M114.606277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sørensen AB, Duch M, Amtoft HW, Jørgensen P, Pedersen FS. 1996. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J Virol 70:4063–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. 2002. New genes involved in cancer identified by retroviral tagging. Nat Genet 32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. 2006. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J 25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. 2004. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res 32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashuba VI, Li J, Wang F, Senchenko VN, Protopopov A, Malyukova A, Kutsenko AS, Kadyrova E, Zabarovska VI, Muravenko OV, Zelenin AV, Kisselev LL, Kuzmin I, Minna JD, Winberg G, Ernberg I, Braga E, Lerman MI, Klein G, Zabarovsky ER. 2004. RBSP3 (HYA22) is a tumor suppressor gene implicated in major epithelial malignancies. Proc Natl Acad Sci U S A 101:4906–4911. doi: 10.1073/pnas.0401238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senchenko VN, Anedchenko EA, Kondratieva TT, Krasnov GS, Dmitriev AA, Zabarovska VI, Pavlova TV, Kashuba VI, Lerman MI, Zabarovsky ER. 2010. Simultaneous down-regulation of tumor suppressor genes RBSP3/CTDSPL, NPRL2/G21 and RASSF1A in primary non-small cell lung cancer. BMC Cancer 10:75. doi: 10.1186/1471-2407-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, Zhang H, Zhang X, Feng D, Luo X, Zeng C, Lin K, Zhou H, Qu L, Zhang P, Chen Y. 2012. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene 31:80–92. doi: 10.1038/onc.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Xiao M, Sun B, Zhang Z, Shen T, Duan X, Yu PB, Feng X, Lin X. 2014. C-terminal domain (CTD) small phosphatase-like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via Smad dephosphorylation. J Biol Chem 289:26441–26450. doi: 10.1074/jbc.M114.568964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brill SJ, Stillman B. 1991. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev 5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 37.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Oh J, Chang KW, Alvord WG, Hughes SH. 2006. Alternate polypurine tracts affect Rous sarcoma virus integration in vivo. J Virol 80:10281–10284. doi: 10.1128/JVI.00361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh J, Chang KW, Hughes SH. 2006. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integrations in Vivo. J Virol 80:451–459. doi: 10.1128/JVI.80.1.451-459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cattoglio C, Pellin D, Rizzi E, Maruggi G, Corti G, Miselli F, Sartori D, Guffanti A, Di Serio C, Ambrosi A, De Bellis G, Mavilio F. 2010. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, Li Y, Crise B, Burgess SM. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 43.Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, Green PL, Wu L, Roth MJ, Bushman FD, Kvaratskhelia M. 2013. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A 110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman F, Landuyt B, Husson S, Busschots K, Gijsbers R, Debyser Z. 2013. The BET family of proteins targets Moloney murine leukemia virus integration near transcription start sites. Cell Rep 5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aiyer S, Swapna GVT, Malani N, Aramini JM, Schneider WM, Plumb MR, Ghanem M, Larue RC, Sharma A, Studamire B, Kvaratskhelia M, Bushman FD, Montelione GT, Roth MJ. 2014. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction. Nucleic Acids Res 42:5917–5928. doi: 10.1093/nar/gku175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaFave MC, Varshney GK, Gildea DE, Wolfsberg TG, Baxevanis AD, Burgess SM. 2014. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res 42:4257–4269. doi: 10.1093/nar/gkt1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba TW, Humphries EH. 1985. Formation of a transformed follicle is necessary but not sufficient for development of an avian leukosis virus-induced lymphoma. Proc Natl Acad Sci U S A 82:213–216. doi: 10.1073/pnas.82.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. 2007. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis 195:716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 49.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pipes GCT, Creemers EE, Olson EN. 2006. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev 20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 51.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19:Unit 19.10.1–19.10.21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goecks J, Nekrutenko A, Taylor J, Galaxy Team . 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A, Galaxy Team . 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26:1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomsen MCF, Nielsen M. 2012. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res 40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two liver tumors exhibited multiple unique integrations within CTDSPL and CTDSPL2. The tumors G-158-L and B-256-L are each represented by a bar chart. Each bar represents a unique viral integration, and the height of the bar corresponds to the number of sonication breakpoints observed for that integration (which is a measure of clonal expansion). Multiple high-breakpoint CTDSPL (red) and CTDSPL2 (blue) integrations can be seen in these tumors. In addition, 5 CTDSPL and 8 CTDSPL2 one- or two-breakpoint integrations were observed for G-158-L, and 1 each CTDSPL and CTDSPL2 single-breakpoint integration was observed for B-256-L (not shown). Download
Gene Ontology analysis. Enriched Gene Ontology terms identified by DAVID for the top 48 common integration sites are shown. Download
Birds infected and neoplasms observed. T, B-cell lymphoma; NF, neoplastic follicles; I, inflammation; H, hemangioma; X, tissue collected and no neoplasm observed. Shaded boxes represent samples that underwent high-throughput sequencing. A superscript 1 indicates MYB integration identified by nested PCR. R794 bursa MYB rearrangement was confirmed by Southern blotting (20). A superscript 2 indicates TERT integration observed via inverse PCR and confirmed via Southern blotting (21).
Expanded statistics for common integration sites. For each common integration site, the number of integrations is shown, followed by the nearest gene. If the integration cluster encompassed two genes, they are both listed. Integration density is the number of integrations observed per kilobase for the area of the integration cluster. “Avg. BP” is the mean number of breakpoints observed per integration in the cluster. “Tissue” refers to the tissue that harbored the integration. “Injection” refers to the age of the chicken embryo when it was infected with ALV: 5- or 10-day embryos were used in this experiment. “Virus” refers to the strain of virus that produced each integration (see Materials and Methods for details). “Orientation” refers to the orientation of the provirus with respect to the nearest gene. If the integration cluster encompasses two genes and they are in opposite orientations, the orientation with respect to each gene in the cluster is shown.