Abstract
Influenza virus infection induces several changes in host miRNA profile, host cell death and tissue damage. Cytochrome c is a regulator of the intrinsic apoptotic pathway and is altered during viral infections. Within the first 3 h of infection with influenza virus, significant down-regulation of hsamiRNA-4276 (miRNA-4276) is followed by a 2-fold increase in cytochrome c oxidase VIC (COX6C) mRNA was found to occur in human alveolar and bronchial epithelial cells. Expression of caspase-9 also increased within the first 3 h of infection, but subsequently decreased. Modulation of miR-4276 using mimic and inhibitor oligonucleotides showed significant down-regulation or up-regulation, respectively, of COX6C expression. Our data suggests that on initial exposure to influenza virus, host cells upregulate COX6C mRNA expression through silencing miR-4276 and repressed viral replication by inducing the apoptotic protein caspase-9. Taken together, these data suggest that miR-4276 may be an important regulator of the early stages of infection by influenza.
Keywords: miRNA, COX6C, Influenza virus, Lung epithelial cells
Introduction
Influenza virus A (H1N1) infection results in a highly contagious respiratory illness that damages the epithelium cells in the human respiratory tract and cause death of infected cells (Thompson et al., 2004; Webster and Rott, 1987). Cell death is an integral part of the host defense against influenza virus infection (Yatim and Albert, 2011). Generation of progeny virus particles by the membrane of the infected cells may also trigger cellular damage. Cellular degeneration can occur even with incomplete replication of the virus in tissue culture cells (Takizawa et al., 1993), however, the mechanism of virus-induced cellular damage that eventually leads to cell death is not well understood. Influenza virus induces cell death through activation of apoptosis, a process regarded as a major contributor to influenza virus pathogenesis resulting in extensive lung tissue damage (Brydon et al., 2005; Ludwig et al., 2006).
Apoptosis or programmed cell death is considered to be a host cellular defense mechanism against the invading pathogens that involve the self-destruction of the cells. Apoptosis plays an important role in the pathogenesis of many infectious diseases including several viruses (Collins, 1995; Ludwig et al., 2006; Razvi and Welsh, 1995; Roulston et al., 1999; Schultz-Cherry et al., 2001). Two types of programmed cell death are known, the extrinsic and intrinsic apoptotic pathways. The extrinsic pathway is initiated from the cell surface death receptors (Fas, TNF) and involves the activation of caspase-8 (Danial and Korsmeyer, 2004), whereas the intrinsic pathway is triggered by changes in mitochondrial integrity and involves the release of cytochrome c oxidase into the cytosol activating caspase-9. The intrinsic pathway is initiated by BcL2, Bcl-XL, Bax, ROS, and p53 (Duprez et al., 2009). Cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, catalyzes the electron transfer from reduced cytochrome c to oxygen. The mammalian cytochrome c oxidase consists of 13 subunits (Grossman and Lomax, 1997). Of these, 3 catalytic subunits are encoded by mitochondrial genes while the multiple structural subunits are encoded by nuclear genes. The mitochondrial encoded subunits function in electron transfer, and the nuclear-encoded subunits appear to be involved in the regulation and assembly of the complex. The specific cytochrome c oxidase subunits involved in apoptosis mediated by influenza virus have not yet been identified and little is known of their involvement in protecting against the virus.
Several studies have shown that influenza virus can manipulate cell death signaling pathways to promote viral replication (Lin et al., 2001; Ludwig et al., 2006; Wurzer et al., 2003). Influenza virus infection induces the up-regulation of several pro-apoptotic factors while inhibition of TRAIL and Fas resulted in increased viral copy numbers (Wurzer et al., 2004). Similarly, respiratory syncytial virus (RSV) and rhinovirus (RV-16) infections are also known to induce anti-apoptotic proteins, such as nerve growth factor (NGF) (Othumpangat et al., 2009; Othumpangat et al., 2012). Earlier studies demonstrated that ectopically overexpressed anti-apoptotic protein BcL-2 resulted in impaired virus production and inhibition of influenza induced apoptosis (Hinshaw et al., 1994). Influenza virus depends on host genes for replication (Karlas et al., 2010). Cellular defense against influenza viral infection involves host proteins that repress viral protein synthesis resulting in reduced infectivity and propagation. Cellular proteins can be regulated by micro-RNAs (miRNA) which are endogenously synthesized in the cell (Bartel, 2004). Target mRNA degradation is the predominant form of miRNA mediated translational repression (Lim et al., 2005). Studies are lacking information regarding the mechanism of regulation of apoptosis in influenza infected cells; especially miRNA mediated host defense mechanisms.
In this study, we investigate the effects of infection with influenza virus on the intrinsic pathway of apoptosis using primary human bronchial epithelial cells (HBEpC) and A549 cells. We observed that cytochrome c oxidase subunit VI c (COX6C) was regulated by the expression of hsa-miR-4276 (miRNA-4276). COX6C in turn regulated the expression of caspase-9, one of the key proteins in apoptosis that was also studied.
Materials and methods
Cell culture
Primary human bronchial epithelial cells (HBEpCs) were purchased from Promo Cells (Hamburg, Germany), and subcultured per manufacturer recommended conditions in bronchial epithelial cell growth media. Undifferentiated HBEpCs were grown in submerged culture and passaged once confluence reached 80%. Low passage cells (P 2–6) were used throughout the experiments. Alveolar epithelial cells, A549 (CCL-34, American Type Culture Collection [ATCC, Manassas, VA] were cultured in standard F12K medium containing 10% heat-inactivated fetal bovine serum (FBS), 100 IU/ml penicillin and 100 μg/ml streptomycin sulfate. Madin-Darby canine kidney (MDCK) cells were used for the propagation of influenza virus. MDCK cells were cultivated in MEM (ATCC) supplemented with 10% FBS and100 IU/ml penicillin and 100 μmg/ml streptomycin sulfate.
Influenza strain A/WS/33 (H1N1, ATCC VR-a) and A/Aichi/2/68 (H3N2- ATCC 1680) were purchased from the ATCC and maintained as described (Blachere et al., 2011; Blachere et al., 2009).
Viral infections
All infections of HBEpCs and A549 cells were performed in 6-well plates at a dose of 1.0 multiplicity of infection (MOI) unless otherwise specified. Control cells were mock infected and referred to as Mock. Six well plates were seeded with 5 × 104 cells per well and grown to 80% confluence. Prior to infection, the cells were rinsed with PBS, and then virus diluted in MHBSS (Modified Hank's Balanced Salt Solution) was added to each well. After 45 min of incubation at 37 °C, excess virus solution was rinsed off using PBS and fresh F12 media was added containing 1 μg/ml of TPCK-trypsin (Sigma-Aldrich, St Louis, MO) and incubated at 37°C and 5% CO2. Cells were harvested at different time intervals and used for protein and RNA studies.
RNA isolation
MicroRNA from A549 cells was isolated using the miRCURY™ RNA extraction kit (Exiqon) and the quantity of total RNA was quantified by Nanodrop ND1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). cDNA was synthesized using the universal cDNA kit (Exiqon) with forward and reverse primer for miRNA4276 purchased from Exiqon.
Total RNA from A549 and HBEpC cells was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA) according to manufacturer's instructions. After quantification by Nanodrop, 1ug of purified RNA was used to generate cDNA using the High capacity ABI-RT PCR kit (Applied Biosystems) in accordance with the provider's protocol.
MicroRNA microarray analysis
A549 cells were infected with influenza virus (3 MOI) for 3 h and RNA was extracted from infected and control cells using the Exiqon miRCURY LNA miRNA extraction kit (Exiqon) and then sent to Exiqon for Microarray analysis (Vedbaek, Denmark). The quality of the total RNA was verified by an Agilent 2100 Bio-analyzer profile. RNA from both sample and reference was labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY LNA™ microRNA Hi-Power Labeling Kit, Hy3™/Hy5™ (Exiqon) following the procedure described by the manufacturer. The Hy3™-labeled samples and a Hy5™-labeled reference RNA sample were mixed pair-wise and hybridized to the miRCURY LNA™ microRNA Array 7th generation, which contains capture probes targeting all microRNAs for human, mouse or rat registered in the miRBASE 16.0. The hybridization was performed according to the miRCURY LNA™ microRNA Array instruction manual using a Tecan HS4800™ hybridization station (Tecan, Mannedorf, Switzerland). After hybridization, the microarray slides were scanned and stored in an ozone free environment (ozone level below 2.0 ppb) in order to prevent potential bleaching of the fluorescent dyes. The miRCURY LNA™microRNA Array slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., Santa Clara, CA) and the image analysis was carried out using the ImaGene® 9 (miRCURY LNA™ microRNA Array Analysis Software, Exiqon). The data obtained were subjected to statistical analysis and differentially regulated miRNAs in infected and uninfected cells were reported.
Quantitative real-time PCR
Real time quantitation of miRNA was done using power SYBR green PCR master mix on a real time PCR 7500 ABI (Applied Biosystems, Foster City, CA). miRNA let-7 was used as the internal control. The fold change in expression of miRNA was calculated using the ddCt method as described for gene analysis (Othumpangat et al., 2012).
Influenza M gene expression was quantified and reported as influenza copy number. QPCR was performed using TaqMan assay using matrix-specific primers, as reported earlier (Blachere et al., 2011; Spackman et al., 2002). All primers and probes were custom synthesized by Applied Biosystems. To determine the relative matrix gene copy number from influenza A strains, a standard curve was generated from the cloned influenza H1N1 matrix gene and analyzed concurrently with RT-PCR reactions (Blachere et al., 2009).
PCR array
PCR array for the expression and identification of different cytochrome oxidases that are targeted by influenza virus was studied by employing the RT2 Profiler™ PCR Array Human Mitochondrial Energy Metabolism (PAHS 008Y, Qiagen) using SYBR green. QPCR data analyzed using the supplier's online software (Qiagen).
RT-PCR
Real time PCR reactions were carried out using the TaqMan FAST PCR reagent (Applied Biosystems) and the ABI 7500 FAST real-time cycler (Applied Biosystems). TaqMan primers for COX6C, Caspase-9, and glyceraldehyde phosphate dehydrogenase (GAPDH) were purchased from Applied Biosystems. Gene expression was normalized using GAPDH as the housekeeping gene, and is reported as fold change. The change in gene expression was calculated using the formula: fold change=2−(ddCt), ddCt=dCt (H1N1 infected) - dCt (mock infected), where dCt=Ct (detected gene) - Ct (GAPDH) and Ct is the threshold number).
Transfection studies
HBEpC and A549 cells were transfected with miRNA-4276 inhibitor oligonucleotide or a miRNA-4276 mimic oligonucleotide (Life Technologies, Carlsbad, CA) using the lipid-based Lipofectamine 2000 reagent diluted in Opti-MEM-I reduced serum medium (Life Technologies) according to the suppliers protocol. Briefly, HBEPC or A549 cells were grown to 80% confluence in 6-well plates and the transfection complexes were directly applied to the cells to a final concentration of 25 nM. After 6 h of transfection, the medium was replaced with fresh complete medium. As a negative control, cells were transfected with the same concentrations of scrambled oligonucleotides (SCR control) (Life Technologies). These cells were then exposed to various MOIs of virus, by following standard infection protocols for influenza virus. Following the incubation, cells were harvested and used for RNA extraction using RNeasy kit (Qiagen) or for Western blot analysis.
Imaging with confocal microscopy
A549 cells were grown to confluence on chamber slides (Chamber slide™, Lab-TekII, Thermo Fisher Scientific, Rochester, NY) and transfected with the miRNA-4276 inhibitor, miRNa-4276 mimic or a non-specific scrambled oligonucleotide (SCR) as a negative control. The transfected cells were exposed to influenza virus (H1N1). After washing with PBS, cells were fixed with 4% methanol-free formaldehyde (Polysciences Inc., Warrington, PA), permeabilized with 0.5% Triton x100 (Sigma) and blocked with Image iTx (Life Technology) for 20 min. Slides were washed with PBS and blocked with 5% bovine serum albumin (BSA). Cells were then stained for 1 h with rabbit anti-COX6C antibody (Santa Cruz Biotechnology, CA), and mouse anti-influenza A nucleoprotein antibody (Millipore, Billerica, MA) followed by Alexa-488 conjugated anti-rabbit secondary antibody and Alexa-546 conjugated anti-mouse antibody (Life technology). The glass slides were mounted with DAPI-Prolong Gold anti-fade reagent (Life technology) and protected with cover slips. Images were obtained using a Zeiss LSM510 confocal microscope with the AxioImager system (Carl Zeiss, Obertochen, AG Germany).
Western immunoblotting
At specific time points after influenza virus infection cells were lysed using RIPA buffer (Thermo scientific) containing complete protease inhibitor cocktail. Thirty micrograms of lysate was solubilized in sample buffer and subjected to electrophoresis on 10% SDS-polyacrylamide gels (SDS-PAGE, Pre-cast gels, BioRad). Gels were then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore) overnight (15 V at 4 °C). Following transfer, immunoblot analysis was performed. Briefly, membranes were blocked using Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE) for 1 h at room temperature and washed (3×10 min) with Tris-buffered saline (pH 7.2) containing 0.5% (v/v) Tween-20 (TBST) and blotted with COX6C anti-mouse monoclonal antibodies to COX6C, p-caspase-9 anti-rabbit polyclonal antibodies (Santa Cruz, CA), and GAPDH anti-mouse monoclonal antibody (Abcam, MA). Immunoblot was then washed with TBST (1 × 5 min; 3 × 10 min) and incubated for 1 h at room temperature with appropriate IRDye 680 or 800 secondary antibodies (LI-COR Biosciences). The membranes were protected by keeping in a closed container to avoid photo-bleaching of the fluorescent dyes. Membranes were washed (3×10 min) in TBST, followed by washes in TBS. Near-infrared fluorescence detection was performed on the Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE), and the fluorescent signal intensities of the individual bands were determined and normalized to the endogenous control GAPDH (36 kDa).
Statistical analysis
One-way analysis of variance (ANOVA) was used to analyze MOI studies as well as studies of mimic and inhibitor on miRNA-4276 and COX6C expression, and post-hoc pairwise multiple comparisons between means were performed using the Holm–Sidak method with a p Value of <0.05 considered statistically significant using Sigma stat version 11.0 for Windows (Systat Software, Chicago, IL).
Results
Microarray screening for miRNA and cytochrome C subunits
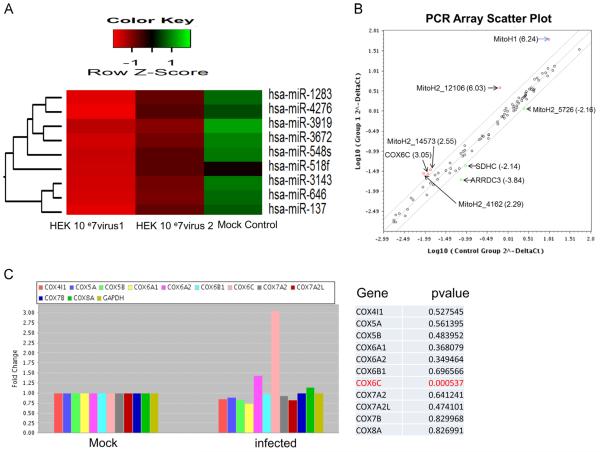
Previous studies from our laboratory (Othumpangat et al., 2013) have shown that the levels of influenza non-structural 1A binding protein (IVNS1ABP) changed significantly in A549 cells exposed to influenza virus for 3 h. Examining early stage infection addresses the primary response of the host cells in defending the invading virus. MicroRNA expression profiling using locked nucleic acid (LNA) based miRNA array on A549 cells infected with influenza virus (MOI 3) showed significantly lower expression of several miRNAs in infected cells (Fig. 1A). Microarray data analysis (Exiqon) provided differential expression of the top 49 miRNAs, of which 10 were significantly downregulated. In parallel, we also analyzed A549 cells infected with influenza virus using the RT2 Profiler™ PCR Array (Human Mitochondrial Energy Metabolism) to analyze 86 genes of mitochondrial metabolism including 11 cytochrome subunits. A scatter plot representing the 86 genes that were analyzed in cells infected with influenza virus compared to the mock controls is shown in Fig. 1B. Fig. 1C shows the expression pattern of cytochrome C subunits on exposure to influenza virus. Only the expression of COX6C was significantly increased (p<0.0005), though COX6A2 showed a slight increase in expression, but was not statistically significant. The data from the microarray and PCR array were analyzed to find which miRNAs are significantly down-regulated as well as correspond to the genes that are overexpressed from the PCR array. We searched the Targetscan database (www.targetscan.org) to identify selected miRNAs that are target for the overexpressing or down-regulating genes from the PCR array. Of the individual miRNAs examined, we found that miRNA-4276 targets COX6C which was downregulated resulting in a corresponding up-regulation of gene expression in PCR array. No associations with any other genes of the PCR array were observed. Till date, no subunits of cytochrome C have been identified as being specifically regulated after influenza virus infection.
Fig. 1.
Influenza virus infection induced changes in miRNA expression: A) Cluster analysis of influenza virus altered miRNA expression in A549 cells. Microarray analysis for miRNA was performed with RNA extracts from influenza virus infected A549 cells for 3 h. The expression index or color key with the ROW-Z-score is on the top representing degree of abundance in each cluster. Green shaded areas indicates higher expression of the specific miRNA, whereas the red shaded areas indicates lower expression and black denotes the relatively similar expression between infected and mock infected cells. HEK 10 07 virus1 & 2 are the replicates of cells infected with influenza virus. B) Scatter plot showing the expression profile of PCR array analysis for Mitochondrial energy metabolism (RT2 Profiler™ PCR Array) was performed with RNA extracts from influenza virus inflected A549 cells. The fold change is given in brackets. MitoH1-Polycistronic H1, MitoH2-12106 - Polycistronic_H2_12106, MitoH2_14573 - Polycistronic H2_57263, MitoH2-5726 - Polycistronic_H2200-5726, MitoH2_4162 - Polycistronic _H2_4162, ARRDC3- Arrestin domain containing 3, COX6C- cytochrome c oxidase subunit VI C, SDHC-Succinate dehydrogenase complex. C) PCR array analysis (RT2 Profiler™ PCR Array) was performed with RNA extracts from influenza virus infected A549 cells. GAPDH was used as the internal control. Data for cytochrome c oxidase subunits are shown with the p Values.
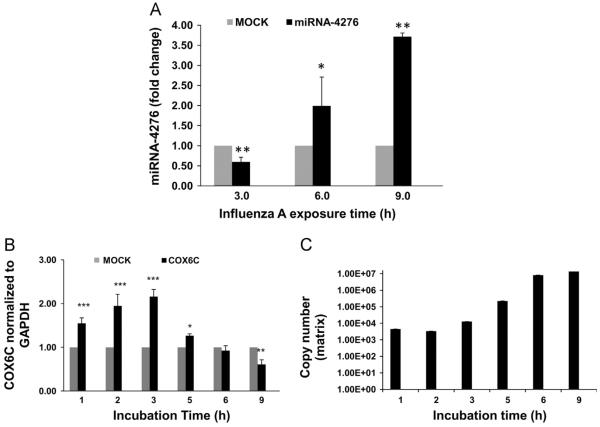
Influenza mediated expression of miRNA-4276 and its role in regulating COX6C was further assessed by infecting A549 cells with influenza virus (H1N1) for 9 h and sampling at 3 h intervals (Fig. 2A). At early stages of infection (3 h), miRNA-4276 expression was significantly downregulated (p<0.01). With increasing exposure time (beyond 3 h) expression of miRNA-4276 gradually increased and peaked at 9 h (3.75 fold). There was a gradual increase in expression of COX6C that correlated with decreased expression of miRNA-4276 (Fig. 2B). At 3 h after infection, there was a 2.2-fold increase (p<0.001) in COX6C mRNA expression which correlated with down-regulation of miR-4276, whereas beyond 3 h COX6C expression declined in concordance with increased miR-4276 expression. Down-regulation of COX6C was significant at 9 h (p<0.01) of exposure in agreement with increased expression of miR-4276. The efficiency of viral replication (matrix gene copy number) gradually increased with down-regulation of COX6C mRNA expression beyond 3 h of exposure (Fig. 2C), suggesting a possible role of the miRNA-4276 and COX6C in influenza virus replication.
Fig. 2.
miRNA 4276 targets COX6C expression: A) A549 cells were infected with influenza virus with MOI of 3 for 9 h were sampled every 3 h, miRNAs extracted and then analyzed by qPCR. Let-7 was used as the internal control. Data from three independent experiments. *=p<0.05, **=p<0.01. B) A549 cells were infected with influenza virus for 9 h and samples were withdrawn at the indicated time points. cDNA synthesized from 1 mg of extracted RNA and used for COX6C qPCR analysis. Data was normalized to GAPDH. Bar graphs represent data from three independent experiments (n=3). *=p<0.05, **=p<0.01. C) Matrix gene copy number was analyzed from the RNA extracted from A549cells infected with influenza virus, (n=3).
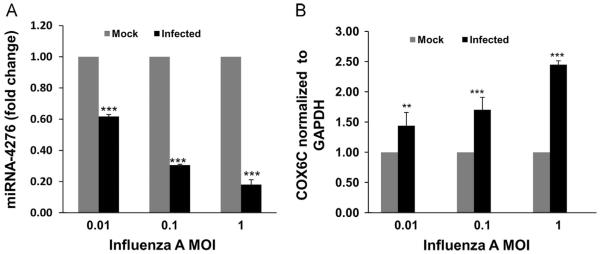
Down-regulation of miRNA-4276 (p<0.001, Fig. 3A) and up-regulation of COX6C mRNA (p<0.001, Fig. 3B) by influenza virus were infectious dose-dependent on the MOI. At the lowest MOI of 0.01, an inverse correlation of miRNA-4276 and COX6C expression was still evident. As the amount of infecting virus was increased to an MOI of 1, the expression of miRNA-4276 declined further with correspondingly increasing COX6C mRNA.
Fig. 3.
Changes in miR-4276 levels alter COX6C mRNA expression: A) miRNA4276 response to increasing MOIs of influenza virus. A549 cells were infected for 3 h with increasing MOIs of influenza. Let-7 was used as the internal control. Data from three independent experiments. ***=p<0.001. B) COX6C expression in response to increasing MOIs of influenza in A549 cells. GAPDH was used as the internal control. Data from three independent experiments. **=p<0.01, ***=p<0.001, (n=3).
COX6C expression regulated by miR-4276
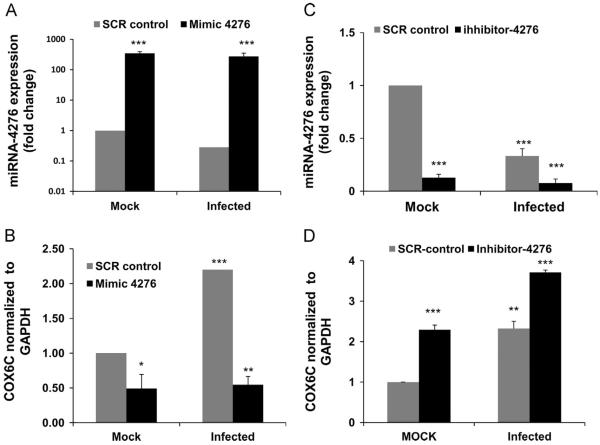
The role of miR-4276 in regulating expression of COX6C was confirmed by transfecting A549 cells with a mimic oligonucleotide specific for miR-4276. Transfection of miR-4276 mimic into mock infected A549 cells significantly increased the fold change in miRNA-4276 (p<0.001), confirming the stability of the miR-4276 mimic within the transfected cells (Fig. 4A). As expected, the level of miRNA-4276 decreases in infected cells transfected with the scrambled oligonucleotide control (SCR). Cells transfected with the mimic and then infected for 3 h showed an intermediate level of total miRNA-4276 consistent with the reduction of endogenous miR-4276 by influenza virus.
Fig. 4.
Down-regulation of miRNA 4276 induces COX6C expression: A) A549 cells were transfected for 48 h with 25 nM miRNA-4276 mimic or a scrambled oligonucleotide (SCR control). The transfected cells were then either mock infected or infected with 1 MOI influenza for an additional 3 h. Let-7 was used as the internal control. Data are expressed as7SEM, ***=p<0.001, (n=3). B) A549 cells transfected with mimic or SCR control were infected with 1 MOI of influenza virus for additional 3 h. The relative abundance of COX6C was measured in infected cells and the data normalized to the abundance of GAPDH. Data are expressed as±SEM, *=p<0.05, **=p<0.01 compared the cells transfected with the scrambled oligonucleotide, (n=3) C) A549 cells were transfected with 25 nM of either miRNA 4276 inhibitor or a SCR control. Relative expression of miRNA4276 was analyzed by qPCR. Data are expressed as±SEM, ***=p<0.001 compared the cells transfected with the scrambled oligonucleotide, (n=3). D) After 48 h, the transfected cells were infected with 1 MOI of influenza virus. The relative COX6C expression was measured in both infected and uninfected cells; GAPDH was used as the internal control. Data are expressed as7SEM, **=p<0.01 compared the cells transfected with the scrambled oligonucleotide, (n=3).
The mimic also significantly reduced (p=0.05) the level of COX6C mRNA in mock transfected cells (Fig. 4B). Consistent with the results shown in Fig. 2C, the level of COX6C mRNA increased ~2-fold (p=0.001) in infected cells transfected with the SCR oligonucleotide. Cells transfected with the mimic and then infected with influenza virus for 3 h show the lowest level of COX6C mRNA (p=0.01).
To further confirm that miRNA-4276 regulates expression of COX6C mRNA, A549 cells were transfected with the inhibitor oligonucleotide specific for miR-4276. Transfection of the inhibitor significantly reduced the expression of endogenous miRNA-4276 in uninfected (mock) cells (Fig. 4C). Expression of miR-4276 was further reduced (p<0.001) during the infection of influenza virus in cells already transfected with inhibitor of miR-4276.
Expression of COX6C mRNA increased 2.2-fold (p=0.001) in mock infected cells transfected with the inhibitor (Fig. 4D). Further, when inhibitor transfected cells were exposed to influenza virus, there was significant (p<0.01) increase (~3.7 fold) in the level of COX6C mRNA relative to in infected cells transfected with the SCR oligonucleotide (Fig. 4D).
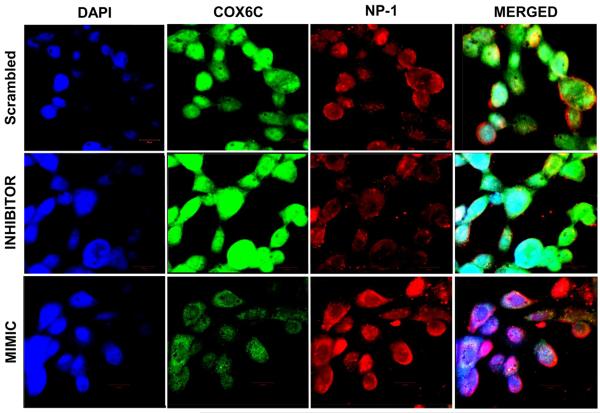
COX6C expression was examined using immunofluorescence microscopy. A549 cells were transfected with SCR, mimic, or inhibitor of miRNA-4276. The cells were then infected with influenza virus at an MOI of 1.0 for 3 h and stained with COX6C or anti-nucleoprotein-1 antibodies (Fig. 5). Regardless of the transfected oligonucleotide, infected A549 cells showed active COX6C expression as green fluorescence with a concomitant virus inflection (red fluorescence). The highest level of COX6C protein was seen in cells transfected with the miRNA inhibitor and the lowest amount of virus nucleoprotein (NP-1) was observed in cells transfected with the inhibitor suggesting a lower survival and/or replication of the virus (Panel 2). In contrast, COX6C expression was lowest and NP-1 expression was highest in cells transfected with the mimic (Panel 3), suggesting that the highest survival and/or replication of the virus is in cells with the lowest expression of COX6C.
Fig. 5.
Immunofluorescence of influenza A virus infected cells: A549 cells were transfected with either the scrambled oligonucleotide (SCR) (top panel), miRNA-4276 inhibitor (middle panel), or miRNA-4276 mimic (bottom panel). Transfected cells were infected with influenza virus and expression of COX6C protein (green) and influenza nucleoprotein (red) was determined by immunofluorescence. DAPI (blue) represents the stained nucleus of the cells.
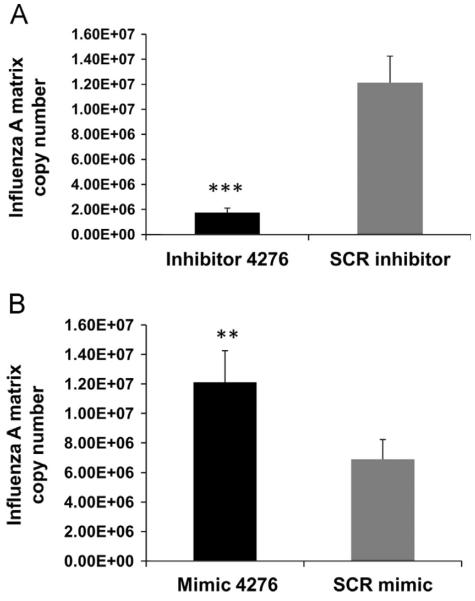
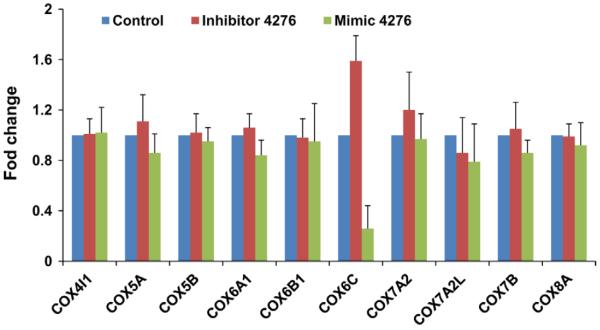
We then analyzed the matrix protein mRNA copy number in cells that were transfected with mimic or inhibitor and then infected with influenza virus. A549 cells transfected with inhibitor showed a ~10-fold reduction in viral copy number (p<0.001) compared to SCR transfected cells (Fig. 6A). The viral copy number in cells transfected with mimic was significantly higher (p<0.01) than the cells transfected with the scrambled oligonucleotide (Fig. 6B), suggesting that virus replication is favored with lower COX6C expression. The effect of miR-4276 appeared to be COX6C-specific as there was a ~3-fold reduction in mimic transfected cells and 1.5-fold increase in inhibitor transfected cells (1.5 fold) but little change was seen in expression of 9 other cytochrome C subunits (Fig. 7).
Fig. 6.
Abundance of COX6C reduced viral replication: A) Matrix copy number in cells transfected with either miRNA-4276 inhibitor or scrambled oligonucleotide is shown. Data are expressed as±SEM, **=p<0.001 (n=4). B) Matrix copy number in cells transfected with either miRNA-4276 mimic or scrambled oligonucleotide is shown. Data are expressed as±SEM, **=p<0.01 compared the cells transfected with the scrambled oligonucleotide, (n=3).
Fig. 7.
Specificity of miR-4276 to COX6C: Other cytochrome oxidase family of proteins was not affected by miR4276. RNA extracted from A549 cells transfected with mimic, inhibitor or a scrambled oligonucleotide used for the PCR array (RT2 Profiler™ PCR Array Human Mitochondrial Energy Metabolism) containing 11 cytochrome family protein was analyzed by qPCR (SYBR green).
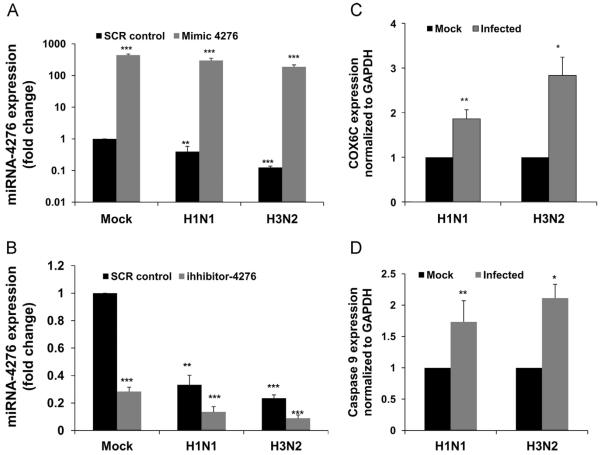
In order to rule out tissue tropism or influenza virus strain specificity for miR-4276 involvement with COX6C expression, we used primary human bronchial epithelial cells. The role of miR-4276 in regulating expression of COX6C was confirmed by transfecting HBEpCs with a mimic oligonucleotide specific for miR-4276. Transfection of miR-4276 mimic into mock infected HBEpCs significantly increased the fold change in miR-4276 expression (p<0.001, Fig. 8A). As expected, the level of miR-4276 decreases in infected cells transfected with the SCR control. Cells transfected with the mimic and then infected for 3 h with H1N1 showed a moderate level of total miR-4276, whereas H3N2 showed highest reduction in miR-4276 level. Consistent and similar results were obtained with inhibitor transfection followed by the infection with a H3N2 influenza strain (Fig. 8 B). We also analyzed the expression of COX6C mRNA in HBEpCs infected with H1N1 and H3N2 strains (Fig. 8C). With H1N1, the increase in COX6C (~2-fold) was comparable to that in A549 cells, and with H3N2, expression of COX6C (3-fold) was the highest. Data presented in Fig. 8C confirms that the COX6C response was consistent regardless of the cell type or influenza strain.
Fig. 8.
Influenza virus H1N1 and H3N2 alter miR-4276 levels in HBEpC cells and increase COX6C and Caspase-9 expression. A) HBEpC cells were transfected with 25 nM of either miRNA 4276 inhibitor or a SCR control. Relative expression of miRNA4276 was analyzed by qPCR. Data are expressed as±SEM, ***=p<0.001 compared the cells transfected with the scrambled oligonucleotide, (n=3). B) HBEpC cells were transfected for 48 h with 25 nM miRNA-4276 mimic or a scrambled oligonucleotide (SCR control). The transfected cells were then either mock infected or infected with 1 MOI influenza for an additional 3 h. Let-7 was used as the internal control. Data are expressed as±SEM, ***=p<0.001, (n=3). C) Primary human bronchial epithelial cells grown to 80% confluency were infected with influenza virus H1N1 or H3N2 of MOI of 1, for 3 h. COX6C expression was analyzed and normalized to GAPDH, Data are expressed as±SEM, *=p<0.05 and **=p<0.01, n=3 independent experiments. D) HBEpC cells grown to 80% confluency were infected with influenza virus H1N1 or H3N2 of MOI of 1, for 3 h, p-caspase-9 expression was analyzed and normalized to GAPDH, Data are expressed as±SEM, *=p<0.05 and **=p<0.01, n=3 independent experiments analyzed in duplicates.
Determining the primary pathway involved in virus replication requires assessment of signaling pathways that are linked to the host genes. In our previous studies (Othumpangat et al., 2013), we found that infection with influenza virus induced apoptosis. We examined 4 apoptotic pathway genes associated with the intrinsic pathway, and found caspase-9 level was significantly (p<0.01) increased in HBEpC cells infected with either H1N1 or H3N2 (Fig. 8D).
COX6C alter the expression of caspase-9
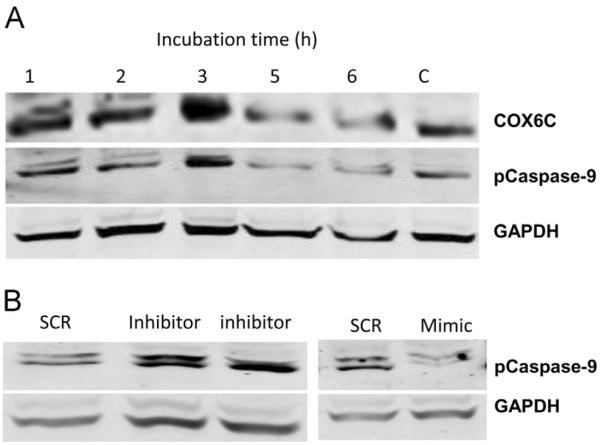
Cytochrome C release from the mitochondria is a hallmark of the intrinsic apoptosis pathway (Kadenbach, 2003; Villani et al., 1998). To determine whether the deficiency of COX6C suppresses the capacity of the influenza virus to induce the intrinsic pathway of apoptosis during infection, we analyzed the levels of caspase-9 in infected and control HBEpCs. COX6C protein level was increased during the early stages of infection (1–3 h) but decreased with greater exposure time. A dramatic increase in the expression of caspase-9 was observed at 1–3 hpi (hours post infection). Pro-caspase -9 levels changed during the time course of exposure to influenza virus and there was a gradual increase by 3 h of exposure and a decline by 6 h (Fig. 9A). To further confirm that COX6C involvement in caspase-9 regulation, we looked at caspase-9 levels in these cells by Western blot analysis. Fig. 9B shows that there was significant decrease in miR-4276 mimic transfected cells and a corresponding increase in cells that were transfected miR-4276 inhibitor, suggesting the possible modulation of apoptosis by cytochrome oxidase 6C.
Fig. 9.
COX6C expression alters the expression of apoptotic protein caspase-9. A) HBEpCs were exposed influenza virus for a time course and the expression of Cox6C and p-caspase-9 were analyzed using Western blot analysis. Primary human bronchial epithelial cells grown to 80% confluency were infected with influenza virus H1N1 or H3N2 of MOI of 1, for 3 h. pro-caspase 9 expression was analyzed and normalized to GAPDH, Data are expressed as±SEM, *=p<0.05 and **=p<0.01, n=3 independent experiments. B) Primary human bronchial epithelial cells grown to 80% confluence were transfected with mimic-4276, inhibitor or SCR control for 48 h and the pattern of p-caspase-9 analyzed by Western immunoblot analysis. GAPDH was used as a loading control.
Discussion
In this study, we evaluated miRNA-mediated regulation of apoptosis as a potential mechanism to eliminate influenza virus infected cells. Influenza virus induces cell death through activation of an apoptotic pathway, a process regarded as a major cause of influenza virus induced lung damage. Here we provide evidence that miRNA-mediated regulation of COX6C mRNA expression in primary human bronchial epithelial cells and lung epithelial cells leads to apoptosis in early stage infection. Influenza viruses failed to replicate or propagate efficiently in cells that overexpress COX6C protein (Fig. 6). Initial screening using microarray and real-time PCR revealed that an infection-dependent decrease in miRNA-4276 expression in A549 cells occurs within 3 h of infection and then gradually increases up to 9 h after infection. Treatment of the A549 cells with miR-4276 inhibitor reduced the virus-induced increase in COX6C expression, whereas transfection of mimic miR-4276 enhanced expression of miRNA-4276 with a concomitant decrease in COX6C expression. Decreased COX6C resulted in decreased caspase-9 protein release and eventually led to increased viral copy number. The inhibitor not only induced caspase-9, but also suppressed viral replication indicated by the reduction of influenza matrix mRNA copies and expression of nucleoproteins (Figs. 5 and 6). The data demonstrate that low expression of COX6C is highly favorable for viral replication, possibly through reduced apoptosis and longer survival of the infected cells.
There is increasing evidence that post-translational regulation of gene expression, mediated by miRNAs plays an important role in the control of apoptosis (Subramanian and Steer, 2010). A recent study showed that influenza virus infection suppressed expression of the anti-apoptotic protein BCL2L2 by inducing the expression of miR-29C (Guan et al., 2012). Our data indicate that miRNA-4276 is reduced in the initial 3 h of infection which leads to induction of COX6C and caspase-9. This suggests that an initial cellular defense is to eliminate infected cells by driving them into an apoptotic state. After this, the virus takes control of the cell, induces miRNA-4276 and represses COX6C and caspase-9 expression, and drives the cell into an anti-apoptotic state to prolong cell survival and promote viral replication. Others have shown that the knockdown of BAD resulted in reduced cytochrome c release and suppression of intrinsic apoptotic pathway during influenza virus replication (Tran et al., 2013).
Micro-RNAs expressed by infected cells can influence the ability of the invading virus to replicate and spread (Triboulet et al., 2007). Both the virus and host are able to manipulate the miRNAome as part of their evolutionary strategies for survival (Ghosh et al., 2009). The involvement of both virus-encoded and cell-encoded miRNAs in prolonging host cell survival, evading immune recognition, and establishing viral latency has also been reported (Gottwein and Cullen, 2008). Thus, a viable alternative to the use of vaccines that may be rendered ineffective due to antigenic shift and drift of the virus, is small non-coding RNA for anti-viral therapy. Our data indicates that the inhibitor of miR-4276 can significantly reduce influenza viral copy numbers. Further, our findings show that changes in miR-4276 and COX6C expression is not viral strain specific and, therefore, inhibitor miRNA might be more broadly applicable unlike that of vaccines which target a specific virus.
Influenza induced apoptosis raises the question as to whether this is advantageous for viral spread or is advantageous to the host for possibly reducing the viral titer. Our data partially answers that question and indicates that apoptosis early in infection is advantageous to the host cells, as it could significantly control viral replication. Moreover, data presented here show that viral replication is regulated by miRNA generated on exposure to the virus. However, prolonged apoptosis of host cells is regarded as a major factor contributing to viral pathogenesis as it results in extensive lung tissue damage (Brydon et al., 2005; Ludwig et al., 2006).
In conclusion, we have shown that COX6C is an important cellular protein required by the host cell for induction of the apoptotic signaling pathway and reduction of viral copy number. Our data shows that influenza infection leads to changes in a host miRNA, miRNA-4276, that regulates expression of COX6C mRNA. Our data also suggests that COX6C is an apoptotic protein that when silenced through degradation of its mRNA by miRNA-4276 up-regulation, allows the influenza virus to replicate and spread to neighboring healthy cells.
Acknowledgments
The findings and the conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health. The authors declare no conflict of interest.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blachere FM, Cao G, Lindsley WG, Noti JD, Beezhold DH. Enhanced detection of infectious airborne influenza virus. J. Virol. Methods. 2011;176:120–124. doi: 10.1016/j.jviromet.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Blachere FM, Lindsley WG, Pearce TA, Anderson SE, Fisher M, Khakoo R, Meade BJ, Lander O, Davis S, Thewlis RE, Celik I, Chen BT, Beezhold DH. Measurement of airborne influenza virus in a hospital emergency department. Clin. Infect. Diseases: an Official Publication Infect. Dis. Soc Am. 2009;48:438–440. doi: 10.1086/596478. [DOI] [PubMed] [Google Scholar]
- Brydon EW, Morris SJ, Sweet C. Role of apoptosis and cytokines in influenza virus morbidity. FEMS Microbiol. Rev. 2005;29:837–850. doi: 10.1016/j.femsre.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Collins M. Potential roles of apoptosis in viral pathogenesis. Am. J. Respir. Crit. Care Med. 1995;152:S20–S24. doi: 10.1164/ajrccm/152.4_Pt_2.S20. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Duprez L, Wirawan E, Vanden Berghe T, Vandenabeele P. Major cell death pathways at a glance. Microbes Infect./Institut Pasteur. 2009;11:1050–1062. doi: 10.1016/j.micinf.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucl. Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman LI, Lomax MI. Nuclear genes for cytochrome c oxidase. Biochim. et Biophys. Acta. 1997;1352:174–192. doi: 10.1016/s0167-4781(97)00025-0. [DOI] [PubMed] [Google Scholar]
- Guan Z, Shi N, Song Y, Zhang X, Zhang M, Duan M. Induction of the cellular microRNA-29c by influenza virus contributes to virus-mediated apoptosis through repression of antiapoptotic factors BCL2L2. Biochem. Biophys. Res. Commun. 2012;425:662–667. doi: 10.1016/j.bbrc.2012.07.114. [DOI] [PubMed] [Google Scholar]
- Hinshaw VS, Olsen CW, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J. Virol. 1994;68:3667–3673. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim. et Biophys. Acta. 2003;1604:77–94. doi: 10.1016/s0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin C, Zimmer SG, Lu Z, Holland RE, Jr., Dong Q, Chambers TM. The involvement of a stress-activated pathway in equine influenza virus-mediated apoptosis. Virology. 2001;287:202–213. doi: 10.1006/viro.2001.1010. [DOI] [PubMed] [Google Scholar]
- Ludwig S, Pleschka S, Planz O, Wolff T. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cellular Microbiology. 2006;8:375–386. doi: 10.1111/j.1462-5822.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- Othumpangat S, Gibson LF, Samsell L, Piedimonte G. NGF is an essential survival factor for bronchial epithelial cells during respiratory syncytial virus infection. PLoS One. 2009;4:e6444. doi: 10.1371/journal.pone.0006444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othumpangat S, Noti JD, Blachere FM, Beezhold DH. Expression of non-structural-1A binding protein in lung epithelial cells is modulated by miRNA-548an on exposure to influenza A virus. Virology. 2013;447:84–94. doi: 10.1016/j.virol.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othumpangat S, Regier M, Piedimonte G. Nerve growth factor modulates human rhinovirus infection in airway epithelial cells by controlling ICAM-1 expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012;302:L1057–L1066. doi: 10.1152/ajplung.00365.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razvi ES, Welsh RM. Apoptosis in viral infections. Adv. Virus Res. 1995;45:1–60. doi: 10.1016/s0065-3527(08)60057-3. [DOI] [PubMed] [Google Scholar]
- Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annu. Rev. Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- Schultz-Cherry S, Dybdahl-Sissoko N, Neumann G, Kawaoka Y, Hinshaw VS. Influenza virus ns1 protein induces apoptosis in cultured cells. J. Virol. 2001;75:7875–7881. doi: 10.1128/JVI.75.17.7875-7881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the Avian H5 and H7 Hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J. Cell. Physiol. 2010;223:289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J. Gen. Virol. 1993;74:2347–2355. doi: 10.1099/0022-1317-74-11-2347. [DOI] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA: J. Am. Med. Assoc. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- Tran AT, Cortens JP, Du Q, Wilkins JA, Coombs KM. Influenza virus induces apoptosis via BAD-mediated mitochondrial dysregulation. J. Virol. 2013;87:1049–1060. doi: 10.1128/JVI.02017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J Biol. Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- Webster RG, Rott R. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- Wurzer WJ, Ehrhardt C, Pleschka S, Berberich-Siebelt F, Wolff T, Walczak H, Planz O, Ludwig S. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J. Biol. Chem. 2004;279:30931–30937. doi: 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- Wurzer WJ, Planz O, Ehrhardt C, Giner M, Silberzahn T, Pleschka S, Ludwig S. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 2003;22:2717–2728. doi: 10.1093/emboj/cdg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim N, Albert ML. Dying to replicate: the orchestration of the viral life cycle, cell death pathways, and immunity. Immunity. 2011;35:478–490. doi: 10.1016/j.immuni.2011.10.010. [DOI] [PubMed] [Google Scholar]