Abstract
Rhenium(V) oxo complexes of general formula [ReO(OMe)(N^N)Cl2], where N^N = 4,7-diphenyl-1,10-phenanthroline, 1, or 3,4,7,8-tetramethyl-1,10-phenanthroline, 2, effectively kill cancer cells by triggering necroptsosis, a non-apoptotic form of cell death. Both complexes evoke necrosome (RIP1-RIP3)-dependent intracellular ROS production and propidium iodide uptake. The complexes also induce mitochondrial membrane potential depletion, a possible downstream effect of ROS production. Apparently, 1 and 2 are the first rhenium complexes to evoke cellular events consistent with programmed necrosis in cancer cells. Furthermore, 1 and 2 display low acute toxicity in C57BL/6 mice and reasonable stability in fresh human blood.
Introduction
Cancer is a leading cause of death and suffering throughout the world and continues to impose a huge socio-economic burden on society. According to the latest statistics released by the World Health Organization, an estimated 8.2 million cancer-related deaths occurred in 2012, a 0.6 million increase from the previous estimation in 2008.1 With the global cancer burden rising, the development of new cancer treatments is crucial. Since the discovery of their antineoplastic properties in 1969, platinum drugs such as cisplatin, oxaliplatin, and carboplatin have become a mainstay chemotherapy for cancer.2,3 Their use, however, is limited by side effects and inherent or acquired resistance.4–6 These limitations have driven the search for new treatments, including investigations of other transition metal compounds. Many ruthenium, osmium, titanium, copper, iron, and other metal compounds have been tested for their anticancer activity, and some of the most promising candidates have been studied clinically.7
Several rhenium-based compounds have been employed as in vitro and in vivo imaging agents, but in-depth studies exploring their anti-proliferative properties are relatively rare.8,9 The most active rhenium compounds reported to date contain Re(I)-carbonyl centers bound to mono-, di-, tri-dentate ligands.10–14 This class of compounds are proposed to induce their cytotoxic effects through covalent interactions with DNA bases and/or protein side chains. A number of photolabile rhenium(I) derivatives that trigger cell death upon irradiation have also been devised.15–17 These complexes offer temporal and spatial control over activation and therefore could be useful in overcoming systemic toxicity. Recently, octahedral Re(IV) complexes bearing polypyridyl ligands were discovered to exhibit potent in vitro antiproliferative activity against breast, ovarian, and prostate cancer cells.18 The complexes were hypothesized to interact with nuclear DNA upon hydrolysis, inducing apoptotic cell death. The anticancer potential of dinuclear rhenium compounds has also been investigated.19–21 In addition to displaying promising anticancer activity, paddle-wheel dirhenate(III) complexes have attractive pharmacological features such as low neuro-, hepato- and nephrotoxicity.21–24 Dirhenate(III) units with propionate ligands and quadruple Re-Re bonds have varying degrees of efficacy in sarcoma-, leukemia- and melanoma-bearing mice models.25 Subsequent studies found that the anticancer activity of the rhenium clusters depends on the identity and orientation of the ligands around the two rhenium(III) centers rather than the presence of a quadruple bond. Remarkably, dinuclear rhenium(III) analogues bearing alkylcarboxylates and zwitterionic aminocarboxylate ligands inhibited tumor growth by up to 60% in Wistar rats inoculated with tumor carcinoma Guerink (T8) cells. Furthermore when combined with cisplatin, the rhenium(III) clusters completely inhibited tumor progression.21–24,26 Dirhenate(III) complexes are also regarded as promising modulators of cisplatin. According to in vitro and in vivo studies, carboxylate-bridges dirhenate(III) complexes stabilized red blood cells (RBC) against haemolysis, thereby diminishing dose-limiting toxicity associated with cisplatin.21–24,27
Many clinically used anticancer agents act by targeting and damaging nuclear DNA, eventually leading to apoptosis.28–31 Cytotoxic compounds may also kill cells through non-apoptotic cell death pathways such as necrosis.32–34 Although necrosis was previously believed to be a random, unregulated process, it is now understood that programmed necrosis, also known as necroptosis, does occur in certain cell types.35 Necroptosis is activated by the formation of a complex between receptor interacting protein (RIP) kinases, RIP1 and RIP3.36 The RIP1-RIP3 complex, also known as a necrosome, is thought to modulate oxidative stress and generate mitochondrial reactive oxygen species (ROS) capable of inducing bioenergetics-related cell death.37 The relationship between ROS production and necroptotic cell death is poorly understood, although some reports link RIP1 and ROS-induced cell death.38 Owing to the persistent use of apoptosis-inducing anticancer drugs, many cancers have evolved resistance to apoptosis.39–41 Therefore chemotherapies capable of inducing non-apoptotic cell death such as necroptosis warrant further investigation. Here we present the synthesis, characterization, and cell-based studies of two necroptosis-inducing rhenium(V) oxo complexes.
Results and Discussion
Synthesis and Characterization
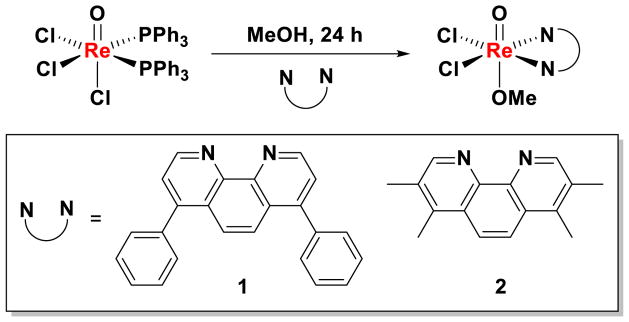
The rhenium(V) oxo complexes 1 and 2 were prepared by the reaction of [ReOCl3(PPh3)2] with 1.5 equivalents of the corresponding bidentate ligand in methanol (Scheme 1). The complexes were isolated in reasonable yields as pale green solids and fully characterized by NMR and IR spectroscopy, and ESI-MS spectrometry. The purity of the complexes was confirmed by elemental analysis. Variable-temperature 1H NMR spectroscopic studies in DMSO revealed the complexes to be stable and remain intact at elevated temperatures (up to 75 °C, see Figure S1).
Scheme 1.
Reaction scheme for preparing rhenium(V) oxo complexes, 1 and 2.
The cyclic voltammograms of 1 and 2 display only a single irreversible reduction within the potential window spanning −1.00 to 1.00 V versus Fc/Fc+ (Figure S2A & B). This event is ascribed to the two-electron reduction of the metal center in each complex to form, initially at least, a Re(III) product. The more negative cathodic peak potential of 2, −1.48 V, compared to that of 1, −1.32 V, is consistent with the more electron donating nature of the 3,4,7,8-tetramethyl-1,10-phenanthroline ligand, which better stabilizes the Re(V) oxidation state. To further rationalize the different redox potentials of the two compounds, DFT calculations were performed at the uPBEPBE/LANL2DZ level of theory in Gaussian 03.42 Solvent effects of DMSO were included implicitly through the self-consistent reaction field approach, as implemented in the default PCM model. The calculations suggested that both compounds preferentially adopt a low-spin configuration, which is significantly lowerer in energy than the corresponding high-spin one (Figure S2C & D). Partial atomic charges were calculated by means of Mulliken population analysis.43 As shown in Table S1, the calculated HOMO energy of 2 is higher than that of 1, which is consistent with observed redox potentials.44 The Mulliken charges on the Re(V) atom also predict that 2 has a lower redox potential than 1. These results can be attributed to the stronger electron-withdrawing effect of the phenyl group (χp = 2.49),45 which can presumably stabilize reduced 1. Overall, the electrochemical studies and calculations shows that the reduction potential of rhenium(V)-oxo complexes such as 1 and 2 can be tuned by subtle ligand modifications.
The lipophilicity of the rhenium(V)-oxo complexes (1 and 2) was determined by measuring the extent to which they partition between octanol and water, Po/w or P. The experimentally determined log P values are 1.20 for 1, and 0.95 for 2. The hydrophobic character of the rhenium(V)-oxo complexes suggests that they will be taken up well by cells.
In Vitro Potency
In vitro effect of rhenium(V) oxo complexes 1 and 2 toward a panel of human cell lines was determined by the colorimetric MTT [[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Cisplatin was also included as a control. The IC50 values or concentration required to induce 50% cell death, were derived from dose-response curves and are summarized in Table 1. Compounds 1 and 2 display nanomolar potency toward cancer cells, with good selectivity over normal fibroblast cells (up to 10-fold). The cytotoxicity of the rhenium complexes was significantly higher than that of cisplatin for the cell lines tested. Notably, 1 displayed 34-fold higher toxicity for lung carcinoma A549 cells compared to cisplatin. Furthermore, the rhenium complexes are not cross-resistant with cisplatin, as demonstrated by their ability to kill cisplatin-resistant ovarian carcinoma cells (A2780CP70) with up to 15-times better efficacy than cisplatin-sensitive cells (A2780).
Table 1.
IC50 values (nM) of 1, 2, and cisplatin against various cancerous and healthy cell lines after 72 h exposure. The errors represent standard deviations.
| Cell line | Cancer type | 1 | 2 | cisplatin |
|---|---|---|---|---|
| A549 | Lung carcinoma | 207 ± 4 | 157 ± 15 | 3230 ± 467 |
| HeLa | Cervical adenocarcinoma | 445 ± 43 | 695 ± 21 | 4100 ± 113 |
| U2OS | Bone osteosarcoma | 274 ± 6 | 209 ± 31 | 4600 ± 600 a |
| NTERA-2 | Testis carcinoma | 230 ± 28 | 255 ± 35 | 385 ± 49 |
| A2780 | Ovarian carcinoma | 670 ± 40 | 150 ± 10 | 700 ± 200 a |
| A2780CP70 | Ovarian carcinoma | 42 ± 15 | 56 ± 2 | 8415 ± 205 |
| MRC-5 | Lung fibroblast | 1351 ± 228 | 709 ± 76 | 5300 ± 600 a |
IC50 values taken from Ref. 46.
Given the ability of 1 and 2 to selectively kill ovarian cisplatin-resistant over sensitive cells, we evaluated their efficacy against other cisplatin-resistant cell lines derived from tissues such as HT-29 (colorectal adenocarcinoma), MDA-MB-231 (breast adenocarcinoma), PC-3 (prostate adenocarcinoma), MCF-7 (breast adenocarcinoma), and DU 145 (prostate carcinoma). The rhenium complexes displayed nanomolar and sub-micromolar toxicities toward the cisplatin-resistant cells (Table 2). Remarkably, 1 and 2 killed colorectal adenocarcinoma HT-29 cells over 300-fold better than cisplatin. Although cisplatin is one of the most successful broad-spectrum anticancer drugs in clinical use, several tumors exhibit resistance. A plethora of molecular mechanisms account for cisplatin resistance, including reduced intracellular accumulation, increased sequestration by scavengers, efficient DNA repair, and deregulation of proteins involved in the DNA damage and apoptotic cell death pathways.47 Therefore compounds such as 1 and 2, which can overcome cisplatin-resistance, hold significant therapeutic potential. To further investigate this potential, cytotoxicity studies were conducted with quiescent A549 cells (Table 2). The potency of 1 and 2 toward quiescent A549 cells was over 40-fold less than toward proliferating A549 cells. This result highlights the ability of 1 and 2 to selectively kill fast growing cancer cells.
Table 2.
IC50 values (nM) of 1, 2, and cisplatin against a panel of cisplatin-resistant cell lines and quiescent lung carcinoma A549 cells after 72 h exposure. The errors represent standard deviations.
| Cell line | Cancer type | 1 | 2 | cisplatin |
|---|---|---|---|---|
| HT-29 | Colorectal adenocarcinoma | 85 ± 11 | 95 ± 20 | 29640 ± 1329 |
| MDA-MB-231 | Breast adenocarcinoma | 475 ± 161 | 1735 ± 275 | 43600 ± 7071 |
| MCF-7 | Breast adenocarcinoma | 285 ± 35 | 805 ± 21 | 9740 ± 537 |
| PC-3 | Prostate adenocarcinoma | 270 ± 14 | 780 ± 10 | 10250 ± 919 |
| DU 145 | Prostate carcinoma | 2840 ± 381 | 1370 ± 84 | > 100000 |
| A549 (quiescent) | Lung carcinoma | 8610 ± 749 | 5245 ± 1986 | 9420 ± 1937 |
Cellular Mechanism of Action and Mode of Cell Death
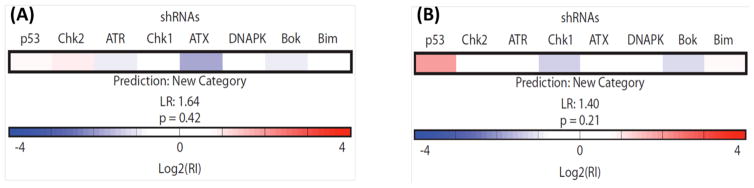
To gain insight into how the rhenium complexes induce cell death, 1 and 2 were analyzed by a recently developed functional strategy employing a RNAi signature assay to predict the mechanism of cytotoxic drug action.48–50 This RNAi-based methodology relies on a fluorescence competition assay with lymphoma cells that are partially infected with eight distinct short hairpin RNAs (shRNAs). shRNA-bearing cells will either enrich or deplete relative to the uninfected population based on the survival advantage or disadvantage conferred by a given shRNA. The responses of these cells compose signatures, which have been obtained from all classes of clinically used cytotoxic agents. These signatures comprise a reference set which is then informatically classified by a probabilistic K-nearest neighbors algorithm to determine whether a new compound belongs to a class in the reference set or requires a new category not yet represented. Neither 1 nor 2 classified as belonging to any category of drug mechanism present in the reference set and thus represent novel mechanisms of drug action (Figure 1).
Figure 1.
RNAi signatures derived from the treatment of Eμ-Mycp19arf−/− lymphoma cells with 1 (A) and 2 (B) at the LD80–90 (1 μM) concentration for each compound. The exposure time was 72 h.
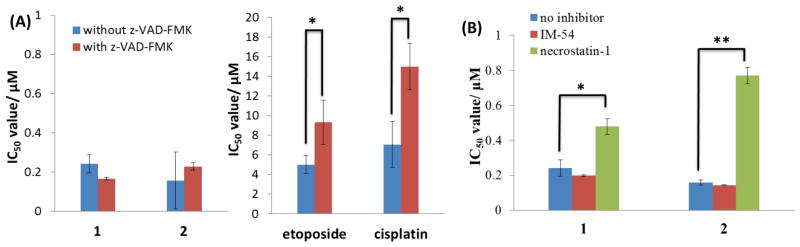
In order to determine the cell killing mechanism of 1 and 2, we carried out cytotoxicity studies in the presence of apoptosis and necrosis inhibitors. Upon addition of z-VAD-FMK, a potent inhibitor of caspase-mediated apoptosis,51 the ability of 1 and 2 to kill A549 cells remained largely unaltered, indicative of a non-apoptotic cell death program (Figure 2A). By contrast, the IC50 values for known apoptosis-inducing agents such as etoposide and cisplatin increased significantly (t test, p < 0.05) in the presence of the inhibitor (Figure 2A). Immunoblotting studies showed that proteins implicated in the apoptotic cell death pathway, namely, cleaved-caspase 7 and 9, were not detected in 1- and 2-treated A549 cells (100 – 400 nM for 72 h; Figure S3), providing further evidence for a non-apoptotic program. We next investigated the possibility that 1- and 2-induce necroptosis. Necroptosis is a well regulated mode of cell death that is different from unregulated necrosis and apoptosis.35 Unlike unregulated necrosis, which can be induced by H2O2 or heat, necroptosis relies on the interaction of protein kinases, RIP1 (receptor-interacting protein 1) and RIP3 (receptor-interacting protein 3), to initiate cell disintegration. This process can be blocked by necrostatin-1, a potent RIP1 kinase inhibitor.52,53 To determine whether 1 and 2 induced necroptosis and/or uncontrolled necrosis, cytotoxicity studies were conducted in the presence of necrostatin-1 (60 μM) and IM-54 (10 μM), an inhibitor of H2O2-induced necrosis.54 Co-incubation with necrostatin-1 markedly decreased the toxicity of 1 and 2 (t test, p < 0.05) against A549, PC-3, and HT-29 cells (Figures 2B and S4). A similar effect was also observed for shikonin, a naturally occurring compound known to induce necroptosis in certain cell types (Figure S4A).38 In contrast, co-treatment with IM-54 did not significantly affect the toxicity of 1 and 2 (Figure 2B). Taken together, the cytotoxicity data suggest that 1 and 2 induce RIP1-RIP3 (necrosome) mediated necroptosis, rather than uncontrolled necrosis or apoptosis. Immunoblotting studies revealed that the overall expressional level of RIP1 and RIP3 in A549 cells remained unchanged with increasing 1 and 2 dosages (Figure S3). Therefore 1- and 2-induced cell death relies on RIP1-RIP3 complex formation and not on the expressional levels of the individual protein kinases. RIP1 can also form a cytosolic complex with Fas-associated death domain (FADD) and caspase 8, known as a ripoptosome, to initiate apoptosis (through caspase 8 cleavage).55–57 Immunoblotting studies showed that FADD and cleaved caspase 8 expression remained unaltered with increasing 1 and 2 concentration (Figure S3), indicating that ripoptosome formation was not responsible for 1- and 2-induced cell death. This finding is consistent with the fact that 1- and 2-treated A549 cells do not undergo apoptosis.
Figure 2.
(A) IC50 values (in μM) of 1, 2, etoposide, and cisplatin against A549 cells in the absence and presence of apoptosis inhibitor, z-VAD-FMK (5 μM) after 72 h incubation. (B) IC50 values (in μM) of 1 and 2 against A549 cells in the absence and presence of H2O2-induced necrosis inhibitor, IM-54 (10 μM), and necroptosis inhibitor, necrostatin-1 (60 μM) after 72 h incubation. Student’s t-test, p < 0.05 or 0.01. Error bars represent standard deviations.
Characterization of Necroptotic Features
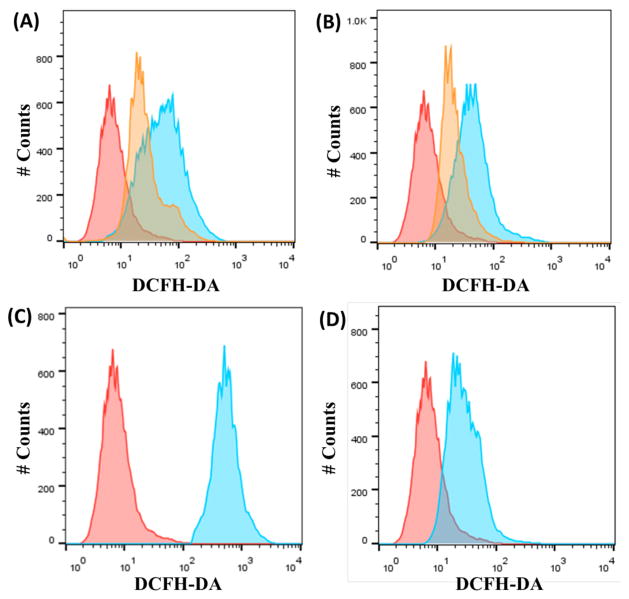
Having established that necrosome formation is a determinant of 1 and 2 activity, we performed additional studies to understand the cascade of events leading from necrosome formation to cell death. Necrosomes generate abnormally high levels of mitochondrial reactive oxygen species (ROS),58–60 leading to ATP depletion and eventual degradation of the mitochondrial membrane potential.61,62 With this fact in mind, we quantified intracellular ROS production by flow cytometry using 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), a well-established ROS indicator. A549 cells incubated with 1 and 2 (20 μM for 12 h) displayed markedly higher levels of ROS than untreated control cells (Figure 3A & B). A549 cells dosed with H2O2 (1 mM for 1 h, ROS-inducer) and shikonin (20 μM for 12 h, necroptosis-inducer) also exhibited significantly higher levels of measurable ROS than untreated cells (Figure 3C & 3D). Remarkably, 1- and 2-induced ROS production was attenuated in the presence of necrostatin-1 (60 μM) (Figure 3A & B), suggesting that the RIP1-RIP3 kinase complex plays a role in modulating intracellular ROS production.
Figure 3.
(A) Representative histograms displaying the green fluorescence emitted by DCFH-DA stained A549 cells (red) and A549 cells treated with 1 (20 μM for 12 h) (blue) or 1 (20 μM for 12 h) with nectrostatin-1 (60 μM for 12 h) (orange). (B) Representative histograms displaying the green fluorescence emitted by DCFH-DA stained A549 cells (red) and A549 cells treated with 2 (20 μM for 12 h) (blue) or 2 (20 μM for 12 h) with nectrostatin-1 (60 μM for 12 h) (orange). (C) Representative histograms displaying the green fluorescence emitted by DCFH-DA stained A549 cells (red) and A549 cells treated with H2O2 (1 mM for 1 h) (blue). (D) Representative histograms displaying the green fluorescence emitted by DCFH-DA stained A549 cells (red) and A549 cells treated with shikonin (20 μM for 12 h) (blue).
The effect of 1 and 2 on the mitochondrial membrane potential was assessed by flow cytometry, using the JC-1 assay (5,5′,6,6′-tetrachloro-1,1′3,3′-tetraethyl benzimidazolyl carbocyanine iodide).63,64 JC-1 is a cationic lipophilic dye that localizes in the mitochondria of healthy cells as red-emitting aggregates. If the mitochondrial membrane potential is disrupted, JC-1 forms green-emitting monomers. A549 cells incubated with 1 and 2 (20 μM for 12 h) displayed increased green fluorescence compared to untreated cells, indicative of mitochondrial membrane disruption (Figure S5A & B). A similar result was observed for A549 cells dosed with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (5 μM for 12 h), a known mitochondrial membrane depolarizer (Figure S5C), and shikonin (20 μM for 12 h), a necroptosis inducing agent (Figure S5D). Notably, 1- and 2-induced mitochondrial membrane depletion was amplified with necrostatin-1, suggesting that 1 and 2 may inherently target mitochondria and induce mitochondrial dysfunction, independent of RIP1-RIP3 formation. This result could explain the high residual toxicity (>1 μM) observed for A549 cells co-incubated with the rhenium complexes (1 and 2) and necrostatin-1 (Figure 2B).
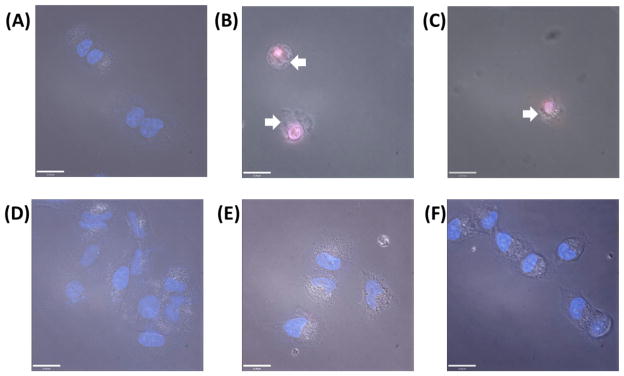
Intracellular ROS production and mitochondrial membrane depletion contribute to necroptosis.61,62 Cells undergoing necroptosis display necrosis-like morphological features such as loss of cell membrane integrity, increase in organelle and cell volume (oncosis), and intact nuclear membrane.65–67 To further test whether 1- and 2-treatment is able to trigger necroptosis, Hoechst 33258/propidium iodide (PI) double staining was carried out to determine nuclear membrane morphology and integrity. Hoechst 33258 is a DNA minor groove binder that is routinely used to visualize the nucleus without the need for cell permeabilization.68 When used without cell permeabilization agents, PI stains the nuclei of necrotic cells.69 Early stage apoptotic and normal cells maintain cell membrane integrity and thus are not stained by PI. A549 cells were treated with 1 and 2 (20 μM for 12 h), incubated with Hoechst 33258 and PI, and imaged using a fluorescent microscope. Untreated A549 cells exhibited bright blue nuclei, owing to Hoechst 33258 uptake (Figure 4A). Cells incubated with 1 and 2 displayed pink nuclei, owing to Hoechst 33258 and PI uptake, which is consistent with necroptosis (Figure 4B & C). Furthermore 1- and 2-treated cells showed clear signs of plasma membrane disintegration with undamaged nuclei. A549 cells co-incubated with 1 or 2 and necrostatin-1 (60 μM for 12 h) were unstained by PI suggesting that necrostatin-1 is able to block 1- and 2-induced necroptosis (Figure 4E & F). Overall, the microscopy data suggest that necrosome formation contributes to the necrosis-like morphological features observed upon 1- and 2-treatment. To further validate this result, A549 cells were treated under the same conditions as above, stained with PI, and analyzed by flow cytometry. Complementary to the microscopy results, 1- and 2-treated cells expressed higher PI uptake compared to untreated control cells, indicative of necrotic cell death (Figure S6A & B). The flow cytometry data also showed that necrostatin-1 could block 1- and 2-mediated PI uptake. Additional studies showed that pre-treatment of A549 cells with N-acetylcysteine (3mM for 1 h), a ROS inhibitor, significantly decreased 1- and 2-induced PI uptake (Figure S6C & D). This result suggests that intracellular ROS generation is an integral part of the necroptotic mechanism of action of 1 and 2.
Figure 4.
Fluorescence microscopy images of A549 cells untreated (A) and treated with 1 (20 μM for 12 h) (B), 2 (20 μM for 12 h) (C), necrostatin-1 (60 μM for 12 h) (D), 1 and necrostatin-1 (E), and 2 and necrostatin-1 (F) and then stained with Hoechst 33258 and PI. Arrows indicate signs of cell membrane disintegration. Scale bar = 21 μM.
PARP-1- and p53-Independent Necroptosis
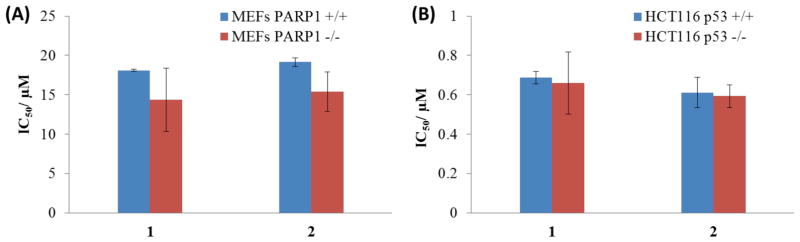
Apart from necrosome formation, necroptosis can also result from the overactivation of poly(ADP–ribose) polymerase (PARP-1).66,67 PARP-1 is a nuclear enzyme that is involved in DNA repair and transcriptional regulation.70 DNA damage can trigger PARP-1 activity, resulting in ATP and NAD depletion and bioenergetic-mediated cell death.71 To determine whether PARP-1 activity is a factor in 1- and 2-mediated cell death, cytotoxicity assays were conducted with wild-type mouse embryonic fibroblast cells (MEFs PARP-1+/+) and the corresponding PARP-1-null cells (MEFs PARP-1−/−). The IC50 values for 1 and 2 were similar for MEFs PARP-1+/+ and MEFs PARP-1−/− cells, indicating that 1 and 2-induced necroptosis is independent of PARP-1 function (Figure 5A). This result is consistent with immunoblotting studies, which revealed that 1- and 2-treatment did not upregulate canonical markers of DNA damage such as the phosphorylated forms of H2AX γH2AX) and CHK2 (Figure S7). Recently, p53 has also been reported to play a role in necroptosis. p53 induces cathepsin Q, a lysosomal protease that cooperates with ROS to execute necrosis.72 To investigate whether p53 might play a role in 1- and 2-mediated necroptosis, cytotoxicity studies were conducted with HCT116 p53+/+ and HCT116 p53−/− cells. The potency of 1 and 2 was statistically similar for HCT116 p53+/+ and HCT116 p53−/− cells, indicating that 1 and 2 induce necroptosis in a manner that is independent of p53 (Figure 5B). This concludion is consistent with the RNAi signatures, which reveal that p53 is not important in the cellular response evoked by the complexes, especially for 1. Apart from the implications of this result on the mechanism of action of 1 and 2, it is clinically very appealing because p53 is mutated, defective, or inactivated in several chemoresistant cancers.
Figure 5.
(A) IC50 values (in μM) of 1 and 2 against MEFs PARP-1+/+ and MEFs PARP-1−/− cells after 72 h incubation. (B) IC50 values (in μM) of 1 and 2 against HCT116 p53+/+ and HCT116 p53−/− cells after 72 h incubation.
Cell Cycle Analysis
To gain a more complete understanding of the cellular response evoked, DNA-flow cytometric studies were conducted to determine the effect of 1 and 2 on the cell cycle. A549 cells were treated with 1 or 2 (2 μM) and the cell cycle was determined over the course of 72 h (Figure S8). After 24 h treatment, both complexes stalled the cell cycle at the G1-phase. Cells treated with 1 remained stalled at the G1-phase after 48 h. Upon further incubation (72 h), large populations of cell debris were detected (32%), indicative of cell death. Cells incubated with 2 for 48 h and 72h also displayed large populations of debris (26 and 38% respectively). G1-phase cell cycle arrest followed by immediate cell death is characteristic of programmed necrosis.73,74
In Vivo Toxicity and Stability in Whole Human Blood
Given the impressive in vitro data acquired for 1 and 2, an in vivo study was conducted with C57BL/6 mice to determine acute toxicity and possible side effects. Single doses of 1 and 2 (3, 7, 11, 15, 20, 36 mg/ kg) in saline, were administered by intra-peritoneal injection. The mice were then monitored for signs of pain, distress, and weight loss for 6 days post-treatment. The compounds exhibited no toxicity in mice, as gauged by a lack of weight loss after treatment. The change in weight of mice after a single dose of 1 and 2 at the maximum solubility of the complexes (36 mg/ kg) is depicted in Figure S9. A single 30 mg/ kg dose of cisplatin induces acute nephrotoxicity in C57BL/6.75 The in vivo data highlight the relatively low toxicity of 1 and 2 compared to cisplatin in C57BL/6 mice. The pharmacological toxicity profile of 1 and 2 is very appealing in terms of further pre-clinical studies.
The stability of biologically active compounds in human blood is vitally important for their potential application in clinical settings. We therefore investigated the stability of 1, in whole human blood using a recently developed protocol.76 This method exploits the ability of octanol to extract hydrophobic metal complexes such as 1. The rhenium complex, 1 (500 μM) was incubated with fresh human blood at 37 °C and aliquots were extracted into octanol at various time points. The amount of 1 in the octanol extracts (corresponding to unreacted 1) was measured by graphite furnace atomic absorption spectroscopy (GFAAS). The data present in Figure S10, revealed that the half-life of 1 in human blood is 29.1 min, comparable to that reported for cisplatin (t1/2 = 21.6 min).77
Conclusion
Two rhenium(V) oxo complexes were prepared and their in vitro properties were investigated. The complexes selectively kill cancer cells over normal cells and display markedly higher cell toxicity than cisplatin. Remarkably, 1 and 2 display two orders of magnitude higher toxicity than cisplatin against colorectal adenocarcinoma cells. Cells treated with 1 and 2 displayed features consistent with programmed necrosis (necroptosis), including RIP1-RIP3 dependent intracellular ROS production, cell membrane disruption, PI uptake, mitochondrial damage, and G1 cell cycle arrest. Given the inherent and/or acquired resistance of tumors toward apoptosis-inducing chemotherapies, compounds such as 1 and 2, capable of killing cancer cells through necroptosis, are highly sought after when selecting preclinical drug candidates for chemoresistant malignancies.
Experimental Details
Material
All synthetic procedures were performed under normal atmospheric conditions without exclusion of oxygen or moisture. The bidentate aromatic ligands 4,7-diphenyl-1,10-phenanthroline and 3,4,7,8-tetramethyl-1,10-phenanthroline were purchased from Sigma Aldrich and used as received. ReOCl3(PPh3)2 was prepared as previously reported.78 The synthesis of 1 has been reported previously,79 but the procedure reported here is novel. Analytical grade acetone and dichloromethane were used as solvents.
Physical Measurements
NMR measurements were recorded on a Bruker 400 MHz spectrometer in the MIT Department of Chemistry Instrumentation Facility (DCIF) at 20 °C. 1H and 13C{1H} NMR chemical shifts were referenced internally to residual solvent peaks or relative to tetramethylsilane (SiMe4, δ = 0.00 ppm). Fourier transform infrared (FTIR) spectra were recorded with a ThermoNicolet Avatar 360 spectrophotometer upon preparation of the samples as KBr disks. The spectra were analyzed using the OMNIC software. Graphite furnace atomic absorption spectrometry was carried out using a Perkin-Elmer AAnalyst600 GFAAS.
Synthesis of [ReO(OMe)(4,7-diphenyl-1,10-phenanthroline)Cl2] (1)
[ReOCl3(PPh3)2] (63.8 mg, 0.08 mmol) was suspended in methanol (15 mL) and heated to 50 °C. To this mixture was added a methanolic solution (5 mL) of 7-diphenyl-1,10-phenanthroline (34.0 mg, 0.10 mmol). The resultant mixture was heated under reflux for 24 h to give a deep purple solution with a pale green precipitate. The precipitate was filtered, washed with hot methanol, cold methanol, and diethyl ether. The rhenium(V) oxo complex was isolated as a pale green solid. Yield: 19.8 mg (37%). Mp 247 °C (dec). 1H NMR (400 MHz, DMSO-d6): δ 10.08 (d, 2H), 8.48 (d, 2H), 8.31 (s, 2H), 7.83 (m, 4H), 7.73 (m, 6H), 2.56 (s, 3H). IR (KBr, cm−1): 941.51 (Re=O), 508.42 (Re-OMe). ESI-MS (MeOH/ DMSO): m/z. 605.0 ([M-OMe]+, calcd. 605.0). Anal. Calcd. for 1, C25H19Cl2N2O2Re: C, 47.17; H, 3.01; N, 4.40. Found: C, 46.79; H, 3.05; N, 4.35.
Synthesis of [ReO(OMe)(3,4,7,8-tetramethyl-1,10-phenanthroline)Cl2] (2)
[ReOCl3(PPh3)2] (50.0 mg, 0.06 mmol) was suspended in methanol (15 mL) and heated to 50 °C. To this mixture, a methanolic solution (5 mL) of 3,4,7,8-tetramethyl-1,10-phenanthroline (21.85 mg, 0.09 mmol) was added. The resultant mixture was heated under reflux for 24 h to give a deep purple solution with a pale green precipitate. The precipitate was filtered, washed with hot methanol, cold methanol, and diethyl ether. The rhenium(V) oxo complex was isolated as a pale green solid. Yield: 14.8 mg (41%). Mp > 276 °C (gradual darkening and decomposition). 1H NMR (400 MHz, DMSO-d6): δ 9.69 (s, 2H), 8.60 (s, 2H), 2.99 (s, 6H), 2.74 (s, 6H), 2.36 (s, 3H). IR (KBr, cm−1): 954.85 (Re=O), 492.85 (Re-OMe). ESI-MS (MeOH/ DMSO): m/z. 509.0 ([M-OMe]+, calcd. 509.0). Anal. Calcd. for 2, C17H19Cl2N2O2Re: C, 37.78; H, 3.54; N, 5.18. Found: C, 37.76; H, 3.63; N, 4.99.
Electrochemistry
Electrochemical experiments were performed at room temperature using a VersaSTAT3 potentiostat from Princeton Applied Research operated with the V3 Studio software. Cyclic voltammetry was performed using a three-electrode system comprising a glassy carbon working electrode, a platinum wire auxiliary electrode, and a silver wire pseudo-reference electrode. Potentials were referenced internally to the ferrocene/ferrocenium redox couple. Solutions of 1 and 2, at concentrations of approximately 2 mM, were prepared in DMSO containing 0.2 M tetra-n-butylammonium hexafluorophosphate. All solutions were sparged with nitrogen for 5 min prior to measurement. Voltammograms were obtained on quiescent solutions at a scan rate of 200 mV/s.
Cytotoxicity MTT assay
The colorimetric MTT assay was used to determine the toxicity of 1, 2, and cisplatin. Cells (2 × 103 cells/well) were seeded in a 96-well plate. After incubating the cells overnight, various concentrations of platinum as determined by GF-AAS (0.3–100 μM) were added and incubated for 72 h (total volume 200 μL). Cisplatin was prepared as a 5 mM solution in PBS and diluted using media. 1 and 2 were prepared as 10 mM solutions in DMSO and diluted using media. The final concentration of DMSO in each well was 0.5% and this amount was also present in the untreated control. After 72 h, the medium was removed, 200 μL of a 0.4 mg/mL solution of MTT in DMEM, RPMI or, McCoy’s 5A was added, and the plate was incubated for an additional 1–2 h. The DMEM/MTT, RPMI/MTT, or McCoy’s 5A/MTT mixture was aspirated and 200 μL of DMSO was added to dissolve the resulting purple formazan crystals. The absorbance of the solution wells was read at 550 nm. Absorbance values were normalized to DMSO-containing control wells and plotted as the concentration of test compound versus % cell viability. IC50 values were interpolated from the resulting dose dependent curves. The reported IC50 values are the average from at least three independent experiments, each of which consisted of six replicates per concentration level.
For specific cell death inhibitor assays, inhibitors of necroptosis (necrostatin-1, 60 μM), H2O2-induced necrosis (IM-54, 10 μM), apoptosis (v-VAD-FMK, 5 μM) were added to cells and incubated for 1 h prior to treatment with the test compounds.
Reactivity of 1 and 2 with necrostatin-1
Mixing the rhenium(V)-oxo complexes (1 and 2) (20 μM) with necrostatin-1 (60 μM) in DMSO and cell culture media (DMEM, RPMI, and McCoy’s 5A) did not result in a precipitate. Incubation of the rhenium(V)-oxo complexes (1 and 2) with necrostatin-1 (1:3 ratio) for up to 6 hr in DMSO-d6 did not lead to a chemical reaction, as determined by 1H NMR analysis (Figure S11 and S12). Despite the presence of sulfur and nitrogen atoms in necrostatin-1, the 1H NMR spectra unequivocally prove that the reactivity/bioactivity of 1 and 2 is not compromised by necrostatin-1.
Intracellular ROS Assay
Untreated and treated A549 cells (1.5 × 106 cells/ well) grown in 6-well plates were incubated with 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (20 μM) for 30 min. The cells were then washed with PBS (1 mL × 3), harvested, and analyzed by using the FACSCalibur-HTS flow cytometer (BD Biosciences) (20,000 events per sample were acquired). The FL2 channel was used to assess intracellular ROS levels. Cell populations were analyzed using the FlowJo software (Tree Star).
JC-1 Assay
The JC-1 Mitochondrial Membrane Potential Assay Kit (Cayman) was used. The manufacturer’s protocol was followed to carry out this experiment. Briefly, to untreated and treated A549 cells (1.5 × 106 cells/ well) grown in 6-well plates was added the JC-1 staining solution (100 μL/ mL of cell media). The cells were incubated for 30 min, harvested, and analyzed by using the FACSCalibur-HTS flow cytometer (BD Biosciences) (20,000 events per sample were acquired). The FL2 channel was used to assess mitochondrial depolarization. Cell populations were analyzed using the FlowJo software (Tree Star).
Propidium Iodide Uptake
Untreated and treated A549 cells (1.5 × 106 cells/ well) grown in 6-well plates, were washed with PBS (1 mL × 3), harvested, incubated with propidium iodide (5 μM), and analyzed by using the FACSCalibur-HTS flow cytometer (BD Biosciences) (20,000 events per sample were acquired). The FL2 channel was used to assess intracellular PI uptake. Cell populations were analyzed using the FlowJo software (Tree Star).
Fluorescence Microscopy
A549 cells (1.5 × 106 cells/ well) were incubated with and without 1 and 2 (20 μM) for 12 h. The media were then removed and the cells were washed with additional media (2 mL × 2). After incubation of the cells with more media containing Hoechst 33258 (7.5 μM) and propidium iodide (5 μM), the nuclear regions were imaged using a fluorescent microscope. Fluorescence imaging experiments were performed using a Zeiss Axiovert 200M inverted epifluorscence microscope with a Hamamatsu EM-CCD digital camera C9100 and a MS200 XY Piezo Z stage (Applied Scientific Instruments, Inc.). An X-Cite 120 metal halide lamp (EXFO) was used as the light source. Zeiss standard filter sets 49 was employed for imaging the nuclear region. The microscope was operated with Volocity software (version 6.01, Improvision). The exposure time for acquisition of fluorescence images was kept constant for each series of images at each channel.
Immunoblotting Analysis
A549 cells (1.5 × 106 cells/ well) grown in 6-well plates were incubated with 1 and 2 (concentrations, μM) for 72 h at 37 °C. Cells were washed with PBS, scraped into SDS-PAGE loading buffer (64 mM Tris-HCl (pH 6.8)/ 9.6% glycerol/ 2% SDS/ 5% β-mercaptoethanol/ 0.01% Bromophenol Blue), and incubated at 95 °C for 10 min. Whole cell lysates were resolved by 4–20 % sodium dodecylsulphate polyacylamide gel electrophoresis (SDS-PAGE; 200 V for 25 min) followed by electrotransfer to polyvinylidene difluoride membrane, PVDF (350 mA for 1 h). Membranes were blocked in 5% (w/v) non-fat milk in PBST (PBS/0.1% Tween 20) and incubated with the appropriate primary antibodies (Cell Signalling Technology and Santa Cruz). After incubation with horseradish peroxidase-conjugated secondary antibodies (Cell Signalling Technology), immuno complexes were detected with the ECL detection reagent (BioRad) and analyzed using an Alpha Innotech ChemiImager™ 5500 fitted with a chemiluminescence filter.
Cell Cycle
In order to monitor the cell cycle, flow cytometry studies were carried out. A549 cells (1.5 × 106 cells/ well) grown in 6-well plates were incubated with and without the test compounds for 24, 48, and 72 h at 37 °C. Cells were harvested from adherent cultures by trypsinization and combined with all detached cells from the incubation medium to assess total cell viability. Following centrifugation at 1000 rpm for 5 min, cells were washed with PBS and then fixed with 70% ethanol in PBS. Fixed cells were collected by centrifugation at 2500 rpm for 3 min, washed with PBS, and centrifuged as before. Cellular pellets were re-suspended in 50 μg/mL propidium iodide (Sigma) in PBS for nucleic acids staining and treated with 100 μg/mL RNaseA (Sigma). DNA content was measured on a FACSCalibur-HTS flow cytometer (BD Biosciences) using laser excitation at 488 nm and 20,000 events per sample were acquired. Cell cycle profiles were analyzed using the ModFit software.
Supplementary Material
Acknowledgments
This work is supported by the NCI under grant CA034992.
Footnotes
Notes: The authors declare no competing financial interests.
Associated Content
Supporting Information: Experimental techniques and data concerning some computational, biophysical, and cellular studies. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cancer Facts & Figures 2013. Atlanta: American Cancer Society; 2013. [Google Scholar]
- 2.Fricker SP. Dalton Trans. 2007:4903. doi: 10.1039/b705551j. [DOI] [PubMed] [Google Scholar]
- 3.Kelland L. Nat Rev Cancer. 2007;7:573. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 4.Brabec V, Kasparkova J. Drug Resist Updates. 2005;8:131. doi: 10.1016/j.drup.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 5.McWhinney SR, Goldberg RM, McLeod HL. Mol Cancer Ther. 2009;8:10. doi: 10.1158/1535-7163.MCT-08-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddik ZH. Oncogene. 2003;22:7265. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 7.Guo ZJ, Sadler PJ. Angew Chem. 1999;38:1513. doi: 10.1002/(SICI)1521-3773(19990601)38:11<1512::AID-ANIE1512>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 8.Lo KKW, Zhang KY, Li SPY. Eur J Inorg Chem. 2011;2011:3551. [Google Scholar]
- 9.Thorp-Greenwood FL, Balasingham RG, Coogan MP. J Organomet Chem. 2012;714:12. [Google Scholar]
- 10.Leonidova A, Gasser G. ACS Chem Biol. 2014;9:2180. doi: 10.1021/cb500528c. [DOI] [PubMed] [Google Scholar]
- 11.Ma DL, Che CM, Siu FM, Yang M, Wong KY. Inorg Chem. 2007;46:740. doi: 10.1021/ic061518s. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Yan YK, Andy Hor TS, Vittal JJ, Wheaton JR, Hall IH. Polyhedron. 2002;21:1991. [Google Scholar]
- 13.Yan YK, Cho SE, Shaffer KA, Rowell JE, Barnes BJ, Hall IH. Pharmazie. 2000;55:307. [PubMed] [Google Scholar]
- 14.Zhang J, Vittal JJ, Henderson W, Wheaton JR, Hall IH, Hor TSA, Yan YK. J Organomet Chem. 2002;650:123. [Google Scholar]
- 15.Kitanovic I, Can S, Alborzinia H, Kitanovic A, Pierroz V, Leonidova A, Pinto A, Spingler B, Ferrari S, Molteni R, Steffen A, Metzler-Nolte N, Wölfl S, Gasser G. Chem Eur J. 2014;20:2496. doi: 10.1002/chem.201304012. [DOI] [PubMed] [Google Scholar]
- 16.Leonidova A, Pierroz V, Rubbiani R, Heier J, Ferrari S, Gasser G. Dalton Trans. 2014;43:4287. doi: 10.1039/c3dt51817e. [DOI] [PubMed] [Google Scholar]
- 17.Leonidova A, Pierroz V, Rubbiani R, Lan Y, Schmitz AG, Kaech A, Sigel RKO, Ferrari S, Gasser G. Chem Sci. 2014;5:4044. [Google Scholar]
- 18.Martinez-Lillo J, Mastropietro TF, Lappano R, Madeo A, Alberto ME, Russo N, Maggiolini M, De Munno G. Chem Commun. 2011;47:5283. doi: 10.1039/c1cc11038a. [DOI] [PubMed] [Google Scholar]
- 19.Clarke MJ, Zhu F, Frasca DR. Chem Rev. 1999;99:2511. doi: 10.1021/cr9804238. [DOI] [PubMed] [Google Scholar]
- 20.Jakupec MA, Galanski M, Arion VB, Hartinger CG, Keppler BK. Dalton Trans. 2008:183. doi: 10.1039/b712656p. [DOI] [PubMed] [Google Scholar]
- 21.Dimitrov NV, GWE . In: Current Chemotherapy. Siegenthaler W, RL, editors. Vol. 2 American Society of Microbiology; Washington, DC: 1978. [Google Scholar]
- 22.Shtemenko N, Shtemenko PCA. In: Metal Ions in Biology and Medicine. Alpoim MC, PVN, Santos MA, Cristovao AJ, Centeno JA, Collery P, editors. Vol. 9. John Libbey Eurotext; Paris, France: 2006. p. 374. [Google Scholar]
- 23.Shtemenko N, Shtemenko PCA. In: Metal Ions in Biology and Medicine. Collery P, IM, Khassanova L, Collery T, editors. Vol. 10. John Libbey Eurotext; Paris, France: 2008. p. 441. [Google Scholar]
- 24.Shtemenko AG, Tretyak S, Shtemenko N, Randarevich M. In: Metal Ions in Biology and Medicine. Collery P, IM, Khassanova L, Collery T, editors. Vol. 10. John Libbey Eurotext; Paris, France: 2008. p. 229. [Google Scholar]
- 25.Shtemenko NI, Zabitskaya ED, Berzenina OV, Yegorova DE, Shtemenko AV. Chem Biodivers. 2008;5:1660. doi: 10.1002/cbdv.200890153. [DOI] [PubMed] [Google Scholar]
- 26.Shtemenko AV, Collery P, Shtemenko NI, Domasevitch KV, Zabitskaya ED, Golichenko AA. Dalton Trans. 2009:5132. doi: 10.1039/b821041a. [DOI] [PubMed] [Google Scholar]
- 27.Shtemenko N, Collery P, Shtemenko A. Anticancer Res. 2007;27:2487. [PubMed] [Google Scholar]
- 28.Fischer U, Schulze-Osthoff K. Cell Death Differ. 2005;12(Suppl 1):942. doi: 10.1038/sj.cdd.4401556. [DOI] [PubMed] [Google Scholar]
- 29.Hickman JA. Cancer Metastasis Rev. 1992;11:121. doi: 10.1007/BF00048059. [DOI] [PubMed] [Google Scholar]
- 30.Jamieson ER, Lippard SJ. Chem Rev. 1999;99:2467. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, Lippard SJ. Nat Rev Drug Discovery. 2005;4:307. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 32.Cepero V, Garcia-Serrelde B, Moneo V, Blanco F, Gonzalez-Vadillo AM, Alvarez-Valdes A, Navarro-Ranninger C, Carnero A. Clin Transl Oncol. 2007;9:521. doi: 10.1007/s12094-007-0096-2. [DOI] [PubMed] [Google Scholar]
- 33.Ernst RJ, Komor AC, Barton JK. Biochemistry. 2011;50:10919. doi: 10.1021/bi2015822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han W, Li L, Qiu S, Lu Q, Pan Q, Gu Y, Luo J, Hu X. Mol Cancer Ther. 2007;6:1641. doi: 10.1158/1535-7163.MCT-06-0511. [DOI] [PubMed] [Google Scholar]
- 35.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Nat Rev Mol Cell Biol. 2010;11:700. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 36.Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, Chan FK, Wu H. Cell. 2012;150:339. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irrinki KM, Mallilankaraman K, Thapa RJ, Chandramoorthy HC, Smith FJ, Jog NR, Gandhirajan RK, Kelsen SG, Houser SR, May MJ, Balachandran S, Madesh M. Mol Cell Biol. 2011;31:3745. doi: 10.1128/MCB.05303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C, Luo Y, Zhao J, Yang F, Zhao H, Fan W, Ge P. PLoS One. 2013;8:e66326. doi: 10.1371/journal.pone.0066326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longley DB, Johnston PG. J Pathol. 2005;205:275. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 40.Pommier Y, Sordet O, Antony S, Hayward RL, Kohn KW. Oncogene. 2004;23:2934. doi: 10.1038/sj.onc.1207515. [DOI] [PubMed] [Google Scholar]
- 41.Simstein R, Burow M, Parker A, Weldon C, Beckman B. Exp Biol Med. 2003;228:995. doi: 10.1177/153537020322800903. [DOI] [PubMed] [Google Scholar]
- 42.Frisch MJ, GWT, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Revision C.02. Gaussian, Inc; Wallingford CT: 2004. [Google Scholar]
- 43.Mulliken RS. J Chem Phys. 1955;23:1833. [Google Scholar]
- 44.Roy LE, Batista ER, Hay PJ. Inorg Chem. 2008;47:9228. doi: 10.1021/ic800541w. [DOI] [PubMed] [Google Scholar]
- 45.Huheey JE. J Phys Chem. 1965;69:3284. [Google Scholar]
- 46.Suntharalingam K, Wilson JJ, Lin W, Lippard SJ. Metallomics: integrated biometal science. 2014;6:437. doi: 10.1039/c3mt00364g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Oncogene. 2012;31:1869. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 48.Jiang H, Pritchard JR, Williams RT, Lauffenburger DA, Hemann MT. Nat Chem Biol. 2011;7:92. doi: 10.1038/nchembio.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritchard JR, Bruno PM, Gilbert LA, Capron KL, Lauffenburger DA, Hemann MT. Proc Natl Acad Sci. 2013;110:E170. doi: 10.1073/pnas.1210419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pritchard JR, Bruno PM, Hemann MT, Lauffenburger DA. Mol BioSyst. 2013;9:1604. doi: 10.1039/c2mb25459j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slee EA, Zhu H, Chow SC, MacFarlane M, Nicholson DW, Cohen GM. Biochem J. 1996;315(Pt 1):21. doi: 10.1042/bj3150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, Hedrick SM, Gerber SA, Lugovskoy A, Yuan J. Nat Chem Biol. 2008;4:313. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Nat Chem Biol. 2005;1:112. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 54.Dodo K, Katoh M, Shimizu T, Takahashi M, Sodeoka M. Bioorg Med Chem Lett. 2005;15:3114. doi: 10.1016/j.bmcl.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 55.Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M. Mol Cell. 2011;43:449. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. Mol Cell. 2011;43:432. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Du F, Wang X. Cell. 2008;133:693. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 58.Cho Y, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FKM. Cell. 2009;137:1112. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis CW, Hawkins BJ, Ramasamy S, Irrinki KM, Cameron BA, Islam K, Daswani VP, Doonan PJ, Manevich Y, Madesh M. Free Radic Biol Med. 2010;48:306. doi: 10.1016/j.freeradbiomed.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. Science. 2009;325:332. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 61.He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Cell. 2009;137:1100. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 62.Temkin V, Huang Q, Liu H, Osada H, Pope RM. Mol Cell Biol. 2006;26:2215. doi: 10.1128/MCB.26.6.2215-2225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reers M, Smith TW, Chen LB. Biochemistry. 1991;30:4480. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- 64.Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW, Steele GD, Chen LB. Proc Natl Acad Sci. 1991;88:3671. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaczmarek A, Vandenabeele P, Krysko DV. Immunity. 2013;38:209. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Sosna J, Voigt S, Mathieu S, Lange A, Thon L, Davarnia P, Herdegen T, Linkermann A, Rittger A, Chan FK, Kabelitz D, Schutze S, Adam D. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu X, Chua CC, Zhang M, Geng D, Liu CF, Hamdy RC, Chua BH. Brain Res. 2010;1343:206. doi: 10.1016/j.brainres.2010.04.080. [DOI] [PubMed] [Google Scholar]
- 68.Latt SA, Stetten G. J Histochem Cytochem. 1976;24:24. doi: 10.1177/24.1.943439. [DOI] [PubMed] [Google Scholar]
- 69.Lecoeur H. Exp Cell Res. 2002;277:1. doi: 10.1006/excr.2002.5537. [DOI] [PubMed] [Google Scholar]
- 70.Schreiber V, Dantzer F, Ame J-C, de Murcia G. Nat Rev Mol Cell Biol. 2006;7:517. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 71.Los M, Mozoluk M, Ferrari D, Stepczynska A, Stroh C, Renz A, Herceg Z, Wang ZQ, Schulze-Osthoff K. Mol Biol Cell. 2002;13:978. doi: 10.1091/mbc.01-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tu HC, Ren D, Wang GX, Chen DY, Westergard TD, Kim H, Sasagawa S, Hsieh JJ, Cheng EH. Proc Natl Acad Sci. 2009;106:1093. doi: 10.1073/pnas.0808173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Greay SJ, Ireland DJ, Kissick HT, Levy A, Beilharz MW, Riley TV, Carson CF. Cancer Chemother Pharmacol. 2010;65:877. doi: 10.1007/s00280-009-1093-7. [DOI] [PubMed] [Google Scholar]
- 74.Sane AT, Bertrand R. Cancer Res. 1999;59:3565. [PubMed] [Google Scholar]
- 75.Pabla N, Dong G, Jiang M, Huang S, Kumar MV, Messing RO, Dong Z. J Clin Invest. 2011;121:2709. doi: 10.1172/JCI45586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng YR, Suntharalingam K, Johnstone TC, Yoo H, Lin W, Brooks JG, Lippard SJ. J Am Chem Soc. 2014;136:8790. doi: 10.1021/ja5038269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erdlenbruch B, Nier M, Kern W, Hiddemann W, Pekrun A, Lakomek M. Eur J Clin Pharmacol. 2001;57:393. doi: 10.1007/s002280100319. [DOI] [PubMed] [Google Scholar]
- 78.Mitsopoulou CA, Dagas C. Bioinorg Chem Appl. 2010:973742. doi: 10.1155/2010/973742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Machura B, Świtlicka A, Wolff M, Kusz J, Kruszynski R. Polyhedron. 2009;28:3999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.