Abstract
Nitrogen and phosphate source sensing, uptake, and assimilation are essential for the growth and development of microorganisms. In this study, we demonstrated that SACE_6965 encodes the phosphate regulator PhoP, which controls the transcription of genes involved in phosphate metabolism in the erythromycin-producing Saccharopolyspora erythraea. We found that PhoP and the nitrogen regulator GlnR both regulate the transcription of glnR as well as other nitrogen metabolism-related genes. Interestingly, both GlnR- and PhoP-binding sites were identified in the phoP promoter region. Unlike the nonreciprocal regulation of GlnR and PhoP observed in Streptomyces coelicolor and Streptomyces lividans, GlnR negatively controls the transcription of the phoP gene in S. erythraea. This suggests that GlnR directly affects phosphate metabolism and demonstrates that the cross talk between GlnR and PhoP is reciprocal. Although GlnR and PhoP sites in the glnR and phoP promoter regions are located in close proximity to one another (separated by only 2 to 4 bp), the binding of both regulators to their respective region was independent and noninterfering. These results indicate that two regulators could separately bind to their respective binding sites and control nitrogen and phosphate metabolism in response to environmental changes. The reciprocal cross talk observed between GlnR and PhoP serves as a foundation for understanding the regulation of complex primary and secondary metabolism in antibiotic-producing actinomycetes.
INTRODUCTION
Soil-dwelling actinomycetes produce many valuable secondary metabolites that exhibit important pharmacological characteristics and have uses as antibiotics, immunosuppressants, anticancer agents, and many other bioactive compounds. Biosynthesis of these secondary metabolites involves complex regulatory interaction networks, which are themselves finely tuned by many intra- and extracellular signals associated with physicochemical stress and the availability of carbon, nitrogen, and phosphate sources (1–10). Concentrations of easily utilizable nitrogen and phosphate sources in the fermentation medium are critical for secondary-metabolite production (11–17).
Nitrogen control of metabolism in several Actinomycetes species is mediated by a response regulator, GlnR, belonging to the OmpR family (10, 18). The GlnR-mediated transcription regulatory network that coordinates the control of expression of genes involved in nitrogen assimilation, nitrogen metabolism, and other metabolic activity has been explored in Streptomyces coelicolor (19, 20) and Streptomyces venezuelae (21). PhoP is a response regulator of the phosphate-sensing two-component system PhoP-PhoR and has been widely investigated in S. coelicolor and Streptomyces lividans (12, 22–24). Recently, it was proposed that PhoP plays a central role in primary and secondary metabolism in S. coelicolor (25). A complete nutrient-sensing signal transduction pathway, PhoP-AfsR-AfsS-SARP (where SARP represents a Streptomyces antibiotic-related protein, such as ActII-orf4 or RedD), was elucidated to demonstrate that the phosphate-regulatory effect on secondary metabolism was exerted via a cross talk between PhoP and the AfsR and AfsS regulators. The studies also observed that the expression of the glnR gene and some other GlnR-regulated genes is repressed by PhoP in S. coelicolor (5, 24–27). PhoP repressed the transcription of these nitrogen genes by its binding to the glnR promoter, encoding the major nitrogen regulator, to the promoters of glnA and glnII, encoding two glutamine synthetases, and to the promoter of the amtB-glnK-glnD operon, encoding an ammonium transporter (28). These findings reveal cross talk between global regulators (PhoP, GlnR, and AfsR) in S. coelicolor that controls the expression of genes associated with secondary metabolite biosynthesis and is governed by a regulatory network of cross-talking global regulators (CTGRs). The CTGR network orchestrating cell nutritional and environmental stress responses modulates primary and secondary metabolism. Interestingly, no phosphate-related gene was found in the GlnR regulon, suggesting that GlnR has no direct effect on phosphate metabolism and demonstrating that the cross talk between GlnR and PhoP is not reciprocal (29).
Saccharopolyspora erythraea, a rare actinomycete species, has been used for industrial-scale production of erythromycin A, a broad-spectrum macrolide antibiotic against pathogenic Gram-positive bacteria. Nitrogen sources and phosphate concentration significantly influence erythromycin production (30); therefore, a detailed investigation to understand the nitrogen/phosphate utilization, metabolism, and regulation will be valuable to S. erythraea research. We previously identified 25 GlnR-dependent and GlnR-controlled transcription units (TUs) involving a total of 82 genes that are directly related to nitrogen utilization in S. erythraea. These genes are involved in ammonium uptake and assimilation, urea utilization, nitrite/nitrate assimilation, glutamate transport, arginine biosynthesis, nitric oxide biosynthesis, and transcriptional regulation and signal transduction associated with nitrogen source type and availability (31).
In this study, we identified the global regulator PhoP (SACE_6965), which controls seven TUs involved in phosphate metabolism in S. erythraea. The results revealed that PhoP and GlnR both collaboratively regulate the transcription of glnR and other nitrogen metabolism-related genes (glnA1, glnA4, csbX, gltP, ureA, and gltB). Furthermore, we found that GlnR negatively controlled phosphate metabolism through its binding to the promoter of phoP-phoR and that the two global regulators played reciprocal regulatory roles associated with nitrogen and phosphate metabolism in S. erythraea. Our findings present a new complex interconnected regulatory network involving GlnR/PhoP and significantly extend the understanding of regulation mechanisms associated with nitrogen/phosphate metabolism in actinomycetes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this work are listed in Table 1. Spores of S. erythraea strains were grown on agar plates in a medium containing 10 g/liter cornstarch, 10 g/liter corn steep liquor, 3 g/liter NaCl, 3 g/liter (NH4)2SO4, 2 g/liter CaCO3, and 20 g/liter agar in distilled H2O (pH 7.2) at 30°C. Escherichia coli strains were grown at 37°C in liquid or on solid LB medium. All media were sterilized by autoclaving at 121°C for 30 min.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics or use | Source or reference |
|---|---|---|
| Strains | ||
| S. erythraea NRRL23338 | Parental strain, wild type | DSM 40517 |
| E. coli DH5α | F− ϕ80dlacZΔM (lacZYA-argF)U169 deoR | Invitrogen |
| E. coli BL21(DE3)-7101 | The strain for expression of GlnR | Lab stock |
| E. coli BL21(DE3) | F′ ompT rB− mB− (λDE3) | Invitrogen |
| S. erythraea phoPO | Overexpression of phoP; NRRL23338 integrated with pIB139-6965 | This study |
| S. erythraea ΔglnR | NRRL23338 glnR::tsr (glnR-null mutant) | Lab stock |
| S. erythraea null mutant | Plasmid-alone negative control strain with pIB139 integrated into the genome | This study |
| S. erythraea ΔglnR/pIB-glnR (glnRC) | glnR-complemented strain; ΔglnR strain carrying pIB-glnR | Lab stock |
| Plasmids | ||
| pET28a(+) | Vector with T7 RNA polymerase-based promoter for expression in E. coli BL21; hexahistidine tag with thrombin cleavage | Thermo Scientific |
| p7101 | pET28a(+) with glnR (SACE-7101) inserted in the NcoI-HindIII site | This study |
| p6965 | pET28a(+) with phoP (SACE-6965) inserted in the NcoI-HindIII site | This study |
| p6965130 | pET28a(+) with phoPDBD (domain for DNA binding, 390 bp of C terminus) inserted in NcoI-HindIII | This study |
| pUC18-tsr | pUC18 with tsr gene for thiostrepton resistance inserted into the BamHI-SmaI site | 32 |
| pIB139 | pSET152 with integrase of phiC31 and PermE, the strong promoter of Streptomyces | 32 |
| pIB139-7101 | pIB139 with glnR inserted into NdeI-EcoRV | This study |
| pIB139-6965 | pIB139 with phoP inserted into NdeI-EcoRV | This study |
Construction of an S. erythraea phoP-overexpressing mutant.
A phoP overexpression (phoPO) mutant strain (YE6965) was generated by protoplast transformation with pIB139-6965 (Table 1) carrying a 681-bp fragment of the phoP open reading frame (SACE_6965) inserted between NdeI and EcoRV restriction sites (32). The plasmid pIB139-6965 was integrated into the S. erythraea genome using the phiC31 integrase. Apramycin resistance was examined following the addition of apramycin at a final concentration of 100 μg/ml, 24 h after transformation. The selected mutants were verified by PCR, real-time quantitative PCR (RT-qPCR), and DNA sequencing (PCR was used to amplify the fragment spanning the inserted phoP gene and its upstream and downstream plasmid regions using primers G6965F and G6965R; RT-qPCR was used to investigate the transcript levels of phoP with primers RT6965F and RT6965R). All primers used in this study are listed in Table 2. The negative control consisted of a plasmid-only construct that underwent protoplast transformation with pIB139 and was labeled “null.” Our attempts to conduct experiments involving a phoP-null mutant strain were unsuccessful.
TABLE 2.
Primers used in this work
| Purpose and primer | Sequence (5′–3′) |
|---|---|
| PCR | |
| RT0175F | CCCCGTGCAGTGGGAGGT |
| RT0175R | GCGCGGAAGCGGTAGAAGTA |
| RT6716F | CTGGCTGCTGGAAGGTTTCC |
| RT6716R | TCAGGTCGTTGGCGTAGTGCT |
| RT7169F | GCCCGAGACCAAGCACCC |
| RT7169R | CGCCGCTGTTGCTGATGAG |
| RT6966F | CGCCAACGTCAGCCACG |
| RT6966R | AACGGCCCAGCGCCTC |
| RT7091F | CCAGATCAAGAAGTGGAACGACC |
| RT7091R | GAGCGGAAGAACGGGACGA |
| RT7099F | CAGATCAAGAAGTGGAACGACCC |
| RT7099R | GAGCGGAAGAACGGGACGA |
| RT6551F | GGCGAGAACGGCGAGC |
| RT6551R | GGGAACCTGATGCGGATGC |
| RT0658F | CCGACCTCGCCGACACCA |
| RT0658R | TACGCCCAGACCCCCTCC |
| RT7101F | GCAGGAGGTCTGGGGCTACG |
| RT7101R | GACGGGCGGACGAACTTGTAG |
| RT6965F | CGGTCAGGAGGCGTTGGAG |
| RT6965R | GCTGCTTGCAGACGTCGGT |
| RT0634F | ACGAGCGCGACAAGCTTCTC |
| RT0634R | ACCTGTTCGGCGGTCAGGAC |
| RT2830F | TGAAGATCCTCGGTGACCTGTT |
| RT2830R | CGACTCCATCTGCGTGATGC |
| RT5355F | GACAACCACGCGCTCTACAAGA |
| RT5355R | ATGAAGTGCTCCATCAGCGGGG |
| RT2507F | CGGCTACACCCCGCTCCCAC |
| RT2507R | GCTCCACACCACGCCCTTCG |
| RT6473F | AGGTGGTGGCGTATCCGAGCAT |
| RT6473R | GCGCGGAGTCGTCGAAAAGCTC |
| RT3798F | GTCTCCACTGTGGCCTCCTACA |
| RT3798R | CACAGTGGAGACGCCCATCT |
| RT8101F | GTTGCGATGCCGTGAGGT |
| RT8101R | CGGGTGTTACCGACTTTCA |
| G6965F | GAGGTACCGGTGATCATGG |
| G6965R | GTTGTGTGGAATTGTGAGCGG |
| PhoP_F | TAAGAATTCGTGACCAGGGTGCTGATCGTGG |
| PhoP_R | TAAAAGCTTTCACACCTCGAACTTGTAGCCGAG |
| PhoDBD_F | CCGGAATTCGAGCTGATCGCCCGG |
| PhoDBD_R | CCCAAGCTTTCACACCTCGAACTTGTAGCCG |
| EMSA | |
| PHO-boxF | GCCTTGTTCACCTGGGGTTCACCTCCAGGGCG |
| PHO-boxR | CGCCCTGGAGGTGAACCCCAGGTGAACAAGGC |
| Bio-primer | Biotin-AGCCAGTGGCGATAAG |
| 7099F | AGCCAGTGGCGATAAGCGGCGATCAAGGTCTA |
| 7099R | AGCCAGTGGCGATAAGGCGAGGAAGCCCAG |
| 6551F | AGCCAGTGGCGATAAGTGCTGGGAGCTCGG |
| 6551R | AGCCAGTGGCGATAAGACCACGACACCGAGG |
| 6473F | AGCCAGTGGCGATAAGGTCGTTGTCGCCGAG |
| 6473R | AGCCAGTGGCGATAAGGGCGTCGATGAGGAT |
| 0175F | AGCCAGTGGCGATAAGCGGAGAGCGAACTGG |
| 0175R | AGCCAGTGGCGATAAGGCCGAGCAGCACGC |
| 6716F | AGCCAGTGGCGATAAGGTGTGACCTCCACCTTC |
| 6716R | AGCCAGTGGCGATAAGGGTCCTGCGGGTCA |
| 6965F | AGCCAGTGGCGATAAGCCAGGGCCGCTGAC |
| 6965R | AGCCAGTGGCGATAAGCGCCCACGAGCAGC |
| 7169F | AGCCAGTGGCGATAAGCGGCACCAATCACGA |
| 7169R | AGCCAGTGGCGATAAGGCCAGCGCCGTCAG |
| 0658F | AGCCAGTGGCGATAAGCATGCCGCGGGCG |
| 0658R | AGCCAGTGGCGATAAGTTGAGGGAGGTGGCG |
| 7169F | AGCCAGTGGCGATAAGCGGCACCAATCACGA |
| 7169R | AGCCAGTGGCGATAAGGCCAGCGCCGTCAG |
| 7101F | AGCCAGTGGCGATAAGCCGGTGTACAGCAAC |
| 7101R | AGCCAGTGGCGATAAGTGCGGCAGCAGACCC |
| 3998F | AGCCAGTGGCGATAAGCGGAGTCGCGTGTGG |
| 3998R | AGCCAGTGGCGATAAGGGTCGTAGAGCCCCC |
| 6473F | AGTGGCGATAAGGAGCCGGACCCCCAGC |
| 6473R | AGTGGCGATAAGGCCGTGGCGTCGATGA |
| 3095F | AGCCAGTGGCGATAAGGGTGAACTACGAGGT |
| 3095R | AGCCAGTGGCGATAAGGTGGCGTCGAGATCG |
| 2507F | AGTGGCGATAAGATTCCGCAAGCCCTCA |
| 2507R | CGATAAGCAGCCGATCCGGTCGC |
| 2830F | AGCCAGTGGCGATAAGCGCCGTCACTGTTTT |
| 2830R | AGCCAGTGGCGATAAGGTGCCGACGACCATC |
| 0634F | AGCCAGTGGCGATAAGATGATGCCTCGTCTC |
| 0634R | AGCCAGTGGCGATAAGACGAGAAGCTTGTCG |
| 5355F | AGCCAGTGGCGATAAGGCGGTGTCGTCGTTGT |
| 5355R | AGCCAGTGGCGATAAGGCGCAGCTCCTCGAC |
Underlining indicates restriction enzyme sites.
Overexpression and purification of the PhoP protein in E. coli.
For heterologous expression of PhoP protein in E. coli, the phoP gene (SACE_6965) was amplified by PCR from the S. erythraea NRRL23338 genome using the primers PhoP_F (with the EcoRI restriction site) and PhoP_R (with the HindIII restriction site), and cloned into a pET-28a(+) vector in order to generate a His tag fusion protein. The pET-28a(+)-phoP plasmid (p6965) was transformed into E. coli BL21(DE3) cells and grown in LB medium at 37°C in an orbital shaker at 250 rpm to an optical density at 600 nm (OD600) of 0.6. The expression of phoP was induced by the addition of isopropyl-β-d-1-thiogalactopyranoside at a final concentration of 0.1 mM and grown for an additional 6 to 8 h. Cells were harvested by centrifugation and washed twice with phosphate-buffered saline (PBS) (pH 8.0), and the cells were lysed by ultrasound. Cell debris and membrane fractions were separated from the soluble fraction by centrifugation at 12,000 × g for 45 min at 4°C. His-tagged PhoP (His6-PhoP) was purified using nickel-nitrilotriacetic acid (Ni-NTA) Superflow columns (Qiagen, Valencia, CA, USA). The protein presented a maximal elution peak at ∼250 mM imidazole in a buffer consisting of 50 mM NaH2PO4 and 300 mM NaCl (pH 8.0). Fractions containing His6-PhoP were pooled and dialyzed in buffer D containing 50 mM Tris, 0.5 mM EDTA, 50 mM NaCl, 20% glycerol, and 1 mM dithiothreitol (DTT) (pH 8.0) at 4°C and stored at −80°C. Purity of the His6-PhoP protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentration was determined using the Bradford reagent. The gene fragment encoding the C-terminal 130 amino acids of PhoP (PhoPDBD) was amplified using the primers PhoDBD_F and PhoDBD_R. The heterologous expression and purification of PhoPDBD protein in E. coli were as described for the PhoP protein.
EMSA.
The upstream regions (−300 to + 50) of phoP, glnR, and other PhoP-targeted genes were amplified by PCR using gene-specific primers containing the universal primer (5′-AGCCAGTGGCGATAAG-3′) sequence (Table 2) and biotin labeled by PCR using the 5′ biotin-modified universal primer. The PCR products were analyzed by agarose gel electrophoresis and purified using a PCR purification kit (Generay Biotech Co., Ltd., Shanghai, China). The concentration of biotin-labeled DNA probes was determined using a microplate reader (Biotek, Winooski, VT, USA). Electrophoretic mobility shift assays (EMSAs) were carried out according to manufacturer protocol for the chemiluminescent EMSA kit (Beyotime Biotechnology, Jiangsu, China). The binding reaction mixture contained 10 mM Tris-HCl (pH 8.0), 25 mM MgCl2, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.01% Nonidet P40, 50 μg/ml poly[d(I-C)], and 10% glycerol. After binding, the samples were separated on a native PAGE gel in an ice bath containing 0.5× Tris-borate-EDTA at 100 V, and bands were detected using BeyoECL Plus (Beyotime Biotechnology). The amount of all DNA probes or double-stranded adapters used in the EMSA experiments was ∼1 pmol, with the protein amount at ∼10 pmol. The reactions were performed at 20°C for 15 min.
Computational analysis.
The MEME/MAST tools (http://meme-suite.org/) and PREDetector (33) software were used to identify the GlnR/PhoP binding motif. Multiple alignment analysis of PhoP homologues was performed using ClustalX2 (34), and the phylogenetic tree was built using MEGA4 (35).
DNase I footprinting assay.
DNase I footprinting assays were performed as previously described (36). The promoter region of phoP was PCR amplified using primers P6965F and P6965R (Table 2), and the amplicon was cloned into the T-vector pUC18B-T (Shanghai Biotechnology Corporation, Shanghai, China). The obtained plasmids were used as templates for further preparation of biotin-labeled probes using the universal primer (bio-Tprimer) (Table 2). After agarose gel electrophoresis, the 6-carboxyfluorescein (FAM)-labeled probes were purified using a QIAquick gel extraction kit (Qiagen) and quantified using a NanoDrop 2000C (Thermo Fisher, Waltham, MA, USA). For each assay, 200 ng of each probe was incubated with different amounts of His-PhoP or His-GlnR in a total volume of 40 μl in EMSA buffer (Beyotime Biotechnology). After incubation for 30 min at 25°C, 10 μl of solution containing ∼0.015 units DNase I (Promega, Madison, WI, USA) and 100 nM freshly prepared CaCl2 was added, and the sample was incubated for 1 min at 25°C. The reaction was stopped by adding 140 μl DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS); the samples were extracted with phenol-chloroform and precipitated using ethanol, and the pellets were dissolved in 30 μl Milli-Q water. Preparation of the DNA ladder, electrophoresis, and data analysis were performed as previously described (36), except that the GeneScan-LIZ500 size standard (Applied Biosystems, Foster City, CA, USA) was used.
Binding assays.
All binding assays were performed in running buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM TCEP [trichloroethyl phosphate], and 0.5 mg/ml bovine serum albumin [BSA]) using an Octet Red instrument (ForteBio, Inc., Menlo Park, CA, USA) at 25°C. Streptavidin-coated biosensors containing immobilized biotinylated double-stranded DNA (dsDNA) (PCR products of the upstream regions of glnR and phoPR obtained with Bio-primer, namely, PglnR and PphoP) were exposed to different concentrations of His6-GlnR or His6-PhoP. The equilibrium dissociation constants (KDs) associated with GlnR and PhoP were calculated and fitted to binding curves using the corresponding analysis software (ForteBio, Inc.).
RNA preparation and RT-PCR.
Cell pellets were collected after 20 min of centrifugation at 4,000 × g. Total RNA was extracted using the RNAprep Pure cell/bacterium kit (Tiangen Biotech Co., Ltd., Beijing, China). RNA integrity was analyzed by 1% agarose gel electrophoresis, and RNA concentration was determined using a microplate reader (BioTek). Reverse transcription was undertaken using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Shiga, Japan). The RT-PCRs were performed in a 20-μl PCR solution from the SYBR premix Ex Taq GC kit (Perfect Real Time; TaKaRa, Japan) using ∼100 ng cDNA as the template. All procedures were performed according to the manufacturer's instructions. PCR was conducted using a CFX96 real-time system (Bio-Rad, Hercules, CA, USA) with PCR conditions of 95°C for 5 min, then 40 cycles at 95°C for 5 s and 58 to 63°C for 30 s, and an extension at 72°C for 10 min.
Erythromycin determination.
The method used for erythromycin determination by high-performance liquid chromatography was previously described (31).
RESULTS
SACE_6965 encodes the putative phosphate utilization regulator PhoP in S. erythraea.
In order to investigate the phosphate control of metabolism, the protein sequences of the S. coelicolor two-component system, PhoP-PhoR (SCO4230-SCO4229), were used as query sequences to aid in the identification of their orthologues in S. erythraea. SACE_6965 and SACE_6966 were identified as the closest homologues of SCO4230-SCO4229 (see Fig. S1 in the supplemental material). The results revealed a high similarity throughout the full-length sequence (especially in DNA recognition helical regions) and, therefore, suggested that SACE_6965 encodes the putative phosphate utilization regulator PhoP, which plays an important role in regulating gene expression in response to phosphate availability in S. erythraea.
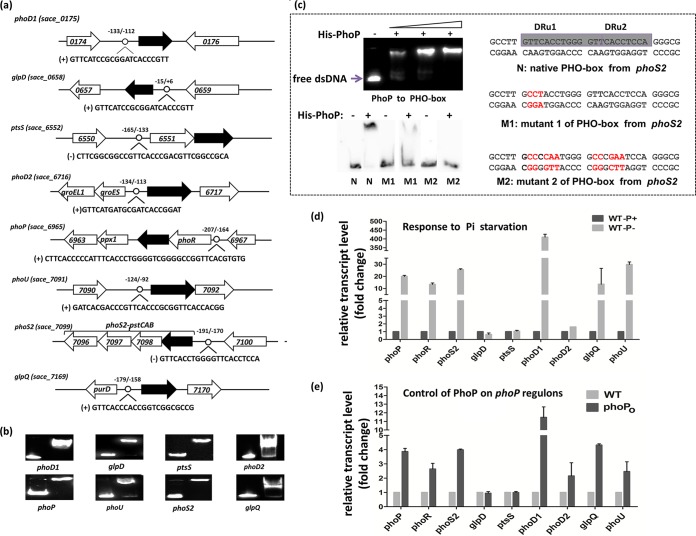
The upstream regions of the genes involved in phosphate metabolism have been reported to contain a putative PhoP-binding motif (PHO box) in different Streptomyces strains, with a typical PHO box consensus being identified. The PHO box is formed by two conserved direct repeat units (DRUs) of 11 nucleotides in actinomycetes, with each one formed by an identical five-nucleotide sequence (GT/GTCA) followed by a six-nucleotide less-conserved tail region, and bound by a protein monomer (12, 22). Based on the Streptomyces PHO box motif, we searched putative PhoP-binding sites in upstream of all the S. erythraea orthologues of known genes involved in the uptake, assimilation, and metabolism of phosphate in actinomycetes. Combined utilization of MAST/MEME tools and the PREDetector software identified putative PHO box sequences in the upstream regions of eight transcription units (TUs) associated with phosphate metabolism (Fig. 1a). These include genes involved in phosphate transport (the phoS2-pstCAB operon; the SACE_6551-SACE_6552 operon, encoding phosphate-binding proteins; and phoU, encoding the phosphate-specific transport system accessory protein PhoU), glycerophospholipid metabolism (glpD, encoding glycerol-3-phosphate dehydrogenase, and glpQ, encoding glycerophosphoryl diester phosphodiesterase), phosphate uptake (phoD1 and phoD2, coding for alkaline phosphatase D), and the two-component system (SACE_6965-SACE_6966). EMSA experiments with purified SACE_6965 were performed, revealing their binding to the upstream promoter regions of eight TUs (Fig. 1b). In order to confirm the PHO box sequences, the 5′ biotin-labeled synthetic dsDNA fragments containing portions of the predicted PhoP-binding motif and mutant motif (in the upstream region of the phoS2-pstCAB operon) were used for the EMSA. The results showed that the native PhoP-binding motif was shifted by purified S. erythraea SACE_6965. However, the shifted bands of the mutant fragments were evidently weakened (Fig. 1c). Taken together, these results indicate that SACE_6965 is the closest homologue of the phosphate regulator PhoP studied in Streptomyces spp., and we propose naming it PhoP.
FIG 1.
PhoP regulates genes involved in phosphate metabolism. (a) PhoP-binding motifs were identified in phosphate-related genes. (b) EMSA results indicating His-PhoP interactions with the predicted motifs. The DNA probes containing the predicted motifs were incubated with 1 μM protein and a 200-fold excess of nonspecific competitor DNA (sperm DNA). (c) EMSA results showing interactions between His-PhoP and the PHO box or mutant PHO box in the phoS2 gene. (d) Transcription profiles of phoPR and PhoP-regulated genes in response to phosphate downshift. (e) Transcription profiles of phoPR and PhoP-regulated genes in response to overexpression of phoP gene.
The transcription levels of genes containing a putative PHO box in response to phosphate availability were investigated using phosphate downshifted cultures. The S. erythraea wild-type strain NRRL23338 was grown in high-phosphate MG medium (MG-10 mM, 60 g/liter starch, 60 mM glutamate, and 10 mM phosphate) for 40 h and then transferred to low-phosphate medium (MG-50 μM) for 12 h. Two culture samples were collected before and after the downshift in order to isolate mRNA for RT-PCR. As expected, the phoP-phoR operon was strongly induced, while expression of most target genes was also upregulated under low-phosphate conditions. The quantitative RT-PCR experiments indicated that transcript levels significantly increased after the downshift: 410-fold for phoD2, 30-fold for phoU, 25.5-fold for phoS2, 20-fold for phoP, 13.4-fold for phoR, and 13.5-fold for glpQ (Fig. 1d). The most strongly induced gene was phoD1 (410-fold increase), which encodes alkaline phosphatase D and is responsible for removing phosphate groups from many types of phosphate-containing molecules prior to uptake. No change in glpD and ptsS transcript levels was observed. To further examine the regulatory role of PhoP on the expression of identified target genes, we constructed a phoP-overexpressing (phoPO) mutant strain exhibiting an ∼4-fold increase in gene expression (see Fig. S2 in the supplemental material). The overexpression of phoP resulted in a 2- to 11-fold (11.5-fold for phoD1, 4-fold for phoS2, 4.3-fold for glpQ, 2.5-fold for phoD, and 2.2-fold for phoD2) increase in induction of target genes (Fig. 1e). A similar result was observed in the wild-type strain responding to phosphate starvation. These results further demonstrate that PhoP exhibits a regulatory function as a transcriptional activator of these target genes involved in phosphate metabolism.
PhoP directly controls the transcription of glnR and other GlnR-regulated genes involved in nitrogen metabolism.
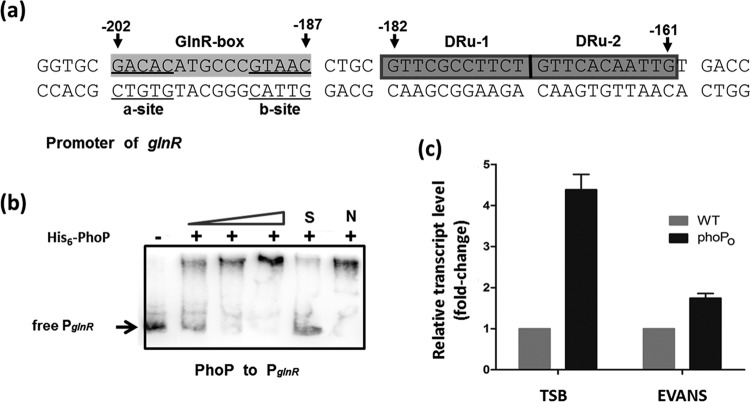
Previously, we reported that the OmpR-type response regulator GlnR (encoded by SACE_7101) was a central global regulator of nitrogen metabolism in S. erythraea (31). A typical PHO box (two DRUs, GTTCGCCTTCT and GTTCACAATTGT) was observed in the upstream region (from position −182 to −161, relative to the translation start site in the antisense strand) of the glnR gene and separated by only 4 bp from the GlnR-binding motif (Fig. 2a). In order to determine whether the PhoP protein binds directly to the promoter regions of the glnR gene, gradient EMSAs were performed using purified His-PhoP protein. To assess binding specificity, an unlabeled specific probe (100-fold) or nonspecific competitor DNA (100-fold; salmon sperm DNA) was used. Obvious shifted bands were observed in all EMSAs following addition of His-PhoP (Fig. 2b). The results indicated that the DNA fragment from the glnR upstream region containing the predicted PhoP binding site was bound by His-PhoP, thus suggesting that global nitrogen regulator GlnR in S. erythraea is subject to transcriptional regulation by the phosphate regulator PhoP.
FIG 2.
PhoP controls glnR transcription. (a) Putative PhoP-binding sites in the glnR promoter region. (b) EMSA results indicating His-PhoP interactions with the glnR upstream promoter region; unlabeled specific probe (100-fold) (S) or nonspecific competitor DNA (100-fold, salmon sperm DNA) (N) was added. (c) The positive regulation of PhoP on the transcription of glnR in TSB and EVANS-15 mM glutamine. Changes represent the level of expression in the phoP overexpression mutant (phoPO) relative to the expression levels observed in the wild-type strain. Results were normalized to 16S rRNA levels. Data are means and standard deviations from three independent experiments, each with triplicate samples, using distinct cDNA preparations for each RNA sample.
To elucidate the regulatory effects of PhoP on the glnR gene in S. erythraea, glnR transcription levels were compared with those of the wild-type and phoPO mutant strains in nitrogen-rich tryptic soy broth (TSB) medium and nitrogen-limited conditions (minimal Evans medium supplemented with 2 mM glutamine). As shown in Fig. 2c, phoP overexpression resulted in a 4.4-fold increase in glnR induction in the nitrogen-rich medium, whereas only a 1.6-fold increase in glnR induction was seen in the nitrogen-limited medium. No changes in erythromycin production or glnR or phoP transcript levels were observed in the plasmid-only negative control (see Fig. S3 in the supplemental material). These results confirm that PhoP enhances glnR expression in S. erythraea.
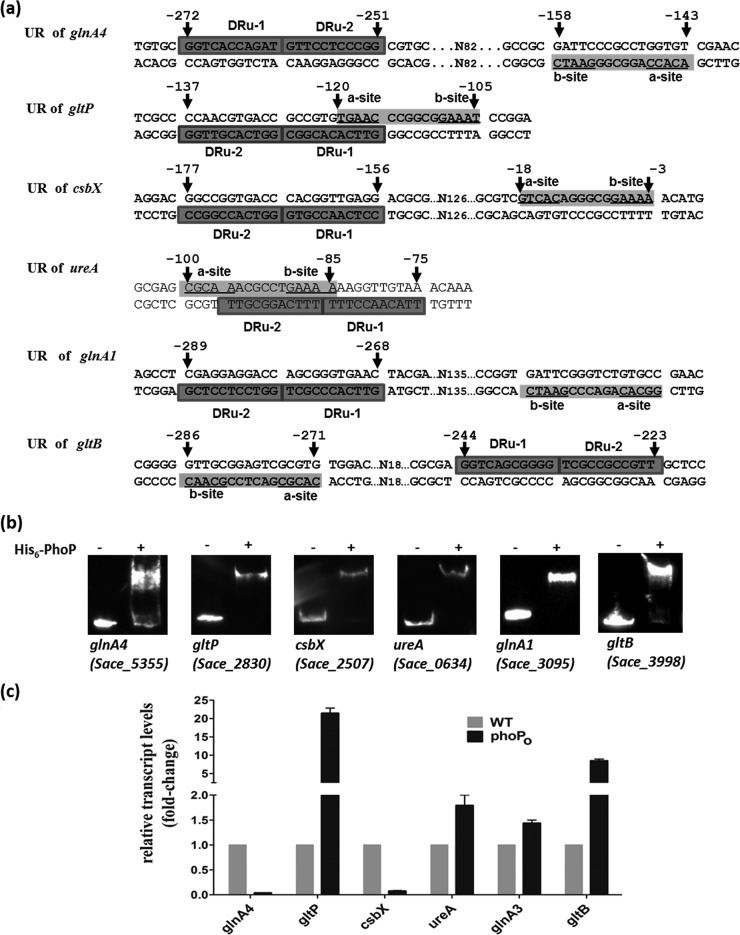
PhoP exerts direct and indirect regulatory effects on nitrogen metabolism in S. coelicolor (25). By searching for promoter regions containing a consensus PhoP-binding motif, we found that many genes involved in nitrogen metabolism are also regulated by PhoP in S. erythraea. The genes or operons having PHO boxes in their promoter regions include glnA3 (SACE_3095; two DRUs [GTTCACCCGCT and GGTCCTCCTCG]) and glnA4 (SACE_5355; two DRUs [GGTCACCAGAT and GTTCCTCCCGG]), which encode two glutamine synthetases; ureABCFGD (SACE_0634 to -0639; two DRUs [TTGCGGACTTT and TTTCCAACATT]), which encode enzymes involved in urea utilization; gltP (SACE_2830; two DRUs [GTTCACACGGC and GGTCACGTTGG]), which encodes a Na+/glutamate:H+ symporter; gltB-gltD (SACE_3998 and SACE_3997; two DRUs [GGTCAGCGGGG and TCGCCGCCGTT]), which encode glutamate synthase (NADPH); and csbX (SACE_2507; two DRUs [CCTCAACCGTG and GGTCACCGGCC]), which encodes α-ketoglutarate permease (Fig. 3a). All of these genes were identified as being targeted by GlnR in our previous study (31). Similar to S. coelicolor PhoP, S. erythraea PhoP can exert control of the nitrogen metabolism through its binding to the promoter of the glnR gene. Moreover, PhoP exerts similar effects on ammonium assimilation, via glutamine synthetase/glutamate synthase (GS/GOGAT), through direct binding to the promoters of glnA, glnA4, and gltB-gltD, whereas PhoP appears to have no effect on the uptake of ammonium (amt operon). Additionally, S. erythraea PhoP exerts a regulatory effect on the uptake of glutamate (gltP) and its precursor α-ketoglutarate (csbX), as well as urea utilization (ureABCFGD).
FIG 3.
PhoP regulates genes involved in nitrogen metabolism in S. erythraea. (a) Predicted GlnR and PhoP binding motifs in the upstream regions (URs) of nitrogen-related genes. (b) EMSA results indicating interactions between His-PhoP and the upstream regions of PHO box-containing genes following incubation with biotin-labeled DNA with 1 μM protein and a 200-fold excess of nonspecific competitor DNA. (c) RT-PCR data. Changes represent expression levels in the wild-type strain relative to the expression levels observed in the phoPO mutant.
To determine whether PhoP binds to the promoter regions of these genes, EMSAs were performed using purified His-PhoP with biotin-labeled DNA probes (representing the upstream regulatory regions of each gene from −250 to +50) containing all of the predicted PhoP motifs. The results confirmed that PhoP binds to these regions (Fig. 3b). RT-PCR experiments indicated that the overexpression of PhoP resulted in the induction of gltP (21.4-fold), gltB (8.5-fold), ureA (1.8-fold), and glnA1 (1.5-fold) in TSB medium. Meanwhile, the transcription levels of glnA4 and csbX significantly decreased in the phoPO mutant by factors of 27-fold and 13-fold, respectively (Fig. 3c). In contrast, S. coelicolor PhoP revealed direct and indirect negative control of glnR, glnA, glnII, and amtB expression, whereas no positive induction of nitrogen-related genes was observed (24, 28).
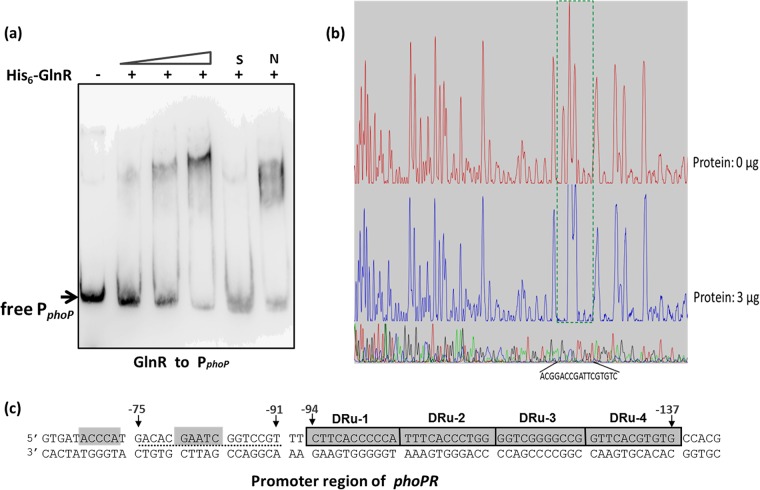
GlnR reciprocally controls the transcription of the phoP gene in S. erythraea.
There is nonreciprocal regulation of the GlnR-regulated genes by the PhoP protein in S. coelicolor and S. lividans (25). PhoP directly controls at least seven nitrogen metabolism-related genes, while GlnR does not regulate the phosphate regulon genes (29). However, we found that S. erythraea GlnR binds to the phoPR operon promoter (Fig. 4a). Shifted bands were observed for all EMSAs when the His-GlnR protein was added. In order to identify the exact DNA sequences that bound GlnR in the phoP promoter region, a DNase I footprinting assay using purified recombinant His-GlnR and a fluorescent FAM-labeled probe was performed. With the addition of His-GlnR (3 μg), a protected region of 17 nucleotides was detected from position −75 to −91, relative to the translation start site in the phoPR promoter coding strand (Fig. 4b). These results revealed that the GlnR-binding site is located in close proximity to the PHO box (separated by only 2 bp), indicating that the two regulators may competitively bind to the adjacent binding sites (Fig. 4c).
FIG 4.
GlnR binds to the phoPR promoter and controls its transcription. (a) EMSA using 1 μM purified S. erythraea His6-GlnR protein and 1 pmol biotin-labeled PCR products from the upstream region of operon phoPR. A 200-fold excess of nonspecific competitor DNA was included in every lane as an internal control to avoid unspecific binding of the protein to the DNA. (b) DNase I footprinting assays for the GlnR-PphoPR complexes. (c) The protected DNA sequences located in upstream region of phoPR. The black dotted line indicates the respective footprints of GlnR.
In order to determine the affinities of GlnR and PhoP for the upstream regions of the phoP and glnR genes, an interferometry assay was performed using the Octet system (ForteBio, Inc.). The KD was determined by immobilizing biotin-modified DNA fragments with increasing amounts of purified regulator proteins. We found that GlnR and PhoP exhibited different affinities (about 2- to 5-fold) for PphoP and PglnR. Purified His-GlnR and His-PhoP bound to PglnR with KDs of ∼203 nM and ∼1.06 μM, respectively, and bound to PphoP with KDs of ∼749 nM and ∼198 nM, respectively. GlnR had a 5-fold-higher affinity for PglnR than PhoP, while PhoP had a 3.8-fold-higher affinity for PphoP than GlnR.
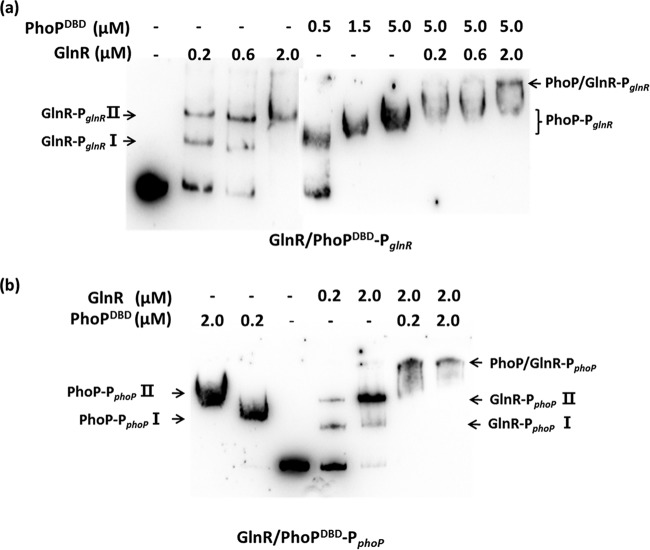
In order to investigate whether GlnR and PhoP interfere with the binding of one another to PglnR and PphoP, competitive EMSAs were conducted using His-GlnR and His-PhoP both together and separately. It was difficult to discriminate between the shifted bands of GlnR and PhoP in EMSA experiments, given their similar molecular weights; however, we found that His-PhoPDBD (protein containing only the DNA binding domain, amino acids 97 to 226) and His-PhoP displayed similar binding affinities for the PHO box (see Fig. S4 in the supplemental material). As shown in Fig. 5a, His-GlnR binds to PglnR to produce shifted bands (Fig. 5a, lanes 2 to 4) in the absence of His-PhoP. In the absence of His-GlnR, the shifted bands representing PglnR-PhoPDBD (Fig. 5a, lanes 5 to 7) were observed with increasing concentrations of His-PhoPDBD. However, the addition of two transcription factors to the in vitro reaction resulted in loss or attenuation of the shifted bands corresponding to PglnR-GlnR and PglnR-PhoPDBD, resulting in an additional band shifted to a greater degree, corresponding to PglnR-GlnR/PhoPDBD complex (a complex of both factors) (Fig. 5a, lanes 8 to 10). Similarly, in the presence of both His-GlnR and His-PhoP, the shifted bands corresponding to PphoP-GlnR and PphoP-PhoPDBD disappeared, whereas the largest shifted band representing PphoP-GlnR/PhoPDBD complex was observed (Fig. 5b). These results demonstrated that both transcription factors were able to simultaneously bind to the promoter regions of their respective genes.
FIG 5.
Interaction of both PhoP and GlnR with promoter regions. (a) Competitive EMSA results indicating the binding of His6-GlnR and His6-PhoPDBD to the glnR promoter (PglnR). (b) Competitive EMSA results indicating the binding of His6-GlnR and His6-PhoPDBD to the phoP promoter (PphoP).
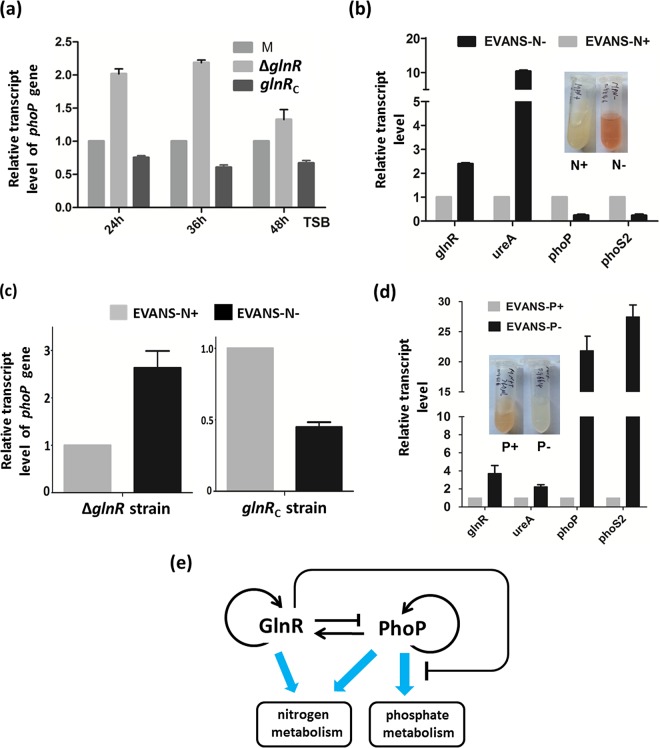
To further elucidate the regulatory effects of GlnR on phoP transcription in S. erythraea, we constructed a glnR deletion mutant (ΔglnR) and a glnR-complemented (glnRC) strain using methods described previously (31, 37). Three strains were cultivated in TSB medium, cells were harvested for RNA extraction at three time points (24 h, 36 h, and 48 h), and phoP transcription was analyzed using RT-PCR. As shown in Fig. 6a, ∼2-fold increases in phoP transcription were observed in the ΔglnR mutant strain, indicating that phoP was repressed by GlnR. Complementation of the glnR gene restored this repressive effect. We also examined the effect of GlnR on phoP transcription in response to nitrogen availability. S. erythraea wild-type strains were grown for 2 days at 30°C in TSB medium and then collected and washed twice with normal saline. The cell pellets were added to Evans medium with glutamine as the nitrogen source. RT-PCR experiments revealed that phoP transcription levels were enhanced 6-fold in the high-nitrogen medium relative to the levels observed in the low-nitrogen medium (Fig. 6b). It was also observed that the nitrogen-mediated effect on phoP expression disappeared in the ΔglnR strain and was recovered in the glnRC strain (Fig. 6c). These results demonstrate that GlnR represses phoP transcription in response to nitrogen availability in S. erythraea. Next, we examined the transcript levels of other genes (glnR, ureA, phoP, and phoS2) involved in nitrogen and phosphate metabolism in nitrogen- and phosphate-limited or -rich media. As expected, nitrogen-related genes (glnR and ureA) were induced about 2.5- to 10-fold under nitrogen-limited conditions (Fig. 6b), while phosphate-related genes (phoP and phoS2) were induced about 22- to 27-fold under phosphate-limited conditions (Fig. 6d). Low phosphate also resulted in the induction of the genes glnR and ureA (about 2- to 4-fold), as the target genes of PhoP. However, we found that the phosphate-related gene phoS2, which is not a direct target gene of GlnR, was repressed under nitrogen-limited conditions (Fig. 6b). Taken together, the results indicate that there appears to be reciprocal regulatory cross talk between GlnR and PhoP in S. erythraea, unlike S. coelicolor and S. lividans, indicating multiple complex regulatory processes associated with response to external nutrition conditions, such as nitrogen and phosphate concentrations. This PhoP-GlnR regulatory circuit, which includes a positive and a negative interaction, leads to homeostasis (Fig. 6e).
FIG 6.
Transcription levels of genes involved in nitrogen/phosphate metabolism in response to nutrient availability. (a) Differences in phoP transcription between the wild-type, ΔglnR, and glnRC strains in TSB medium. RNAs were extracted from S. erythraea NRRL2338 and the ΔglnR and glnRC strains grown in TSB at 24 h, 36 h, and 48 h. Results were normalized to 16S rRNA levels. (b) Transcription profiles of genes (glnR, ureA, phoP, and phoS2) under nitrogen-rich conditions (i.e., in Evans medium with 30 mM glutamine [Evans-N+]) or nitrogen-limited conditions (i.e., in Evans with 2 mM glutamine [Evans-N−]). (c) Transcription profiles of phoP in ΔglnR and glnRC strains in Evans-N+ or Evans-N−. (d) Transcription profiles of genes (glnR, ureA, phoP, and phoS2) under phosphate-rich conditions (i.e., in Evans medium with 10 mM K2HPO4 [Evans-P+]) or phosphate-limited conditions (i.e., in Evans medium with 40 μM K2HPO4 [Evans-P−]). Fold changes represent gene expression levels in Evans-N+ or -P+ relative to the expression levels observed in Evans-N− or -P−. Data are means and standard deviations from three independent experiments, each with triplicate samples, using distinct cDNA preparations for each RNA sample. (e) The PhoP-GlnR regulatory circuit, which includes a positive and a negative interaction.
Impact of nitrogen and phosphate status on antibiotic biosynthesis.
Finally, we investigated the effect of GlnR and PhoP on S. erythraea growth and erythromycin production. The growth curves observed for the wild type (WT), the glnR deletion (ΔglnR) mutant, the glnR-complemented (glnRC) strain, the phoP-overexpressing mutant (phoPO), and the phoP-overexpressing ΔglnR (ΔglnR phoPO) strain in TSB medium are presented in Fig. S5A in the supplemental material. No change of growth in all five strains was observed. Notably, both regulators exerted an impact on erythromycin production; specifically, production of erythromycin in the ΔglnR and phoPO mutants increased by ∼23% and ∼40%, respectively, compared to the wild-type strain, with further improvement by ∼56% observed in the ΔglnR phoPO strain (see Fig. S5B in the supplemental material). The results revealed that GlnR repressed whereas PhoP activated erythromycin production, thereby demonstrating the importance of nitrogen/phosphate source sensing/utilization in S. erythraea antibiotic production.
DISCUSSION
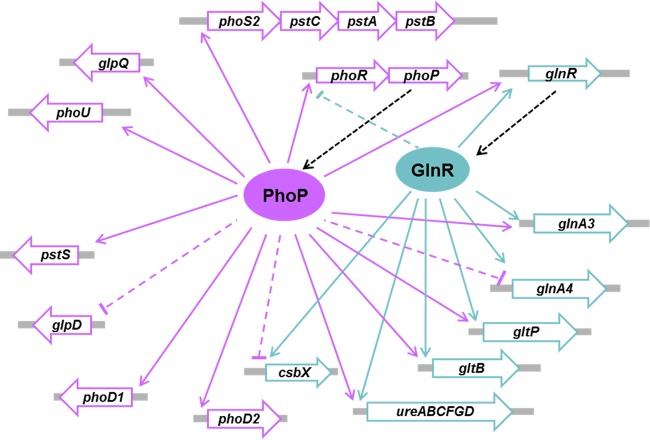
Here, we identified PhoP as a global regulator of phosphate utilization in S. erythraea and showed the reciprocal cross talk between PhoP and the nitrogen regulator GlnR, suggesting a reciprocal regulatory GlnR/PhoP network in coordinating metabolic response according to nitrogen/phosphate availability. PhoP negatively controls ammonium uptake and assimilation by repressing the transcription of glnR, glnA, glnII, and the amtB-glnK-glnD operon in S. coelicolor (24). The regulatory effects of S. erythraea PhoP on nitrogen metabolism revealed three distinct features: (i) S. erythraea PhoP can exert positive control of nitrogen metabolism through its binding to the promoter of glnR gene; (ii) PhoP exerts enhanced ammonium assimilation of glutamine synthetase/glutamate synthase (GS/GOGAT) through direct binding to the promoters of glnA, glnA4, and gltB-gltD, while PhoP has no effect on the uptake of ammonium (amt operon); (iii) S. erythraea PhoP exerts regulatory effects on the uptake of glutamate (gltP) and its precursor α-ketoglutarate (csbX), as well as utilization of urea (ureABCFGD). Interestingly, GlnR- and PhoP-binding sites were identified in the promoter regions of both glnR and phoP in S. erythraea. Unlike the nonreciprocal regulation of GlnR and PhoP in S. coelicolor, S. erythraea phoP and phoR genes were found in the GlnR regulon, suggesting that GlnR exerts a direct regulatory effect on phosphate metabolism and demonstrating that the cross talk between GlnR and PhoP is reciprocal. It is worth noting that two binding sites in the glnR and phoP promoter regions are located in close proximity to one another (separated by only 2 to 4 bp), suggesting that the two proteins may competitively bind to their adjacent binding sites. However, the experimental results revealed that the binding of GlnR and PhoP to their respective sites occurred independently of one another without any interference. Both regulators collaboratively controlled nitrogen and phosphate metabolism in response to environmental changes. In summary, these observations demonstrated that S. erythraea has an extensive cross talk mechanism between regulatory systems involved in the global coordination of nitrogen and phosphate metabolism (Fig. 7).
FIG 7.
Networks of GlnR and PhoP in S. erythraea.
Phosphorus and nitrogen are essential components of microbial nutrition. Soil microorganisms frequently encounter limited supplies of phosphate and nitrogen. The regulation of GlnR-related metabolism in soil microorganisms is under PhoP control, indicating that phosphate starvation triggers the PhoP/GlnR-mediated response, resulting in the expression of genes involved in nitrogen and phosphate metabolism. However, GlnR directly represses phoP transcription, indicating that nitrogen starvation results in the repression of genes associated with phosphate metabolism. The reciprocal cross-regulation of phosphate and nitrogen metabolism may be explained as a mechanism for saving cell resources in order to ensure that S. erythraea obtains nitrogen (may nitrogen control be implemented first in S. erythraea?). However, PhoP negatively regulates glnR and other nitrogen-related genes (glnA, glnII, and amtB-glnK-glnD) in S. coelicolor, suggesting that phosphate limitation reduces transcription of genes involved in nitrogen utilization (may phosphate control be implemented first in S. coelicolor?). The differential control of PhoP (positive and negative) on nitrogen metabolism in S. erythraea and S. coelicolor indicates that actinomycetes have evolved several nutritional regulation networks to adapt to changes in the availability of nitrogen and phosphate nutrients in their various ecological niches.
Knowledge about nitrogen and phosphate metabolism and control in the erythromycin producer S. erythraea is sparse. The choice of nitrogen/phosphate source and its concentration have a great influence on the erythromycin production (30). Reeve and Baumberg investigated the effect of glucose, nitrogen, and phosphorus sources on the timing and extent of erythromycin production (30). The erythromycin production and erythromycin biosynthesis gene (ery) mRNAs were detectable in low-phosphate (<1 mM) cultures. High levels of phosphate (10 to 100 mM) in cultures repressed erythromycin synthesis and the transcription of ery genes. These results suggest that ammonium and phosphate impact the transcription of ery cluster genes and that nitrogen/phosphate metabolism and synthesis of erythromycin are deeply interconnected. This conclusion was also supported by results of recent experiments showing that erythromycin production is strongly inhibited by ammonium (38). These observations provide evidence that S. erythraea may possess a molecular mechanism involving cross talk between nitrogen/phosphate metabolism and erythromycin synthesis.
In our study, the deletion of glnR increased erythromycin production; however, when the glnR gene was reintroduced into the ΔglnR mutant, production of erythromycin decreased to levels comparable to that of the wild-type strain (see Fig. S5B in the supplemental material). The deletion of the glnR gene in S. coelicolor and Amycolatopsis mediterranei results in reduced antibiotic production (19, 39). The PhoR-PhoP system is involved in regulating the production of actinorhodin in S. lividans (12), undecylprodigiosin in S. coelicolor (40), and pimaricin in Streptomyces natalensis (41). Martin et al. found that PhoP regulatory effect on antibiotic biosynthesis may be exerted through signaling cascades of PhoP-AfsS-AfsR-SARP (ActII-orf4 or RedD) in Streptomyces (25). However, the gene homologous to afsS was not found in the S. erythraea genome, and no SARP was identified as being responsible for erythromycin biosynthesis. Interestingly, erythromycin production in the phoPO strain increased by 40% relative to the wild-type strain, suggesting that PhoP also regulates erythromycin biosynthesis in S. erythraea. Such observations suggest that GlnR and PhoP both may play a role in the regulation of S. erythraea antibiotic production; however, it remains unclear how GlnR/PhoP-mediated regulation is connected to antibiotic production. These nutrient-sensing regulators may be important for the induction of a general stress response, triggered by nutrient limitation, which finally activates antibiotic biosynthesis. Deeper knowledge of nitrogen/phosphate metabolism could provide a better understanding of the link between primary and secondary metabolism in erythromycin-producing microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the China NSF (21276079), SRFDP (20120074110009) of the Chinese Ministry of Education, the National Key Technologies R&D Programs (2014AA02150 and 22007AA02Z331), and Fundamental Research Funds for the Central Universities.
Funding Statement
This work was supported by grants from the China NSF (21276079), SRFDP (20120074110009) of the Chinese Ministry of Education, the National Key Technologies R&D Programs (2014AA02150 and 22007AA02Z331), and Fundamental Research Funds for the Central Universities.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02960-15.
REFERENCES
- 1.Martín JF, Demain AL. 1980. Control of antibiotic biosynthesis. Microbiol Rev 44:230–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol Rev 59:604–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodgson DA. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv Microb Physiol 42:47–238. [DOI] [PubMed] [Google Scholar]
- 4.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin JF. 2004. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol 186:5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Divecha N, Irvine RF. 1995. Phospholipid signaling. Cell 80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 7.Antelmann H, Scharf C, Hecker M. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J Bacteriol 182:4478–4490. doi: 10.1128/JB.182.16.4478-4490.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laakel M, Lebrihi A, Khaoua S, Schneider F, Lefebvre G, Germain P. 1994. A link between primary and secondary metabolism: malonyl-CoA formation in Streptomyces ambofaciens growing on ammonium ions or valine. Microbiology 140:1451–1456. doi: 10.1099/00221287-140-6-1451. [DOI] [Google Scholar]
- 9.Laakel M, Lebrihi A, Khaoua S, Schneider F, Lefebvre G, Germain P. 1994. Relationship between valine, fatty acids, and spiramycin biosynthesis in Streptomyces ambofaciens.. Can J Microbiol 40:672–676. doi: 10.1139/m94-106. [DOI] [PubMed] [Google Scholar]
- 10.Amon J, Bräu T, Grimrath A, Hänssler E, Hasselt K, Höller M, Jessberger N, Ott L, Szököl J, Titgemeyer F, Burkovski A. 2008. Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J Bacteriol 190:7108–7116. doi: 10.1128/JB.00855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martín JF, Marcos AT, Martín A, Asturias JA, Liras P. 1994. Phosphate control of antibiotic biosynthesis at the transcriptional level, p 140–147. In Torriani-Gorini A, Yagil E, Silver S (ed), Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 12.Sola-Landa A, Moura RS, Martín JF. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans.. Proc Natl Acad Sci U S A 100:6133–6138. doi: 10.1073/pnas.0931429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doull JL, Vining LC. 1989. Culture conditions promoting dispersed growth and biphasic production of actinorhodin in shaken cultures of Streptomyces coelicolor A3(2). FEMS Microbiol Lett 53:265–268. [DOI] [PubMed] [Google Scholar]
- 14.Doull JL, Vining LC. 1990. Physiology of antibiotic production in actinomycetes and some underlying control mechanisms. Biotechnol Adv 8:141–158. doi: 10.1016/0734-9750(90)90010-9. [DOI] [PubMed] [Google Scholar]
- 15.Liras P, Asturias JA, Martín JF. 1990. Phosphate control sequences involved in transcriptional regulation of antibiotic biosynthesis. Trends Biotechnol 8:184–189. doi: 10.1016/0167-7799(90)90170-3. [DOI] [PubMed] [Google Scholar]
- 16.Lounes A, Lebrihi A, Benlismane C, Lefebvre G, Germain P. 1996. Regulation of spiramycin synthesis in Streptomyces ambofaciens: effects of glucose and inorganic phosphate. Appl Microbiol Biotechnol 45:204–211. doi: 10.1007/s002530050671. [DOI] [PubMed] [Google Scholar]
- 17.Maharjan S, Park JW, Yoon YJ, Lee HC, Sohng JK. 2010. Metabolic engineering of Streptomyces venezuelae for malonyl-CoA biosynthesis to enhance heterologous production of polyketides. Biotechnol Lett 32:277–282. doi: 10.1007/s10529-009-0152-9. [DOI] [PubMed] [Google Scholar]
- 18.Amon J, Titgemeyer F, Burkovski A. 2010. Common patterns—unique features: nitrogen metabolism and regulation in Gram-positive bacteria. FEMS Microbiol Rev 34:588–605. doi: 10.1111/j.1574-6976.2010.00216.x. [DOI] [PubMed] [Google Scholar]
- 19.Tiffert Y, Supra P, Wurm R, Wohlleben W, Wagner R, Reuther J. 2008. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol Microbiol 67:861–880. doi: 10.1111/j.1365-2958.2007.06092.x. [DOI] [PubMed] [Google Scholar]
- 20.Fink D, Weisschuh N, Reuther J, Wohlleben W, Engels A. 2002. Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol Microbiol 46:331–347. doi: 10.1046/j.1365-2958.2002.03150.x. [DOI] [PubMed] [Google Scholar]
- 21.Pullan ST, Chandra G, Bibb MJ, Merrick M. 2011. Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12:175. doi: 10.1186/1471-2164-12-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sola-Landa A, Rodríguez-García A, Franco-Domínguez E, Martín JF. 2005. Binding of PhoP to promoters of phosphate regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol Microbiol 56:1373–1385. doi: 10.1111/j.1365-2958.2005.04631.x. [DOI] [PubMed] [Google Scholar]
- 23.Ghorbel S, Kormanec J, Artus A, Virolle MJ. 2006. Transcriptional studies and regulatory interactions between the phoR-phoP operon and the phoU, mtpA, and ppk genes of Streptomyces lividans TK24. J Bacteriol 188:677–686. doi: 10.1128/JB.188.2.677-686.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allenby NNE, Laing E, Bucca G, Kierzek AM, Smith CP. 2012. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res 41:1152–1158. doi: 10.1093/nar/gks766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin JF, Sola-Landa A, Santos-Beneit F, Fernandez-Martinez LT, Prieto C, Rodriguez-Garcia A. 2011. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb Biotechnol 4:165–174. doi: 10.1111/j.1751-7915.2010.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin JF, Liras P. 2010. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces.. Curr Opin Microbiol 13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Santos-Beneit F, Rodriguez-Garcia A, Sola-Landa A, Martin JF. 2009. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol Microbiol 72:53–68. doi: 10.1111/j.1365-2958.2009.06624.x. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-García A, Sola-Landa A, Apel K, Santos-Beneit F, Martín JF. 2009. Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res 37:3230–3242. doi: 10.1093/nar/gkp162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sola-Landa A, Rodríguez-García A, Amin R, Wohlleben W, Martín JF. 2013. Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor.. Nucleic Acids Res 41:1767–1782. doi: 10.1093/nar/gks1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeve LM, Baumberg S. 1998. Physisological controls of erythromycin production by Saccharopolyspora erythraea are exerted at least in part at the level of transcription. Biotechnol Lett 20:585–589. doi: 10.1023/A:1005357930000. [DOI] [Google Scholar]
- 31.Yao LL, Liao CH, Huang G, Zhou Y, Rigali S, Zhang BC, Ye BC. 2014. GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea.. Appl Microbiol Biotechnol 98:7935–7948. doi: 10.1007/s00253-014-5878-1. [DOI] [PubMed] [Google Scholar]
- 32.Yin X, Xu X, Wu H, Yuan L, Huang X, Zhang B. 2013. SACE_0012, a TetR-family transcriptional regulator, affects the morphogenesis of Saccharopolyspora erythraea.. Curr Microbiol 67:647–651. doi: 10.1007/s00284-013-0410-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiard S, Marée R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Rigali S. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun 357:861–864. doi: 10.1016/j.bbrc.2007.03.180. [DOI] [PubMed] [Google Scholar]
- 34.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Cen XF, Zhao GP, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao CH, Yao LL, Ye BC. 2014. Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators GlnR, DasR, and CRP. Mol Microbiol 94:1065–1084. doi: 10.1111/mmi.12818. [DOI] [PubMed] [Google Scholar]
- 38.Flores ME, Sanchez S. 1985. Nitrogen regulation of erythromycin formation in Streptomyces erythreus.. FEMS Microbiol Lett 26:191–194. doi: 10.1111/j.1574-6968.1985.tb01589.x. [DOI] [Google Scholar]
- 39.Yu H, Peng WT, Liu Y, Wu T, Yao YF, Cui MX, Jiang WH, Zhao GP. 2006. Identification and characterization of glnA promoter and its corresponding trans-regulatory protein GlnR in the rifamycin SV producing actinomycete, Amycolatopsis mediterranei U32. Acta Bioch Bioph Sin 38:831–843. doi: 10.1111/j.1745-7270.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 40.Santos-Beneit F, Rodríguez-García A, Martín JF. 2012. Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor.. Microbiol Res 167:532–535. doi: 10.1016/j.micres.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Mendes MV, Tunca S, Anton N, Recio E, Sola-Landa A, Aparicio JF, Martin JF. 2007. The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab Eng 9:217–227. doi: 10.1016/j.ymben.2006.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.