Abstract
Allosteric regulation, the most direct and efficient way of regulating protein function, is induced by the binding of a ligand at one site that is topographically distinct from an orthosteric site. Allosteric Database (ASD, available online at http://mdl.shsmu.edu.cn/ASD) has been developed to provide comprehensive information featuring allosteric regulation. With increasing data, fundamental questions pertaining to allostery are currently receiving more attention from the mechanism of allosteric changes in an individual protein to the entire effect of the changes in the interconnected network in the cell. Thus, the following novel features were added to this updated version: (i) structural mechanisms of more than 1600 allosteric actions were elucidated by a comparison of site structures before and after the binding of an modulator; (ii) 261 allosteric networks were identified to unveil how the allosteric action in a single protein would propagate to affect downstream proteins; (iii) two of the largest human allosteromes, protein kinases and GPCRs, were thoroughly constructed; and (iv) web interface and data organization were completely redesigned for efficient access. In addition, allosteric data have largely expanded in this update. These updates are useful for facilitating the investigation of allosteric mechanisms, dynamic networks and drug discoveries.
INTRODUCTION
Allosteric regulation fine-tunes most biological processes, including signal transduction, enzyme activity, metabolism and transport (1–3). Allostery, an intrinsic property of a protein, is referred to as the regulation of activity at one site (also known as an orthosteric site) in a protein by a topographically and spatially distant site; the latter is designated as an allosteric site (4–6). Allosteric regulation occurs through binding of a modulator (e.g., small molecule) at an allosteric site to engender a conformational change that affects function at the orthosteric site (7–11). This effect may cause the re-distribution of the conformational ensemble by either stabilizing an active conformation or destabilizing an inactive conformation in response to allosteric perturbations (12,13). Traditionally, the repertoire of allostery was primarily confined to determining the allosteric effects or mechanisms in individual multi-subunit or monomer proteins by conformational transitions (14–17). Recently, increasing evidence has indicated that allosteric signals can propagate across several or numerous proteins to sculpt allosteric networks (18–22). A quintessential example of allosteric propagation is pertinent to the identification of interconnected proteins that govern the reversible switch between gluconeogenesis and glycolysis in human metabolite dynamics (18). Thus, focusing on the fundamental role of allostery in a cellular network is an instrumental strategy for dissecting the consequences of pathological allosteric events (7,20–22).
Despite the increasing interest in the development of allosteric drugs as a new tactic in drug discovery, the structural mechanisms underlying allosteric drug action represent a key challenge. The hotspots of conformational changes in the allosteric site are of particular importance to understanding the underpinnings underlying allostery, because the efficacy of an allosteric drug is often determined by specific conformational transitions in the allosteric site (23). Identification of triggering motions in an allosteric site is highly helpful to uncovering structural mechanisms of allosteric drug action and contributes to drug discovery.
In drug discovery, protein kinases and G protein-coupled receptors (GPCRs) are two of the largest targets. Due to the remarkable structural conservation of orthosteric sites in the kinome and homologous GPCRs, there has been a long-standing challenge in the development of highly specific kinase and GPCR orthosteric inhibitors or activators (24–27). Fortunately, breakthroughs in GPCR structural determination have resulted in a fast-growing number of GPCR structures obtained in complex with allosteric modulators (28–34), and biochemical studies, such as disulfide trapping (35), high-throughput screening (36) and fragment-based screening (37), promote the identification of allosteric sites in protein kinases. Thus, elucidating the family profile of allosteric sites in protein kinases and GPCRs provides a promising new paradigm for the discovery of novel allosteric sites and the design of potent modulators with improved protein subtype selectivity, adverse effects and pathway-biased signaling (38–43).
The Allosteric Database (ASD) has been developed to provide comprehensive information characterizing allosteric regulation since 2009 (44,45), ranging from allosteric proteins, modulators to interactions, sites, pathways, functions and related diseases. Here, in a current update of the resource, in addition to the significant expansion of data, we focused on the characterization of structural mechanisms of allosteric drug action, allosteric networks, and protein kinases and GPCRs allosteromes. These updates include the elucidation of >1600 allosteric actions in 308 allosteric proteins, the identification of 261 endogenous/exogenous allosteric networks and the construction of protein kinases and GPCRs allosterome. In addition, the web interface and data organization have been completely redesigned for efficient access on the basis of community demands. Cumulatively, these updates have the benefit of prompting an investigation of allosteric mechanism, allosteric related disease and allosteric drug discovery.
DATABASE GROWTH AND STATISTICS
Allosteric molecules and features, including structures, sites, pathways, functions, related diseases and external links, were collected using previously described methods (44,45). A detailed content comparison between the current and previous versions of ASD is provided in Table 1. The most exciting enhancement in ASD v3.0 is the number of allosteric modulators. Currently, ASD v3.0 contains 71538 allosteric modulators consisting of 25339 activators, 33604 inhibitors and 13462 regulators, which have increased by >200% since the ASD v2.0 update. The definition of the three classes was provided in our previous publication (44). This increase is primarily a result of the significant expansion of allosteric drug discovery. Among the increased number of allosteric modulators, 10.5% bind to new allosteric targets that have been discovered within the last 2 years, 2.80% were found to have more than one allosteric target and 1.43% were found to regulate allosteric targets with different allosteric effects, dual activators/regulators (0.58%), dual inhibitors/regulators (0.54%), dual activators/inhibitors (0.28%) and multiple activators/inhibitors/regulators (0.03%). Importantly, regardless of the potential off-target binding to orthosteric sites, the growth of modulators interacting with multiple allosteric sites from ∼1.5% in ASD v2.0 to nearly 3% in ASD v3.0 reveals a potentially dangerous situation about the so-called selectivity of allosteric sites.
Table 1. Data statistics for allosteric proteins and modulators in updated ASD 3.01.
| Data category | ASD 3.0 | ASD 2.0 |
|---|---|---|
| Number of all modulators | 71 538 | 22 003 |
| Number of activators | 25 339 | 15 144 |
| Number of inhibitors | 33 604 | 6205 |
| Number of regulators | 13 462 | 854 |
| Number of dual activators/regulators | 334 | 50 |
| Number of dual inhibitors/regulators | 320 | 55 |
| Number of dual activators/inhibitors | 257 | 125 |
| Number of multiple activators/inhibitors/regulators | 44 | 30 |
| Number of all allosteric sites | 1930 | 907 |
| Number of all proteins | 1473 | 1261 |
| Number of kinases | 207 | 187 |
| Number of GPCRs | 118 | 109 |
| Number of ion channels | 134 | 119 |
| Number of peptidases | 59 | 55 |
| Number of phosphatases | 30 | 27 |
| Number of transcription factors | 55 | 46 |
| Number of nuclear receptors | 26 | 24 |
| Number of E-proteins | 5 | 5 |
| Number of other proteins | 839 | 689 |
| Number of protein–modulator interactions | 75 462 | 23 120 |
| Number of all protein pathways | 56 | 48 |
| Number of allosteric related diseases | 3350 | 565 |
1The number of allosteric proteins in ASD 3.0 was counted as the sum of all reported species of each allosteric protein.
In addition to the substantially increasing number of allosteric modulators, we have also increased the associated allosteric features and annotations, including more interactions (∼226% increase), more sites (∼113% increase), more pathways (∼17%) and a greater number of related diseases (∼493% increase) (Table 1). As the identification of allosteric sites increases, proteins with up to four allosteric sites are observed in ASD v3.0. For example, both Human α-amylase (46) and glycogen phosphorylase (47) can be regulated by endogenous/exogenous modulators on four distinct allosteric sites. Due to the key roles of glycogen phosphorylase in the process of anti-diabetes, various classes of inhibitors were discovered on each of the four allosteric sites and the combination of inhibitors on different sites demonstrate synergistic effects on the enzyme activity (48). These findings indicate a novel strategy of synergistic regulation on allosteric drug design by targeting more than one allosteric site on the same protein.
NEW FEATURES AND FUNCTIONALITIES
Web interface and access
With collaboration with interested users, we have designed a completely new web interface for ASD v3.0 that is more user-friendly and better integrated (Figure 1). Allosteric data were restructured into three categories with clearly demarcated menus ‘MOLECULES’, ‘FEATURES’ and ‘DOWNLOAD’. The ‘MOLECULES’ menu provides allosteric proteins in ‘PROTEIN’ and allosteric modulators in ‘MODULATOR’, where all molecules are curated by experimental confirmation and annotated with allosteric features. The new flat layout of ‘PROTEIN’ and ‘MODULATOR’ pages allows streamlined access to the data. We believe this should make the data easier to view and browse. Allosteric molecules and related annotations are also downloadable from the ‘DOWNLOAD’ menu for off-line study. In the ‘FEATURES’ menu, there are four auxiliary allosteric datasets that were originally identified by literature and then computationally generated, including conventional ‘SITE’ for binding pockets of allosteric modulators and ‘PATHWAY’ for remote communications from allosteric site to orthosteric site, newly introduced ‘NETWORK’ for allosteric signal propagations in interconnected cellular pathways and ‘ALLOSTEROME’ for the relationship of allostery in a protein family (see below).
Figure 1.
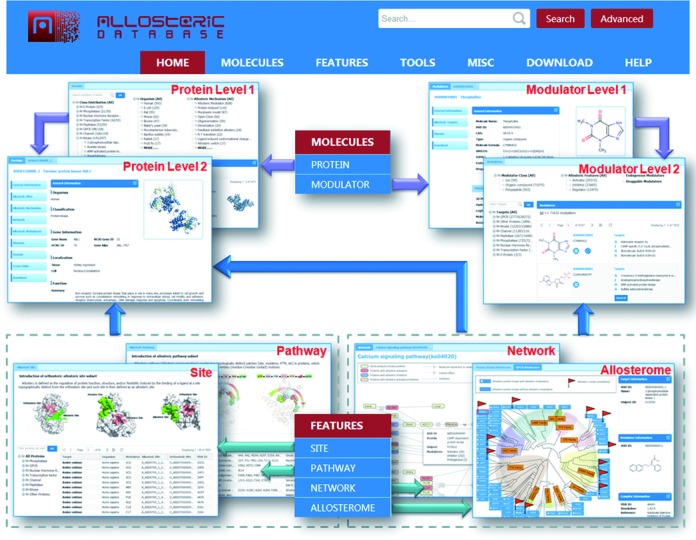
The new web interface and data architecture in ASD 3.0 are depicted.
In addition to the data, several computational tools were enhanced and added to allow efficient access to the ASD. First, through text-mining design based on the characters of allosteric information, the precision and time of search engine in ASD v3.0 was heavily improved and more accurate search results can be retrieved by performing a quick ‘Search’ or ‘Advanced’ search in the upper right corner of ASD home page. Second, interactive classification trees and filters were added to the browse pages of allosteric data in ‘PROTEIN’, ‘MODULATOR’, ‘SITE’ and ‘NETWORK’, allowing users to precisely restrict the browse by multiple options. Third, an internal crosslink between different datasets in ASD was carefully generated, such as links from nodes in ‘NETWORK’ and ‘ALLOSTEROME’ to entries in ‘PROTEIN’ or ‘MODULATOR’, integrating the knowledge of allosteric data by sequence, structure, evolution and cellular functions. Fourth, all java-based plugins in the previous version of ASD were replaced with javascript codes, making the visualization of the data much simpler and much faster. Fifth, the high-quality data of allosteric sites in ASBench was integrated in ASD for the development of efficient algorithms on the prediction of unknown allosteric sites, which is located in the ‘TOOL’ menu of ASD (49,50). ‘MISC’ is a collection of allosteric knowledge, research and applications, such as ‘GLOSSARY’, ‘DIAGRAM’, ‘LITERATURE’, ‘DRUG’ and ‘EXPERT’, which have been built in the previous version and carefully refined in the current version. Finally, the new web interface has been extensively tested to support Firefox 5+, Chrome 9+, Safari 4+ and Internet Explorer 11 and more FAQs can be found in the ‘HELP’ menu of the ASD homepage.
Allosteric modulator action
Bioactive modulators can exploit functional pockets in protein targets to exert action on regulation. Unlike orthosteric modulators, for which the key mechanism of small molecule action is trying to compete with the natural ligand in active site, in the case of allosteric modulators, the extent of conformational transitions into functional state can be pivotal in specifying the modulator action in allosteric sites (23). Identification of the origin of the conformational transitions in protein by the modulator is highly dependent on comparing the structure of the allosteric sites before binding (apo structure) with that after binding (holo structure) (23,51,52), which could not only account for the underlying mechanism of specific allosteric triggering action but may also be used to improve the triggering efficacy in allosteric drug design. Ruth et al. proposed an alignment strategy to successfully illustrate a series of allosteric modulator action in the regulation of regulatory subunit of protein kinase A (allosteric agonist: cAMP), Ras (allosteric agonist: GTP) and PDK1 (allosteric agonist: P47, etc.) (23). Likewise, using allosteric apo/holo structural alignment, we investigated the action of endogenous adenosine triphosphate (ATP) as a modulator in distinct allosteric sites of 13 proteins and found that a majority of the allosteric sites (80%) were induced by the triphosphate in ATP as a trigger, whereas a smaller number of allosteric sites (20%) were triggered by the adenine in ATP (53). These studies showed a fundamentally different mechanistic foundation of allosteric modulator action compared with the orthosteric modulator. To better understand modulator mechanism in allosteric site, a large-scale dataset revealing allosteric modulator action is more desirable than ever and was thus built in the ASD in the current release.
In ASD v3.0, 1688 allosteric apo/holo paired structures for allosteric modulator action in 308 proteins from 107 organisms were constructed using the same protocol described in ATP action (53) (see Supporting Information). All allosteric actions for a protein are listed in the ‘Allosteric Mechanism’ section of the protein page from the ‘PROTEIN’ submenu of ‘MOLECULES’ in the ASD home page. Clicking the modulator button under an action of interest in the list, a new window opens to show details of the apo/holo structure pair with basic information, including the bound allosteric modulator, the source of the apo and holo structures and the extent of the conformational transition, etc. As shown in Figure 2, the superimposition of allosteric apo/holo paired structures in ‘Glutamate racemase (Helicobacter pylori)’ is displayed. Conformational transitions in allosteric site upon binding of the corresponding allosteric modulator ‘KRH’ are identified in ‘backbone’ and highlighted in color. The alignment and identified conformational transitions can also be downloaded as a PyMOL session (PSE) file in the page for further analysis. Of the 1688 allosteric apo/holo paired structures, 92.6% and 5.9% allosteric sites showed local motions in backbone and in sidechain, respectively upon the binding of modulators. Remarkably, 20 allosteric proteins can be regulated by the actions of both agonism and antagonism on the same allosteric site, such as glutamate dehydrogenase (bovine) (54,55). The apo/holo results revealed that the opposite mechanisms into one allosteric site occur by triggering different hotspots of the site, raising great challenges in allosteric drug design.
Figure 2.
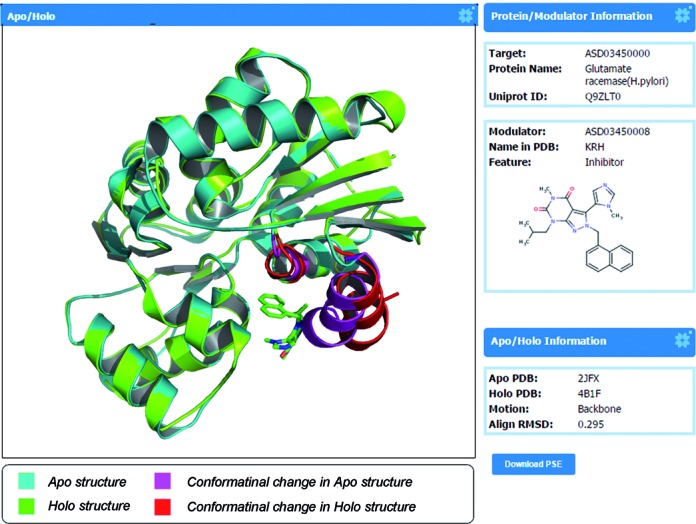
Conformational changes of the allosteric site comparing the protein structure of apo and holo. Example of Glutamate racemase in Helicobacter pylori (ASD03450000_1) with the binding of the allosteric modulator ‘KRH’ (ASD03450008).
Allosteric network
Because proteins function through a molecular network that is highly interconnected by cellular pathways, a single allosteric action on the given protein in the network can strengthen, weaken or switch the signal propagation toward a specific pathway, triggering various favorable or unfavorable destinies of the cell (56). More generally, cellular functions are determined and dynamically regulated by multiple co-occurring allosteric actions in the network. An illustration of the allosteric map at the network level is highly useful to understand how allostery controls physiological and pathological effects by endogenous/exogenous action and can subsequently reveal critical targets for allosteric related diseases (57,58). To integrate the dynamic regulation of allosteric proteins and reveal their relationships in functional networks, great efforts have been made to build allosteric networks through the Kyoto Encyclopedia of Genes and Genomes (KEGG) reference pathway maps (59,60) (see Supporting Information). In terms of experimental allosteric knowledge, 261 allosteric networks, in which allosteric proteins function as perturbed nodes and allosteric modulators act as perturbants, were built through crosslinking and manual calibration. The networks can be accessed under the ‘NETWORK’ submenu of ‘FEAUTRES’ in the ASD home page. By clicking the selected network, the details of allosteric actions in the network are shown in a separate window. As an example, ‘Calcium signaling pathway’ in Figure 3 demonstrates that the network contains 19 known allosteric proteins, where 13 and 11 can be regulated to produce enhanced and reduced signals to downstream partners for the cellular function. More importantly, endogenous allosteric ligands were found in 13 targets, such as ‘Phosphorylase kinase (activator: cAMP)’ (61), ‘Protein kinase Cα (activator: arachidonic acid)’ (62), ‘5-hydroxytryptamine receptor 7 (inhibitor: c-9,10-octadecenoamide)’ (63), ‘Phospholipase Cγ1 (activator: phosphatidic acid)’ (64) and ‘1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase δ1 (activator: glycerophosphoinositol)’ (65), based on published literature, suggesting that normal physiological effect of Ca2+ concentration in the cell is achieved by a complicated balance of these endogenous allosteric regulations and that the dysregulation of allostery could disrupt the balance into a pathological state and related disease.
Figure 3.
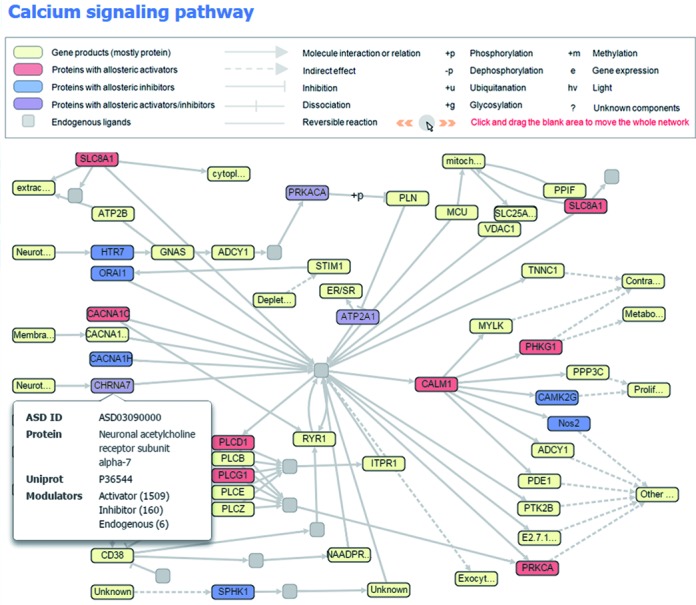
Example of the allosteric network ‘Calcium signaling pathway’.
Allosterome
Allosteric sites, which differ from highly conserved orthosteric sites, offer intrinsic opportunities for drug target by providing higher selectivity, fewer side effects and lower toxicity (7,66,67). Analysis of the ASD collection reveals that protein kinases and GPCRs (68) are two of the largest families of allostery that have been successfully exploited in drug discovery. Dissection of the map of the less evolutionary conserved allosteric sites in both families could not only reveal the interrelatedness and specificity among the sites, but be used to guide the rational discovery of allosteric sites in other challenging families, such as nuclear receptors, ion channels and transcription factors, etc. In ASD v3.0, two allosteromic maps in humans, protein kinases and GPCRs, were thoroughly constructed based on multiple sequence alignment of sequences and dendrogram (see Supporting Information) under the menu of ‘ALLOSTEROME’ in the ASD home page. In Figure 4, the allosteric modulators of 51 of 76 human allosteric protein kinases have been discovered (white circle), and the binding allosteric sites of 12 of these modulators have been resolved using X-ray crystallography and nuclear magnetic resonance (red flag). These sites actually cluster into nine locations throughout classical kinase structures in the family. Likewise, all five allosteric sites in human GPCRs show three different locations by structural superimposition. These results indicate that despite the high specificity of allosteric sites, the location of novel sites could still be inferred by the known sites of the same family.
Figure 4.
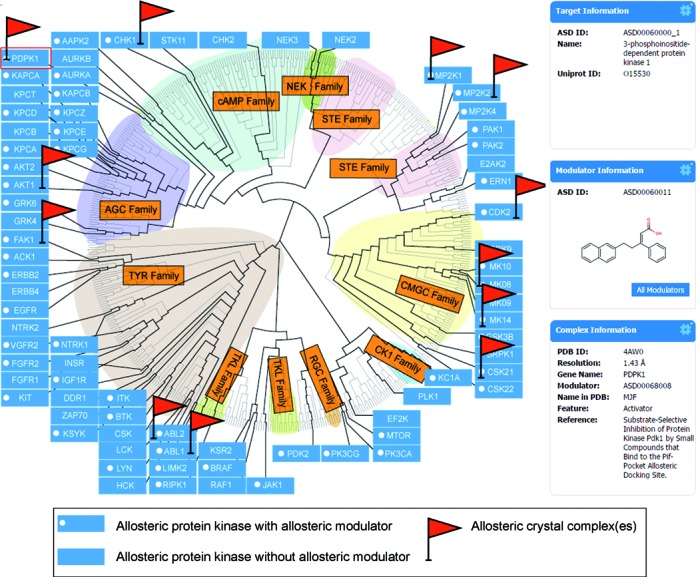
The allosteric distribution in the protein kinase allosterome.
CONCLUSION AND FUTURE DIRECTIONS
The ASD is a comprehensive and integrated allosteric platform that consolidates manually curated chemical, biological and clinical data about all experimentally confirmed allosteric evidences. Building on previous versions, we have significantly expanded the allosteric molecules and features, particularly modulators, sites, interactions and related diseases, which reflect the recent considerable growth of allosteric studies in both pharmaceuticals and medicine. We also introduced three novel datasets for the investigation of allosteric mechanism including (i) allosteric modulator action; (ii) allosteric network and (iii) protein kinase and GPCR allosteromes in this update. The ASD site was completely redesigned, with greatly improved performance of data access, a cleaner appearance and improved usability. In the future, we aim to continue to curate allosteric data from the primary biomedical literature and to refine the quality of the data according to the research progress of allostery. The ASD will also continue to improve database architecture for easy and intuitive use based on the response from community. In addition to our current knowledge, we will focus on introducing allosteric evolution by interspecies navigation and comparison in order to trace to the beginning of allostery. With the increasing number of clinical mutations discovered in patients using next-generation sequencing, the effects and mechanism of the mutations around the allosteric site could provide new insights in syndromes, physiological abnormalities and diseases, and this is another aspect in which we are planning to curate in the future.
Acknowledgments
We thank Mr. Zhaoliang at the Shanghai Jiaotong University School of Medicine for fruitful discussions on the allosteric modulators.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (973 Program) [2015CB910403, in part]; National Natural Science Foundation of China [81322046, 81473137, 81302698, in part]; Shanghai Rising-Star Program [13QA1402300, in part]; Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning and the Program for New Century Excellent Talents in University [NCET-12-0355, in part]. Funding for open access charge: National Basic Research Program of China (973 Program) [2015CB910403, in part]; National Natural Science Foundation of China [81322046, 81473137, 81302698, in part]; Shanghai Rising-Star Program [13QA1402300, in part]; Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning and the Program for New Century Excellent Talents in University [NCET-12-0355, in part].
Conflict of interest statement. None declared.
REFERENCES
- 1.Goodey N.M., Benkovic S.J. Allosteric regulation and catalysis emerge via a common route. Nat. Chem. Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 2.Changeux J.-P., Edelstein S.J. Allosteric mechanism of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 3.Changeux J.-P. Allosteric receptors: from electric orange to cognition. Annu. Rev. Pharmacol. Toxicol. 2010;50:1–38. doi: 10.1146/annurev.pharmtox.010909.105741. [DOI] [PubMed] [Google Scholar]
- 4.Fenton A.W. Allostery: an illustrated definition for the ‘second secret of life’. Trends Biochem. Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motlagh H.N., Wrabl J.O., Li J., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu S., Huang W., Zhang J. Recent computational advances in the identification of allosteric sites in proteins. Drug Discov. Today. 2014;19:1595–1600. doi: 10.1016/j.drudis.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Nussinov R., Tsai C.-J. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 8.Lu S., Li S., Zhang J. Harnessing allostery: a novel approach to drug discovery. Med. Res. Rev. 2014;34:1242–1285. doi: 10.1002/med.21317. [DOI] [PubMed] [Google Scholar]
- 9.Raman S., Taylor N., Genuth N., Fields S., Church G.N. Engineering allostery. Trends Genet. 2014;30:521–528. doi: 10.1016/j.tig.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Smet F., Christopoulos A., Carmeliet P. Allosteric targeting of receptor tyrosine kinases. Nat. Biotechnol. 2014;32:1113–1120. doi: 10.1038/nbt.3028. [DOI] [PubMed] [Google Scholar]
- 11.Peracchi A., Mozzarelli A. Exploring and exploiting allostery: models, evolution, and drug targeting. Biochem. Biophys. Acta. 2011;1814:922–933. doi: 10.1016/j.bbapap.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Gunasekaran K., Ma B., Nussinov R. Is allostery an intrinsic property of all dynamic proteins. Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 13.Tsai C.-J., Nussinov R. A unified view of “how allostery works”. PLoS Comput. Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaya C., Armutlulu A., Ekesan S., Haliloglu T. MCPath: monte carlo path generation approach to predict likely allosteric pathways and functional residues. Nucleic Acids Res. 2013;41:W249–W255. doi: 10.1093/nar/gkt284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goncearenco A., Mitternacht S., Yong T., Eisenhaber B., Eisenhaber F., Berezovsky I.N. SPACER: server for predicting allosteric communication and effects of regulation. Nucleic Acids Res. 2013;41:W266–W272. doi: 10.1093/nar/gkt460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das A., Gur M., Cheng M.H., Jo S., Bahar I., Roux B. Exploring the conformational transition of biological systems using a simple two-state anisotropic network model. PLoS Comput. Biol. 2014;10:e1003521. doi: 10.1371/journal.pcbi.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.General I.J., Liu Y., Balckburn M., Mao W., Gierasch L., Bahar I. ATPase subdomain IA is a mediator of interdomain allostery in Hsp70 molecular chaperones. PLoS Comput. Biol. 2014;10:e1003624. doi: 10.1371/journal.pcbi.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Link H., Kochanowski K., Sauer U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat. Biotechnol. 2013;4:357–361. doi: 10.1038/nbt.2489. [DOI] [PubMed] [Google Scholar]
- 19.Nussinov R. The spatial structure of cell signaling systems. Phys. Biol. 2013;10:045004. doi: 10.1088/1478-3975/10/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussinov R., Tsai C.-J., Ma B. The underappreciated role of allostery in the cellular network. Annu. Rev. Biophys. 2013;42:169–189. doi: 10.1146/annurev-biophys-083012-130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussinov R., Tsai C.-J., Liu J. Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 2014;136:17692–17701. doi: 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard P.A., Moody C.L., Murali R. Allosteric modulation of Ras and the PI3K/AKT/mTOR pathway: emerging therapeutic opportunities. Front. Physiol. 2014;5:478. doi: 10.3389/fphys.2014.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussinov R., Tsai C.-J. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Cowan-Jacob S.W., Jahnke W., Knapp S. Novel approaches for targeting kinases: allosteric inhibition, allosteric activation and pseudokinases. Future Med. Chem. 2014;6:541–561. doi: 10.4155/fmc.13.216. [DOI] [PubMed] [Google Scholar]
- 25.Foda Z.H., Seeliger M.A. An allosteric add-on. Nat. Chem. Biol. 2014;10:796–797. doi: 10.1038/nchembio.1630. [DOI] [PubMed] [Google Scholar]
- 26.Langmead C.J., Christopoulos A. Functional and structural perspectives on allosteric modulation of GPCRs. Curr. Opin. Cell Biol. 2014;27:94–101. doi: 10.1016/j.ceb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Mol. Pharmacol. 2014;86:463–487. doi: 10.1124/mol.114.094342. [DOI] [PubMed] [Google Scholar]
- 28.Kruse A.C., Ring A.M., Manglik A., Hu J., Hu K., Eitel K., Hübner H., Pardon E., Valant C., Sexton P.M., et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan Q., Zhu Y., Li J., Chen Z., Han G.W., Kufareva I., Li T., Ma L., Fenalti G., Li J., et al. Structure of the CCR5 chemokine receptor―HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollenstein K., Kean J., Bortolato A., Cheng R.K.Y., Doré A.S., Jazayeri A., Cooke R.M., Weir M., Marshall F.H. Structure of class B GPCR corticotrophin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 31.Wu H., Wang C., Gregory K.J., Han G.W., Cho H.P., Xia Y., Niswender C.M., Katritch V., Meiler J., Cherezov V., et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava A., Yano J., Hirozane Y., Kefala G., Gruswitz F., Snell G., Lane W., Ivetac A., Aertgeerts K., Nguyen J., et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014;513:124–127. doi: 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- 33.Lee C.-H., Lü W., Michel J.C., Goehring A., Du J., Song X., Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D., Gao Z.-G., Zhang K., Kiselev E., Crane S., Wang J., Paoletta S., Yi C., Ma L., Zhang W., et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015;520:317–321. doi: 10.1038/nature14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldman T., Kabaleeswaran V., Jang S.B., Antczak C., Djaballah H., Wu H., Jiang X. A class of allosteric caspase inhibitors identified by high-throughput screening. Mol. Cell. 2012;47:585–595. doi: 10.1016/j.molcel.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahnke W., Rondeau J.-M., Cotesta S., Marzinzik A., Pellé X., Geiser M., Strauss A., Götte M., Bitsch F., Hemmig R., et al. Allosteric non-bisphosphonate FPPS inhibitors identified by ferment-based discovery. Nat. Chem. Biol. 2010;6:660–666. doi: 10.1038/nchembio.421. [DOI] [PubMed] [Google Scholar]
- 38.Taly A., Corringer P.J., Guedin D., Lestage P., Changeux J.P. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat. Rev. Drug Discov. 2009;8:733–750. doi: 10.1038/nrd2927. [DOI] [PubMed] [Google Scholar]
- 39.Müller C.E., Schiedel A.C., Baqi Y. Allosteric modulators of rhodopsin-like G protein-coupled receptors: opportunities in drug development. Pharmacol. Ther. 2012;135:292–315. doi: 10.1016/j.pharmthera.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Wootten D., Christopoulos A., Sexton P.M. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat. Rev. Drug. Discov. 2013;12:630–644. doi: 10.1038/nrd4052. [DOI] [PubMed] [Google Scholar]
- 41.Matschinsky F.M. Assessing the potential of glucokinase activator in diabetes therapy. Nat. Rev. Drug Discov. 2009;8:399–416. doi: 10.1038/nrd2850. [DOI] [PubMed] [Google Scholar]
- 42.Shonberg J., Lopez L., Scammells P.J., Christopoulos A., Capuano B., Lane J.R. Biased agonism at G protein-coupled receptors: the promise and the challenges―a medicinal chemistry perspective. Med. Res. Rev. 2014;34:1286–1330. doi: 10.1002/med.21318. [DOI] [PubMed] [Google Scholar]
- 43.Fang Z., Grütter C., Rauh D. Strategies for the selective regulation of kinases with allosteric modulators: exploiting exclusive structural features. ACS Chem. Biol. 2013;8:58–70. doi: 10.1021/cb300663j. [DOI] [PubMed] [Google Scholar]
- 44.Huang Z., Zhu L., Cao Y., Wu G., Liu X., Chen Y., Wang Q., Shi T., Zhao Y., Wang Y., et al. ASD: a comprehensive database of allosteric proteins and modulators. Nucleic Acids Res. 2011;39:D663–D669. doi: 10.1093/nar/gkq1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Z., Mou L., Shen Q., Lu S., Li C., Liu X., Wang G., Li S., Geng L., Liu Y., et al. ASD v2.0: updated context and novel features focusing on allosteric regulation. Nucleic Acids Res. 2014;42:D510–D516. doi: 10.1093/nar/gkt1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rydberg E.H., Li C., Maurus R., Overall C.M., Brayer G.D., Withers S.G. Mechanistic analyses of catalysis in human pancreatic alpha-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids. Biochemistry. 2002;41:4492–4502. doi: 10.1021/bi011821z. [DOI] [PubMed] [Google Scholar]
- 47.Johnson L.N. Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J. 1992;6:2274–2282. doi: 10.1096/fasebj.6.6.1544539. [DOI] [PubMed] [Google Scholar]
- 48.Tsitsanou K.E., Skamnaki V.T., Oikonomakos N.G. Structural basis of the synergistic inhibition of glycogen phosphorylase a by caffeine and a potential antidiabetic drug. Arch. Biochem. Biophys. 2000;384:245–254. doi: 10.1006/abbi.2000.2121. [DOI] [PubMed] [Google Scholar]
- 49.Huang W., Wang G., Shen Q., Liu X., Lu S., Geng L., Huang Z., Zhang J. ASBench: benchmarking sets for allosteric discovery. Bioinformatics. 2015;31:2598–2600. doi: 10.1093/bioinformatics/btv169. [DOI] [PubMed] [Google Scholar]
- 50.Huang W., Lu S., Huang Z., Liu X., Mou L., Luo Y., Zhao Y., Liu Y., Chen Z., Hou T., Zhang J. Allosite: a method for predicting allosteric sites. Bioinformatics. 2013;29:2357–2359. doi: 10.1093/bioinformatics/btt399. [DOI] [PubMed] [Google Scholar]
- 51.Daily M.D., Gray J.J. Local motions in a benchmark of allosteric proteins. Proteins. 2007;67:385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 52.Chang D.T., Yao T.J., Fan C.Y., Chiang C.Y., Bai Y.H. AH-DB: collecting protein structure pairs before and after binding. Nucleic Acids Res. 2012;40:D472–D478. doi: 10.1093/nar/gkr940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu S., Huang W., Wang Q., Shen Q., Li S., Nussinov R., Zhang J. The structural basis of ATP as an allosteric modulator. PLoS Comput. Biol. 2014;10:e1003831. doi: 10.1371/journal.pcbi.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.George A., Bell J.E. Effects of adenosine 5’-diphosphate on bovine glutamate dehydrogenase: diethyl pyrocarbonate modification. Biochemistry. 1980;19:6057–6061. doi: 10.1021/bi00567a017. [DOI] [PubMed] [Google Scholar]
- 55.Li C., Li M., Chen P., Narayan S., Matschinsky F.M., Bennett M.J., Stanley C.A., Smith T.J. Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site. J. Biol. Chem. 2011;286:34164–34174. doi: 10.1074/jbc.M111.268599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nussinov R., Ma B., Tsai C.J., Csermely P. Allosteric conformational barcodes direct signaling in the cell. Structure. 2014;21:1509–1521. doi: 10.1016/j.str.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korcsmáros T., Farkas I.J., Szalay M.S., Rovó P., Fazekas D., Spiró Z., Böde C., Lenti K., Vellai T., Csermely P. Uniformly curated signaling pathways reveal tissue-specific cross-talks and support drug target discovery. Bioinformatics. 2010;26:2042–2050. doi: 10.1093/bioinformatics/btq310. [DOI] [PubMed] [Google Scholar]
- 58.Nussinov R., Ma B., Tsai C.J. A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochim. Biophys. Acta. 2013;1834:820–829. doi: 10.1016/j.bbapap.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 59.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nadeau O.W., Anderson D.W., Yang Q., Artigues A., Paschall J.E., Wyckoff G.J., McClintock J.L., Carlson G.M. Evidence for the location of the allosteric activation switch in the multisubunit phosphorylase kinase complex from mass spectrometric identification of chemically crosslinked peptides. J. Mol. Biol. 2007;365:1429–1445. doi: 10.1016/j.jmb.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacEwan D.J., Mitchell R., Johnson M.S., Thomson F.J., Lutz E.M., Clegg R.A., Connor K. Evidence that protein kinase C alpha has reduced affinity towards 1, 2-dioctanoyl-sn-glycerol: the effects of lipid activators on phorbol ester binding and kinase activity. Eur. J. Pharmacol. 1993;246:9–18. doi: 10.1016/0922-4106(93)90003-r. [DOI] [PubMed] [Google Scholar]
- 63.Hedlund P.B., Carson M.J., Sutcliffe J.G., Thomas E.A. Allosteric regulation by oleamide of the binding properties of 5-hydroxytryptamine7 receptors. Biochem. Pharmacol. 1999;58:1807–1813. doi: 10.1016/s0006-2952(99)00274-9. [DOI] [PubMed] [Google Scholar]
- 64.Jones G.A., Carpenter G. The regulation of phospholipase C-gamma 1 by phosphatidic acid. Assessment of kinetic parameters. J. Biol. Chem. 1993;268:20845–20850. [PubMed] [Google Scholar]
- 65.Wu Y., Perisic O., Williams R.L., Katan M., Roberts M.F. Phosphoinositide-specific phospholipase C delta1 activity toward micellar substrates, inositol 1, 2-cyclic phosphate, and other water-soluble substrates: a sequential mechanism and allosteric activation. Biochemistry. 1997;36:11223–11233. doi: 10.1021/bi971039s. [DOI] [PubMed] [Google Scholar]
- 66.Conn P.J., Christopoulos A., Lindsley C.W. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat. Rev. Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis J.A., Lebois E.P., Lindsley C.W. Allosteric modulation of kinases and GPCRs: design principles and structural diversity. Curr. Opin. Chem. Biol. 2008;12:269–280. doi: 10.1016/j.cbpa.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 68.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]


