Abstract
Purpose
Radiolabeled methionine (Met) promises to be useful in the positron emission tomography (PET) imaging of hepatocellular carcinoma (HCC). However, its metabolic routes in HCC have not yet been fully understood. In this study, the metabolic pathway(s) of radiolabeled Met in HCC were investigated.
Procedures
To simulate the rapid blood clearance of radiolabeled Met, pulse–chase experiments were conducted. L-[methyl-3H]-Met or L-[1-14C]-Met was pulsed over control or cycloheximide- treated WCH17 cells and rat hepatocytes for 5 min and chased with cold media. The water-soluble, lipid-soluble, DNA, RNA, and protein phases were subsequently extracted and measured from the acid-precipitable and acid-soluble fractions of whole cells. The radioactive metabolites Met, S- adenosylmethionine (SAM), S-adenosylhomocysteine, Met sulfoxide, and Met sulfone were further separated by radio thin layer chromatography.
Results
(1) The uptake of L-[methyl-3H]-Met in both cell types was higher than that of L-[1-14C]-Met. In rat hepatocytes, the uptake of L-[methyl-3H]-Met was significantly higher than that of L-[1-14C]-Met, which may contribute to its physiologic accumulation in surrounding hepatic tissues seen in PET imaging of HCC using L-[methyl-11C]-Met. Compared to rat hepatocytes, WCH17 cells had significantly higher uptake of both radiotracers. (2) For L-[methyl-3H]-Met, the major intracellular uptake was found mostly in the protein phase and, to a lesser degree, in the phosphatidylethanolamine (PE) methylation pathway, which is fairly stabilized within the 55-min chase period (the main metabolites were SAM, Met, Met sulfoxide, and Met sulfone). In contrast, the uptake of Met in rat hepatocytes mainly points to phosphatidylcholine (PC) synthesis through the PE methylation pathway (the main metabolite was PC). (3) Both cell types incorporated L-[1-14C]-Met predominantly into protein synthesis. (4) Finally, when the protein synthesis pathway was inhibited, the incorporation of SAM derived from L-[methyl-3H]-Met to lipid class (PC was the main metabolite) occurred at a reduced rate in WCH17 cells, suggesting that the route may be impaired in HCC.
Conclusions
This study demonstrated that different metabolic pathways of radiolabeled Met exist between HCC and surrounding hepatic tissue and contribute to the patterns of increased uptake of radiolabeled Met in HCC.
Keywords: Hepatocellular carcinoma, Radiolabeled methionine, Protein synthesis, Lipid synthesis, Phosphoethanolamine methylation pathway, Phosphatidylcholine, Phosphoethanolamine N- methyltransferase 2
Introduction
In our previous study (unpublished data), L-[methyl-14C]-methionine (Met) and L-[1- 14C]-Met demonstrated different uptake patterns in the liver of a woodchuck model of hepatocellular carcinoma (HCC). Regions of HCC showed higher uptake of both radiolabeled Met compared to surrounding hepatic tissues. However, L-[methyl-14C]-Met also contributed to a substantial background uptake in the surrounding hepatic tissues, which compromised its contrast resolution. Conversely, L-[1-14C]-Met showed minimal background uptake and a higher tumor to liver (T/L) uptake ratio, suggesting that it may have a better potential for imaging HCC. Different uptake patterns imply different metabolic fates for these radiolabeled Mets in HCC. The study that explores the exact uptake metabolism(s) of L-[methyl-14C]-Met and L-[1-14C]-Met in HCC is urgently needed to facilitate the interpretation of positron emission tomography (PET) images of HCC using both tracers.
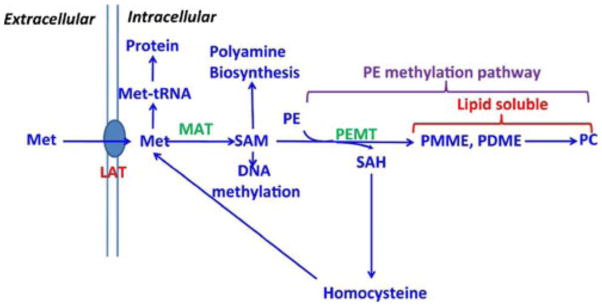
L-[methyl-11C]-Met has been shown to be useful for PET studies in a variety of malignant tumors [1–7]. Enhanced demand for Met in tumor cells is caused by increased fluxes in various Met metabolic pathways [8]. The major metabolic pathway of Met is shown in Fig. 1. After transport into cells, Met can either be incorporated into protein synthesis or be converted into S-adenosylmethionine (SAM) via catalysis by the key enzyme Met adenosyltransferase. In the phosphatidylethanolamine (PE) methylation pathway, SAM reacts with PE to form S-adenosylhomocysteine (SAH), phosphatidyl- monomethylethanolamine (PMME), and phosphatidyl- dimethylethanolamine (PDME) via catalysis by the key enzyme PE N-methyltransferase (PEMT); SAH can be decomposed back into Met, while PMME and PDME can immediately convert to phosphatidylcholine (PC). In normal brain and brain tumor tissue, the metabolic fate of L-[methyl-11C]-Met seems to be relatively simple, as it is assumed that contribution to the PE methylation pathway will be negligible and most of the Met will be incorporated into protein synthesis [9].
Fig. 1.
Metabolism of Met. Met methionine, SAM S-adenosyl- methionine, SAH S-adenosylhomocysteine, PMME phos- phatidylmonomethylethanolamine, PDME phosphatidyldi- methylethanolamine, PE phosphatidylethanolamine, PC phosphatidylcholine, MAT methionine adenosyltransferase, PEMT phosphatidylethanolamine N-methyltransferase, LATL- amino acid transporter.
The quality of PET imaging of HCC using L-[1-11C]-Met or L-[methyl-11C]-Met will depend on the metabolic fate of 11C. In normal liver cells, the two radiolabeled function groups in Met follow different metabolic pathways (Supplemental Fig. 1). L-[methyl- 11C]Met is thought to reflect PC synthesis via the PE methylation pathway [10], while L-[1-11C]Met mainly tracks incorporation into protein synthesis [11]. In the case of HCC, the situation is more complex and the metabolic fates of radiolabeled Met are far from being well identified. Therefore, it is of critical importance to identify the exact mechanism(s) of radiolabeled Met metabolism in primary liver cancer, to better understand how the uptake of Met impacts the interpretation of the PET imaging data.
In the present study, the metabolism of radiolabeled Met was characterized in a woodchuck HCC cell line, WCH17 [12], and in freshly derived rat hepatocytes. Due to the very short physical half-life of 11C (20 min), 14C-labeled 1-Met and 3H-labeled methyl-Met were used for this study. Radiolabeled Met-derived metabolites were analyzed by radio thin layer chromatography (TLC). Detailed analysis of radiotracer metabolites enabled the elucidation of the mechanism underlying the higher uptake of L- [methyl-3H]-Met and L-[1-14C]-Met in regions of HCC observed in the woodchuck model.
Materials and Methods
Materials
All chemical reagents used were obtained from Sigma Chemicals (St. Louis, MO, USA) unless stated otherwise. L-[methyl-3H]-Met (specific activity, 2.96 GBq/mmol), and L-[methyl-14C]-Met (specific activity, 2.035 GBq/mmol), were obtained from American Radiochemical Inc. (St. Louis, MO, USA). Hank's balanced salt solution (HBSS), Dulbecco's modified Eagle's medium (DMEM), and penicillin–streptomycin were obtained from Invitrogen Co. (Carlsbad, CA, USA). WCH17 cells were purchased from American Type Culture Collection (Manassas, VA, USA). All organic solvents were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Cell Cultures
WCH17 cells in the exponential phase were trypsinized and were plated (1 × 107 cells/75 cm2 flask) in 10 ml DMEM (contains 28.5 μM CHOL choride, 4,500 mg/l D-glucose, L-glutamine, and 110 mg/l sodium pyruvate; GIBCO/Invitrogen, Grand Island, NY, USA) supplemented with 10% FBS and 1% penicillin–streptomycin solution (10,000 U/ml penicillin and 10 μg/ml streptomycin) under a 10% CO2-humidified atmosphere at 37 °C. Freshly derived rat hepatocytes were purchased from Lonza Walkersville, Inc. (Walkersville, MD, USA) and maintained as the vendor's instruction. All experiments were carried out in triplicate, 24 h after plating.
Pulse–Chase Study
In order to mimic in vivo conditions, pulse–chase experiments were used to investigate the transient change of intracellular metabolites derived from radiolabeled Met. Cells were preincubated with a protein synthesis inhibitor 500 μg/ml cycloheximide (treated) or without cycloheximide (untreated) for 2 h [10, 13–15]. Cells were then incubated in 10 ml of HBSS containing 370 KBq of either L-[methyl-3H]-Met or L-[1-14C]-Met (American Radiolabeled Chemicals, Inc., St. Louis, MO, USA) for 5 min at 37 °C (pulse). After incubation, the media were removed and the cells were washed three times with PBS. The cells were then incubated in 10 ml of HBSS containing nonradioactive L-Met at the same concentration as radiolabeled Met for 10, 25, 40, and 55 min (chase); the treated condition had 500 μg/ml cycloheximide supplemented into the chase media, while the untreated group did not. After the chase, the cells were rinsed twice with PBS, trypsinized with 3 ml trypsin, and neutralized with 3 ml of ice-cold DMEM. Cell suspensions were centrifuged at 1,500×g at 4 °C for 10 min and the supernatants subsequently discarded. Cell pellets were resuspended in 1.2 ml ice-cold 0.4 M perchloric acid (HClO4) containing 100 mg/l BHT and incubated on ice for 30 min. The cell pellets were homogenized on ice for 3 min and the homogenate used for the extraction of radiolabeled metabolites.
Extraction of Radiolabeled Metabolites
From each 1.2 ml sample of homogenate, 0.1 ml was used to measure the total radioactivity with liquid scintillation counting using a Beckman LS-6500 Liquid Scintillation Counter (Beckman Coulter Inc., Fullerton, CA, USA) and Bio-Safe II (Fisher Scientific Inc., Pittsburgh, PA, USA) as the scintillation fluid. Disintegrations per minute were obtained by correcting for background activity and counter efficiency based on counts of calibrated standards. Another 0.1 ml of homogenate was used for protein quantification using the Bradford method (Bio-Rad Laboratories, Inc., Hercules, CA, USA) [16].
The remaining 1 ml homogenate was used for the extraction of radiolabeled metabolites following a previously described procedure [17] with modification (Supplemental Fig. 2). Briefly, 1 ml homogenate was centrifuged at 3,000×g for 5 min and separated into acid-precipitable fraction (APF) and acid-soluble fraction (ASF) (step 1). The major radioactive components in the APF include lipids (PMME, PDME, and PC), RNA, DNA, protein, etc. The major radioactive metabolites of the ASF include Met, SAM, SAH, etc. The APF and ASF were further extracted in 2:1 chloroform/methanol and centrifuged at 3,000×g for 10 min to divide the whole mixture into water-soluble and lipid-soluble fractions (step 2). The water-soluble fraction was further separated into individual radioactive metabolites (Met, SAM, and SAH) as described in the next paragraph.
The precipitate from APF at step 2 was dissolved in 1 ml 0.3 M KOH and incubated at 37 °C for 1 h, after which 0.32 ml 3 N perchloric acid was added. The mixture sat on ice for 5 min. At this step, the supernatant at this step is an alkaline-labile fraction containing RNA hydrolysate (step 3). The precipitate from step 3 was resuspended in 1 ml of 0.5 M perchloric acid and heated at 90 °C for 15 min. The supernatant at this step is an acid-labile fraction containing DNA hydrolysate (since there is no pathway to derive DNA from Met, this might be derived from basic protein, such as chromosomal histone) (step 4). The final precipitate is protein.
Analysis of Radioactive Metabolites in Water-Soluble Fraction
The water-soluble radioactive metabolites were analyzed by Ricerca Biosciences, LLC (Cleveland, OH, USA) with a contract service. Briefly, silica gel G plate with concentration zone (Merck KGaA, Gibbstown, NJ, USA) was used for TLC analysis. Samples or standards were spotted into pre-adsorbent zones of individually labeled lanes and allowed to air-dry. Eluant was made fresh and allowed to pre-equilibrate in the TLC tank for ~10 min prior to elution. The mobile phase consisted of butanol/acetic acid/water (2:1:2, v/v/v). The water-soluble metabolites were separated and identified via comparison of the retention factor (Rf) with those of the cold standards. The Rf is defined as the ratio of moving distance of the compound to the moving distance of the solvent from the origin [18]. The plate was eluted to a distance of 15 cm with an elapsed time of ~3 h. The plate was then air-dried and visualized using short wave UV light (handheld lamp), and sample lanes were scanned using the Bioscan AR 2000 (Bioscan Inc., Washington, DC, USA) to quantify the incorporation of radioactivity in each metabolite. The Rf values were 0.29 for SAM, 0.435 for Met sulfoxide and Met sulfone, 0.65 for SAH, and 0.77 for Met. Supplemental Fig. 3 shows a representative radio-TLC chromato- gram from the water-soluble phase of WCH17 cells after a 5-min pulse with L-[methyl-3H]-Met.
Statistical Analysis
The results are reported as the average ± standard deviation. The data were compared using one-way analysis of variance (ANOVA) or ANOVA on ranks, when appropriate. All pairwise multiple comparison procedures used Tukey's honestly significant difference test. Differences were regarded as statistically significant for p values < 0.05.
Results
Radiolabeled Met has a rapid circulation and clearance during PET imaging. In order to mimic this in vivo condition, pulse–chase experiments were used to investigate the transient change of the intracellular metabolites derived from L-[methyl-3H]-Met and L-[1-14C]-Met.
Radiolabeled Met Metabolism
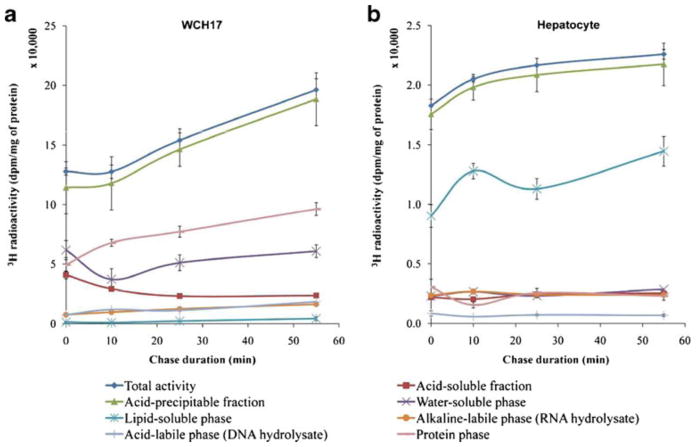
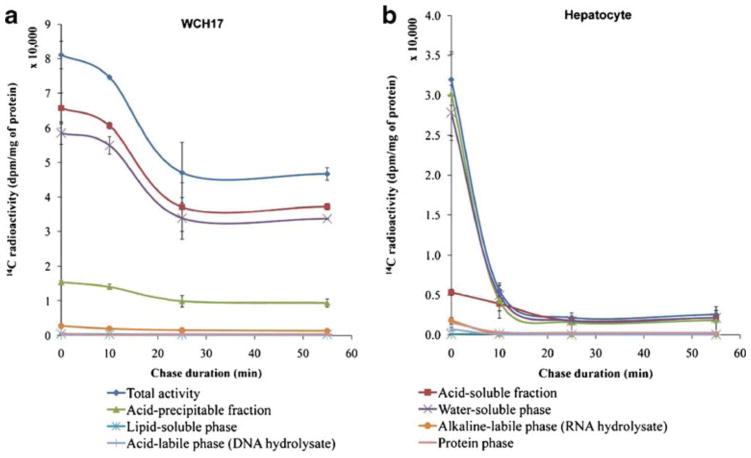
For L-[methyl-3H]-Met, rat hepatocytes and WCH17 cells were pulsed for 5 min and then chased for various durations (Fig. 2). For both cell types, the total radioactivity content within the collected cells increased with the duration of the chase; however, the total radioactivity retained into WCH17 cells was around tenfold higher than that in rat hepatocytes during the whole pulse and chase period (p < 0.001) (Fig. 2).
Fig. 2.
Fraction of metabolites in the WCH17 HCC cells and rat hepatocytes during the pulse and chase period with L-[methyl-3H]-Met. a WCH17 HCC cells, b rat hepatocytes. Chase 0 min means only pulse of 5 min, no chase.
In WCH17 cells, the levels of 3H in the water-soluble phase were the highest at the early pulse period, and then the levels of 3H in the protein phase derived from the APF started to increase and exceed the levels of 3H in the water-soluble phase at the late pulse and whole chase periods (Fig. 2a). A certain amount of 3H in the RNA and DNA phases also accumulated throughout the pulse–chase period. Interestingly, very low levels of 3H were observed in the lipid-soluble phase during the pulse–chase period. On the other hand, in rat hepatocytes, the levels of 3H in the lipid-soluble phase were significantly higher than other phases during the whole pulse–chase period (Fig. 2b).
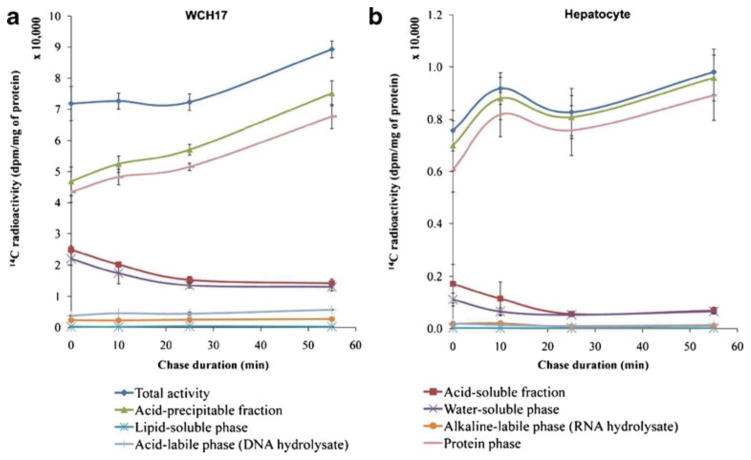
For both cell types, the uptake patterns of L-[1-14C]-Met differed from that of L-[methyl-3H]-Met (Fig. 3): (1) In rat hepatocytes, the uptake of L-[1-14C]-Met was twofold less than that of L-[methyl-3H]-Met; (2) in WCH17 cells, the level of 14C derived from the APF in the protein phase was much higher than that in the water-soluble phase during the pulse–chase period; (3) in rat hepatocytes, the greatest accumulation of 14C was found in the protein phase, as opposed to the lipid-soluble phase; and (4) in both cell types, the lipid-soluble phase had negligible levels of radiolabeled metabolites.
Fig. 3.
Fraction of metabolites in the WCH17 HCC cells and rat hepatocytes during the pulse and chase period with L-[1-14C]-Met. a WCH17 HCC cells, b rat hepatocytes. Chase 0 min means only pulse of 5 min, no chase.
Effect of Cycloheximide on Tracer Uptake, Metabolism, and Protein Incorporation
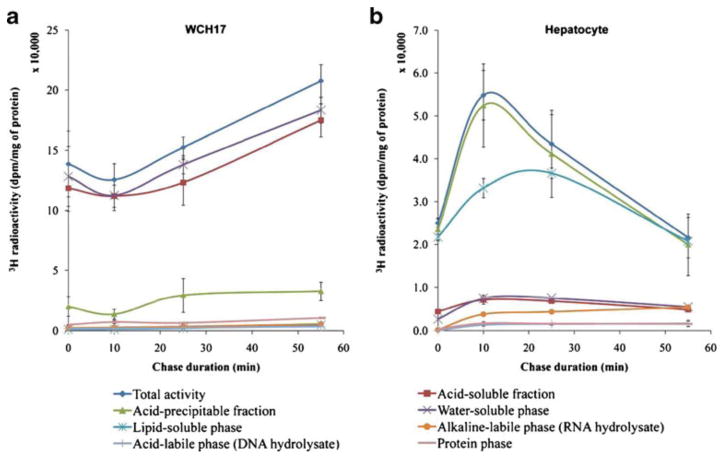
The effects of inhibiting protein synthesis with cycloheximide on liver metabolism were also investigated. Fig. 4 illustrates the effect of cycloheximide treatment on the uptake of L-[methyl-3H]-Met in WCH17 cells and rat hepatocytes. In WCH17 cells, total radioactivity increased with the duration of the chase period. Total radioactivity retained in WCH17 cells during the whole pulse and chase period was significantly higher than that in rat hepatocytes (p < 0.001). The largest accumulation of 3H in WCH17 cells was in the water-soluble fraction, while the radioactivity in the protein, DNA, and RNA phases were negligible (Fig. 4a). In contrast, the radioactivity in rat hepatocytes increased for the first 10 min and decreased gradually (Fig. 4b). The largest accumulation of 3H in rat hepatocytes was in the lipid-soluble phase.
Fig. 4.
Fraction of metabolites in the WCH17 HCC cells and rat hepatocytes with cycloheximide treatment during the pulse and chase period with L-[methyl-3H]-Met. a WCH17 HCC cells, b rat hepatocytes. Chase 0 min means only pulse of 5 min, no chase.
Fig. 5 shows the effect of treatment with cycloheximide on the uptake of L-[1-14C]-Met in WCH17 cells and rat hepatocytes. As compared to the uptake pattern with untreated cells, total radioactivity in both cells decreased with the duration of the chase (Fig. 3 vs. Fig. 5). In both cell types, the distribution of radioactivity was similar: (1) the greatest levels of radioactivity were found in the water- soluble phase; (2) the radioactivity contained in the lipid- soluble phase was negligible; and (3) due to the effect of cycloheximide inhibition, radioactivity in the protein phase of treated cells was very low.
Fig. 5.
Fraction of metabolites in the WCH17 HCC cells and rat hepatocytes with cycloheximide treatment during the pulse and chase period with L-[1-14C]-Met. a WCH17 HCC cells, b rat hepatocytes. Chase 0 min means only pulse of 5 min, no chase. 14C-labeled metabolites in the water-soluble phase, an opposite result than that observed with L-[methyl-3H]-Met.
Radiolabeled Metabolites Extracted from the Water-Soluble Phase
Table 1 compares the distribution of radiolabeled metabolites derived from L-[methyl-3H]-Met and L-[1-14C]-Met in WCH17 cells and rat hepatocytes. In WCH17 cells, the major L-[methyl-3H]-Met metabolites in the water-soluble phase were 3H-SAM, 3H-Met sulfone, 3H-Met sulfoxide, and 3H-Met. However, in rat hepatocytes, the major 3H- labeled metabolites in the water-soluble phase were undetectable due to their rapid conversion to lipids (PC, PMME, and PDME) that is generally observed in hepatocytes.
Table 1.
Radiolabeled metabolites extracted from the water-soluble phase of radiolabeled Met metabolism Uptake (dpm/mg of protein)
| Uptake (dpm/mg of protein)
|
|||||
|---|---|---|---|---|---|
| 0 min | 10 min | 25 min | 55 min | ||
|
|
|||||
| L-[methyl-3H]-Met | |||||
| HCC cells | SAM | 20,330.30±2,616.42 | 12,503.05±2,987.16 | 16,610.13±2,138.09 | 21,565.50±1,907.79 |
| Met sulfoxide+Met sulfone | 14,954.91±1,924.63 | 8,207.80±1,960.96 | 11,731.70±1,510.13 | 14,006.65±1,239.10 | |
| Met | 25,298.83±255.84 | 13,101.26±3,130.08 | 18,559.46±2,389.01 | 21,170.86±1,872.88 | |
| Rat hepatocytes | SAM | ND | |||
| Met sulfoxide+Met sulfone | ND | ||||
| Met | ND | ||||
| L-[1-14C]-Met | |||||
| HCC cells | SAM | 1,684.62±162.53 | 3,267.29±73.04 | 3,547.40±222.44 | 2,899.98±285.29 |
| Met sulfoxide+Met sulfone | 4,023.27±388.16 | 5,457.63±122.01 | 4,754.08±298.10 | 4,498.85±442.59 | |
| SAH | 10,173.79±981.55 | 5,823.55±130.19 | 3,733.04±234.08 | 3,416.53±336.11 | |
| Met | 5,293.89±510.75 | 1,276.39±28.54 | 519.26±32.56 | 1,139.28±112.08 | |
| Rat hepatocytes | SAM | 142.19±31.55 | 68.78±14.19 | 35.67±4.88 | 66.68±7.99 |
| Met sulfoxide+Met sulfone | 311.71±69.16 | 167.25±34.50 | 122.23±16.73 | 182.70±21.89 | |
| SAH | 257.27±57.08 | 189.66±39.20 | 130.71±17.88 | 160.54±19.24 | |
| Met | 348.45±77.31 | 173.62±35.81 | 93.26±12.76 | 178.27±21.36 | |
Values are the mean±SD (n93). HCC vs. rat hepatocytes for each metabolite at each time point: p<0.05. The time is the chase time after pulse of the cells for 5 min; chase 0 min means only pulse of 5 min, no chase ND nondetectable due to very low radioactivity in the water-soluble phase
In WCH17 cells, the major L-[1-14C]-Met metabolites in the water-soluble phase were 14C-SAM, 14C-SAH, 14C-Met sulfone, 14C-Met sulfoxide, and 14C-Met. The radioactivity of all metabolites was less than that detected of their L- [methyl-3H]-Met-derived counterparts. The level of each metabolite was less than that from L-[methyl-3H]-Met. Interestingly, rat hepatocytes showed the accumulation of 14C-labeled metabolites in the water-soluble phase, an opposite result than that observed with L-[methyl-3H]-Met.
Table 2 compares the distribution of radiolabeled metabolites derived from L-[methyl-3H]-Met and L-[1-14C]-Met in WCH17 cells and rat hepatocytes when protein synthesis was inhibited by cycloheximide. In both WCH17 cells and rat hepatocytes, greater levels of radioactivity were detected in the water-soluble phase compared to all other phases when protein synthesis was blocked.
Table 2.
Radiolabeled metabolites extracted from the water-soluble phase of radiolabeled Met metabolism with the inhibition of protein synthesis Uptake (dpm/mg of protein)
| Uptake (dpm/mg of protein)
|
|||||
|---|---|---|---|---|---|
| 0 min | 10 min | 25 min | 55 min | ||
|
|
|||||
| L-[methyl-3H]-Met | |||||
| HCC cells | SAM | 30,306.88±5,896.17 | 25,122.31±2,810.35 | 41,240.96±2,235.72 | 44,459.63±7,437.17 |
| Met sulfoxide+Met sulfone | 39,523.46±7,689.25 | 36,777.81±4,114.21 | 22,276.74±1,207.65 | 37,770.53±6,318.22 | |
| Met | 54,272.55±1,055.67 | 47,105.74±5,269.56 | 71,619.58±3,882.59 | 99,878.39±1,677.57 | |
| Rat hepatocytes | SAM | 197.26±15.20 | 681.70±54.38 | 1,396.43±104.99 | 1,165.78±106.74 |
| Met sulfoxide+Met sulfone | 846.41±65.22 | 2,526.95±201.74 | 1,570.61±118.08 | 1,053.03±96.42 | |
| Met | 1,601.81±123.42 | 3,961.04±351.98 | 4,404.37±331.12 | 3,346.26±306.40 | |
| L-[1-14C]-Met | |||||
| HCC cells | SAM | 17,568.71±984.62 | 16,450.71±760.17 | 9,190.51±1,628.26 | 10,648.85±1,168.62 |
| Met sulfoxide+Met sulfone | 20,127.03±1,128.00 | 19,001.04±878.02 | 10,452.68±1,851.88 | 12,516.19±1,180.65 | |
| SAH | 13,962.47±782.51 | 12,817.60±592.29 | 8,726.92±546.13 | 7,836.08±550.49 | |
| Met | 6,416.30±359.60 | 6,007.56±277.60 | 4,923.49±872.28 | 2,018.74±513.01 | |
| Rat hepatocytes | SAM | 7,230.98±1,673.03 | 696.43±90.22 | 134.86±14.1 | 141.29±29.75 |
| Met sulfoxide+Met sulfone | 10,660.13±2,466.43 | 1,659.78±218.15 | 693.79±72.98 | 510.87±107.55 | |
| SAH | 6,026.74±1,394.40 | 1,377.83±181.09 | 611.86±64.36 | 478.59±100.76 | |
| Met | 2,405.69±556.60 | 1,256.79±165.18 | 317.89±33.44 | 594.23±125.10 | |
Values are mean±SD (n93). HCC vs. rat hepatocytes for each metabolite at each time point: p<0.05. The time is the chase time after pulse of the cells for 5 min; chase 0 min means only pulse of 5 min, no chase
Discussion
This study demonstrated the existence of different metabolic fates for radiolabeled Met in WCH17 cells and rat hepatocytes in addition to distinct uptake mechanisms for L-[methyl-3H]-Met and L-[1-14C]-Met. The findings in the present study support the following views:
Uptake of L-[methyl-3H]-Met in both cell types was significantly higher than that of L-[1-14C]-Met. This finding is consistent with the results of autoradiography in previous studies using the woodchuck model of HCC. L- [methyl-3H]-Met also displayed a nonnegligible amount of uptake in the rat hepatocytes, which may contribute to the noticeable background contrast seen in our previous PET imaging of the woodchuck model of HCC using L- [methyl-11C]-Met. Conversely, the relative uptake of L- [1-14C]-Met in the rat hepatocytes was lower, which corresponded with the clear background and higher T/L uptake ratio seen in the L-[1-14C]-Met autoradiograph of liver slices from a woodchuck with HCC.
Uptake of L-[methyl-14C]-Met was significantly greater in WCH17 cells compared to rat hepatocytes. Protein synthesis and, to a lesser degree, incorporation into SAM were the major metabolic fates in WCH17 cells; as such, radiolabeled protein and SAM were the predominant L-[methyl-14C]-Met-derived metabolites. In rat hepatocytes, lipid synthesis was the major metabolic fate of L-[methyl-14C]-Met, and the predominant L-[methyl-14C]-Met-derived metabolites were radiolabeled phospho- lipids (PC, PMME, and PDME). A noticeable phenomenon for L-[methyl-14C]-Met uptake in WCH17 cells was the rapid incorporation of Met into SAM and negligible conversion of SAM into phospholipids.
Uptake of L-[1-14C]-Met was significantly greater in WCH17 cells compared to rat hepatocytes. For both cell types, protein synthesis was the major metabolic fate and radiolabeled protein was the predominant L-[1-14C]-Met- derived metabolite. In WCH17 cells, a small amount of radiolabeled Met, SAM, and SAH was also observed.
When protein synthesis was inhibited by cycloheximide, the PE methylation pathway became the predominant metabolic pathway for both L-[methyl-3H]-Met and L-[1-14C]-Met in both cell types. The total uptake of 14C decreased in a time-dependent manner in both cell types, which suggests that PE methylation is a minor pathway for L-[1-14C]-Met metabolism. In WCH17 cells, most L- [methyl-3H]-Met-derived radioactivity accumulated in the water-soluble phase. The conversion of metabolites from the water-soluble phase to the lipid-soluble phase was negligible, which suggests that conversion of SAM to PC was impaired in WCH17 cells. In contrast, the PC synthesis pathway is the predominant metabolic pathway in primary hepatocytes.
The distribution of metabolites in the water-soluble phase of both cell types confirmed the different metabolic fates of radiolabeled Met between WCH17 cells and primary hepatocytes.
Total uptake of L-[methyl-3H]-Met was higher in both cell types compared to that of L-[1-14C]-Met. One reason for this is explained by the different metabolic fates of these two radiotracers. A second possibility is that decarboxylation of branched-chain amino acids occurred as a minor metabolic pathway; as a result, the L-[1-14C]-Met was washed out as 14C-CO2, while the decarboxylated L-[methyl-3H]-Met, still retaining its radiolabel, remained [19].
Distinct Metabolic Fates of L-[methyl-11C]-Met in HCC Cells and Primary Hepatocytes
SAM reacts with PE and converts to PC in the liver via the enzyme PEMT (Supplemental Fig. 1a). PEMT has two isoforms. An isoform, PEMT2, has been found in a mitochondria-associated membrane in rat liver. Interestingly, PEMT2 might have an exclusive role in the regulation of hepatoma cell division, and its expression in rat hepatoma cells can specifically suppress cell division [20]. In this study, the PE methylation pathway in HCC was disrupted, and conversion of L-[methyl-3H]-Met-derived SAM into lipid (PC, PMME, and PDME) occurred slowly even when protein synthesis was inhibited. These data suggest that PEMT activity may be much lower in WCH17 cells compared to primary hepatocytes, implying a state of high cell division and DNA replication occurring in HCC cells. Low PEMT activity in HCC cells could divert the metabolic pathway of L-[methyl-3H]-Met towards protein synthesis and, to a lesser degree, cause the intermediate metabolites to accumulate in the water-soluble phase. Similarly, conversion of metabolites from the water-soluble phase to the lipid- soluble phase may be impaired.
In contrast, PEMT activity was high in rat hepatocytes and the route from water-soluble phase to lipid-soluble phase was clear. Thus, the predominant pathway for metabolites derived from L-[methyl-3H]-Met in hepatocytes was lipid synthesis. It is still unknown why lipid synthesis overwhelms protein synthesis during the metabolism of L- [methyl-3H]-Met in primary hepatocytes. One possible explanation is physiological adaption of primary hepatocytes. Lipids (PC, PMME, and PDME) derived from the cytidine diphosphate (CDP)-choline and PE methylation pathways possess inverse physiological functions. PC derived from the PE methylation pathway will suppress cell division and DNA replication, while PC derived from the CDP–choline pathway will accelerate the rate of cell division of hepatoma cells grown in culture and of hepatocytes in intact animals [20, 21]. As such, a high level of PEMT2 expression and the preference for lipid synthesis over protein synthesis in primary hepatocytes may represent a physiological adaption, i.e., a mechanism possessed by normal hepatocytes that controls the rate of cell division. Within the same metabolic context, it stands to reason that there is a lack of PEMT activity in HCC.
Superior Contrast Uptake of L-[1-11C]-Met Compared to L-[methyl-11C]-Met Despite Common Incorporation into Protein Synthesis
Normal liver tissue synthesizes many Met-derived secretory proteins, notably albumin, immunoglobulin G (IgG), high-molecular-weight materials, and low-molecular-weight materials [10]. In normal liver, within 1 h postinjection, 80% of the radioactivity released into the plasma from L-[methyl-3H]-Met metabolism was detected in the albumin fraction, while radioactivity in the IgG and high-molecular-weight fractions was minor [10]. Albumin biosynthesis and secretion are characteristic functions of normal rat liver, with albumin being the most prominent protein synthesized by the adult organ. Serum albumin is responsible for maintaining the osmotic equilibrium of body fluids and plays an important role in the transport of various essential metabolites and cellular constituents [22, 23].
During hepatic neoplasm, genetic regulation of albumin synthesis may be altered. Studies have shown that hepatoma cells lose their capacity to synthesize albumins, which causes up to a fourfold decrease in albumin release compared to primary hepatocytes [22, 23]. The question remains: why does L-[1-11C]-Met have a better contrast uptake (T/L ratio) than L-[methyl-11C]-Met? One possibility is that the suppression of secretory protein synthesis in HCC reduces the overall amount of secretory proteins released, thus producing an increase in the retention of radiolabeled intermediate metabolites. Conversely, although both HCC cells and hepatocytes predominantly incorporate L-[1- 11C]-Met into protein synthesis, the reduced secretory protein synthesis and increased rate of protein synthesis might provide a better contrast uptake in regions of HCC in PET imaging with L-[1-11C]-Met. Meanwhile, the rapid synthesis and secretion of secretory proteins that occurs within 1 h post-injection may wash out the intracellular radioactive protein, contributing to a lower background uptake.
Implication for In Vivo PET Imaging of HCC with Radiolabeled Met
The present study investigating the metabolism and uptake of radiolabeled Met in HCC provides insights to aid the quantitative analysis of PET imaging of HCC with radiolabeled Met and the interpretation of image results, e.g., studies on tracer kinetics via compartment modeling analysis. In our previous imaging study, WCH17 cells displayed a faster and greater accumulation of radiolabeled Met compared to surrounding hepatic tissues. This implies that, to model the rate of Met transport, investigators should anticipate a greater rate of transport in regions of HCC compared to the surrounding hepatic tissues during parameter estimation.
Radiolabeled Met transported into HCC cells was rapidly incorporated into protein or converted into SAM. Thus, the parameter for the influx rate between the free Met compartment and the metabolized Met compartment in HCC cells should represent a combination of protein synthesis rate and Met transmethylation rate; due to the suppression of secretory protein synthesis in HCC, the parameter for the rate of outflux from the metabolized Met compartment in HCC cells can be ignored. In contrast, the patterns observed in primary hepatocytes point to the PE methylation pathway. As such, the parameter for the influx rate between the free Met compartment and the metabolized Met compartment in primary hepatocytes should predominantly represent the Met transmethylation rate. In the context of PET imaging using L-[1-11C]-Met, although both WCH17 cells and primary hepatocytes incorporate radiolabeled Met in protein synthesis, a significantly higher rate of protein synthesis is expected in WCH17 cells as compared to surrounding hepatic tissues.
Taken together, this metabolism study will be useful for the development and validation of a compartment model for kinetic analysis of Met radiotracers. Such a model would enable the estimation of the higher rate of Met transport into HCC and the higher rate of protein synthesis and/or Met transmethylation based on different labeling positions compared to surrounding hepatic tissues. The resulting parameters could be quantitatively useful, for example, for the early detection of HCC and/or monitoring the treatment response in HCC patients.
This in vitro metabolism study obtained basic information on the metabolism of radiolabeled Met in HCC cells without confounding influences, such as microenvironments, non- neoplastic cells, precancerous cells, competing plasma substrates, and heterogeneity of cancers. Validation of the applicability of these in vitro observations to in vivo conditions requires in vivo evaluation of the patterns of Met metabolism in both experimental and human HCC tumors.
Conclusion
This study demonstrated that distinct metabolic fates exist for radiolabeled Met in HCC cells and hepatocytes. In the context of our previous study, these patterns are likely to be responsible for the enhanced uptake of Met in PET imaging of HCC. In HCC cells, the major metabolic fate of L- [methyl-3H]-Met was protein synthesis and, to a lesser degree, the PE methylation pathway. However, in primary hepatocytes, lipid synthesis was the predominant metabolic fate of L-[methyl-3H]-Met; these 3H-radiolabeled lipids (PC, PMME, and PDME) that were produced contribute to the background contrast observed in PET imaging of HCC using L-[methyl-11C]-Met. Both HCC cells and primary hepatocytes incorporated L-[1-14C]-Met into protein synthesis. Of particular note, primary hepatocytes had significantly lower uptake of L-[1-14C]-Met than that of L-[methyl-3H]-Met, which may imply a better contrast resolution (T/L ratio) during PET imaging of HCC using L-[1-11C]-Met.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jim Basilion for sharing the laboratory resources. This work was supported by an NIH/NCI R01 grant CA095307.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11307-013-0678-z) contains supplementary material, which is available to authorized users.
Conflict of Interest. The authors declare that they have no conflict of interest.
References
- 1.Leskinen-Kallio S, Lindholm P, Lapela M, et al. Imaging of head and neck tumors with positron emission tomography and [11C]methionine. Int J Radiat Oncol Biol Phys. 1994;30:1195–1199. doi: 10.1016/0360-3016(94)90328-x. [DOI] [PubMed] [Google Scholar]
- 2.Jager PL, Vaalburg W, Pruim J, et al. Radiolabeled amino acids: basic aspects and clinical applications in oncology. J Nucl Med. 2001;42:432–445. [PubMed] [Google Scholar]
- 3.Leskinen-Kallio S, Ruotsalainen U, et al. Uptake of carbon-11- methionine and fluorodeoxyglucose in non-Hodgkin's lymphoma: a PET study. J Nucl Med. 1991;32:1211–1218. [PubMed] [Google Scholar]
- 4.Nettelbladt OS, Sundin AE, Valind SO, et al. Combined fluorine-18-FDG and carbon-11-methionine PET for diagnosis of tumors in lung and mediastinum. J Nucl Med. 1998;39:640–647. [PubMed] [Google Scholar]
- 5.Inoue T, Kim EE, Wong FC, et al. Comparison of fluorine-18- fluorodeoxyglucose and carbon-11-methionine PET in detection of malignant tumors. J Nucl Med. 1996;37:1472–1476. [PubMed] [Google Scholar]
- 6.Deloar HM, Fujiwara T, Nakamura T, et al. Estimation of internal absorbed dose of L-[methyl-11C]methionine using whole-body positron emission tomography. Eur J Nucl Med. 1998;25:629–633. doi: 10.1007/s002590050265. [DOI] [PubMed] [Google Scholar]
- 7.Plathow C, Weber WA. Tumor cell metabolism imaging. J Nucl Med. 2008;49(Suppl 2):43S–63S. doi: 10.2967/jnumed.107.045930. [DOI] [PubMed] [Google Scholar]
- 8.Singhal T, Narayanan TK, Jain V, et al. 11C-L-methionine positron emission tomography in the clinical management of cerebral gliomas. Mol Imaging Biol. 2008;10:1–18. doi: 10.1007/s11307-007-0115-2. [DOI] [PubMed] [Google Scholar]
- 9.Bustany P, Chatel M, Derlon JM, et al. Brain tumor protein synthesis and histological grades: a study by positron emission tomography (PET) with C11-L-methionine. J Neurooncol. 1986;3:397–404. doi: 10.1007/BF00165590. [DOI] [PubMed] [Google Scholar]
- 10.Ishiwata K, Enomoto K, Sasaki T, et al. A feasibility study on L- [1-carbon-11]tyrosine and L-[methyl-carbon-11]methionine to assess liver protein synthesis by PET. J Nucl Med. 1996;37:279–285. [PubMed] [Google Scholar]
- 11.DeLong CJ, Hicks AM, Cui Z. Disruption of choline methyl group donation for phosphatidylethanolamine methylation in hepatocarcinoma cells. J Biol Chem. 2002;277:17217–17225. doi: 10.1074/jbc.M108911200. [DOI] [PubMed] [Google Scholar]
- 12.Kuang Y, Salem N, Corn DJ, et al. Transport and metabolism of radiolabeled choline in hepatocellular carcinoma. Mol Pharm. 2010;7:2077–2092. doi: 10.1021/mp1001922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Ashwell MA, Work TS. Contrasting effects of cycloheximide on mitochondrial protein synthesis “in vivo” and “in vitro”. Biochem Biophys Res Commun. 1968;32:1006–1012. doi: 10.1016/0006-291x(68)90129-0. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Fang Z, Liang Y, et al. Overexpression of integrin beta1 inhibits proliferation of hepatocellular carcinoma cell SMMC-7721 through preventing Skp2-dependent degradation of p27 via PI3K pathway. J Cell Biochem. 2007;102:704–718. doi: 10.1002/jcb.21323. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Ishiwata K, Vaalburg W, Elsinga PH, et al. Comparison of L-[1- 11C]methionine and L-methyl-[11C]methionine for measuring in vivo protein synthesis rates with PET. J Nucl Med. 1988;29:1419–1427. [PubMed] [Google Scholar]
- 18.Meyers CL, Meyers DJ. Thin-layer chromatography. Curr Protoc Nucleic Acid Chem Appendix:3D. 2008 doi: 10.1002/0471142700.nca03ds34. [DOI] [PubMed] [Google Scholar]
- 19.Ishiwata K, Kubota K, Murakami M, et al. A comparative study on protein incorporation of L-[methyl-3H]methionine, L-[1-11C]leucine and L-2-[18F]fluorotyrosine in tumor bearing mice. Nucl Med Biol. 1993;20:895–899. doi: 10.1016/0969-8051(93)90089-d. [DOI] [PubMed] [Google Scholar]
- 20.Vance DE, Houweling M, Lee M, Cui Z. Phosphatidylethanol- amine methylation and hepatoma cell growth. Anticancer Res. 1996;16:1413–1416. [PubMed] [Google Scholar]
- 21.Cui Z, Houweling M, Vance DE. Expression of phosphatidyl- ethanolamine N-methyltransferase-2 in McArdle-RH7777 hepatoma cells inhibits the CDP–choline pathway for phosphatidylcholine biosynthesis via decreased gene expression of CTP:phosphocholine cytidylyltransferase. Biochem J. 1995;312(Pt 3):939–945. doi: 10.1042/bj3120939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capetanaki YG, Flytzanis CN, Alonso A. Repression of the albumin gene in Novikoff hepatoma cells. Mol Cell Biol. 1982;2:258–266. doi: 10.1128/mcb.2.3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse TP, Morris HP, Taylor JM. Molecular basis of reduced albumin synthesis in Morris hepatoma 7777. Biochemistry. 1978;17:3121–3128. doi: 10.1021/bi00608a028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.