Abstract
The sequences of the full set of pepper genomes including nuclear, mitochondrial and chloroplast are now available for use. However, the overall of simple sequence repeats (SSR) distribution in these genomes and their practical implications for molecular marker development in Capsicum have not yet been described. Here, an average of 868,047.50, 45.50 and 30.00 SSR loci were identified in the nuclear, mitochondrial and chloroplast genomes of pepper, respectively. Subsequently, systematic comparisons of various species, genome types, motif lengths, repeat numbers and classified types were executed and discussed. In addition, a local database composed of 113,500 in silico unique SSR primer pairs was built using a homemade bioinformatics workflow. As a pilot study, 65 polymorphic markers were validated among a wide collection of 21 Capsicum genotypes with allele number and polymorphic information content value per marker raging from 2 to 6 and 0.05 to 0.64, respectively. Finally, a comparison of the clustering results with those of a previous study indicated the usability of the newly developed SSR markers. In summary, this first report on the comprehensive characterization of SSR motifs in pepper genomes and the very large set of SSR primer pairs will benefit various genetic studies in Capsicum.
Simple sequence repeats (SSRs), also termed microsatellites, consist of tandemly arranged repeats of short DNA motifs (1–6 bp in length). Since they were first documented in the 1980s1 and their abundance and ubiquity has been confirmed in prokaryotic and eukaryotic genomes in subsequent studies2,3, SSRs have become one of the most attractive markers for plant genetics and breeding4. As molecular markers, SSRs present several important advantages, such as being locus-specific and multi-allelic, exhibiting co-dominant transmission, their ease of detection by PCR and their high rates of transferability across species5. SSRs have been extensively involved in a variety of applications including cultivar identification6, the determination of ‘hybridity’7, genetic diversity assessment8, genetic mapping9, gene tagging10, gene flow11 and molecular evolution12 in various plant and animal systems.
In general, SSR markers for plant studies have been discovered by cross-species amplification13, screening either SSR-enriched cDNA or genomic libraries14 and searching public databases15. With the decreasing cost of next generation sequencing (NGS), which is a powerful and convenient tool for marker discovery16, SSRs can now be identified on a large scale from the de novo assembled transcriptome or whole genome17. In the latter case, the whole picture of SSR frequency and distribution can be concurrently described18 and provide practical implications for their use as molecular markers19. Genome-wide SSR identification has been performed in many organisms including humans20, marine animals21, insects22, medicinal fungi23 and certain economically valuable plants24,25,26. However, except for cucumber27 and Chinese cabbage28, the effort in this area for important vegetable species such as Capsicum spp. lags far behind.
In addition, similar to the nuclear genomes, SSRs in organelle genomes are common29 and it is generally accepted that polymorphisms due to variations in SSR motif length in the chloroplast or mitochondrial genomes would also be of considerable practical value for monitoring gene flow30, population differentiation31 and cytoplasmic diversity32. To our knowledge, a complete analysis of SSR loci in the mitochondrial or chloroplast genomes has been performed only in a relatively limited set of species, such as bryophytes33, rice34 and soybean29. Thus far, this analysis has rarely been fully conducted in horticultural crops including pepper.
Pepper (Capsicum spp.) belongs to the Solanaceae family and is one of the most economically important vegetable crops with versatile applications for food, spice, ornamental and medicinal purposes35. A variety of marker systems, such as restriction fragment length polymorphisms (RFLPs), amplified fragment length polymorphisms (AFLPs), random amplified polymorphic DNA (RAPDs), single nucleotide polymorphisms (SNPs), insertion/deletion (InDel) polymorphisms and SSRs have been adopted in pepper molecular genetics research35,36,37,38,39,40,41,42,43,44,45,46. However, the total number of publicly available PCR-based anchored markers, including SSR markers, is still insufficient46,47.
In addition, the development of SSR markers of pepper in previous studies mainly focused on mining from either selected SSR-enriched libraries39,48,49 or public database50,51,52. The massive amount of RNA-seq data has facilitated the in silico generation of SSR information with unprecedented dimensions53,54,55,56,57,58. Nevertheless, a systematic survey of SSRs in the pepper, including nuclear, mitochondrial and chloroplast genomes, has not yet been conducted despite the recent availability of the relevant information35,59,60,61,62. Furthermore, the utilization of these SSR loci to develop molecular markers for genetic applications such as diversity assessment, positional cloning, genome assembly and breeding activities such as marker-assisted selection (MAS) will increase continuously63,64,65.
Consequently, the aim of this study was to perform a genome-wide identification of SSRs in the pepper and evaluate them for marker development. We initially detected all SSR motifs in a total of six pepper genomes including nuclear, mitochondrial and chloroplast genomes, from two independent sources. Simultaneously, for comparison purposes, we performed the same analysis for another five species: tomato (Solanum lycopersicum), potato (Solanum tuberosum), cucumber (Cucumis sativus), Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Then, a comprehensive characterization and comparison of SSR frequency and distribution within different genomes were performed, and a large number of unique SSR loci were identified using bioinformatics. Finally, a collection of 21 pepper genotypes with extensive representations was selected to verify the availability of the primer pairs of those unique SSR loci. The information and primer pairs for the very large set of SSRs distributed throughout the genome would benefit pepper research and the breeding community in the future.
Results and Discussion
Content of SSR motifs in the pepper nuclear genome and its comparison with related plant species
In this study, a total of 11 genomes (~9.18 Gb of sequence in total) from various species and types were collected from different databases and submitted for SSR motif identification (Supplementary Table S2). To our knowledge, unlike for Arabidopsis66, rice67 and cucumber27, there are no reports on the characterization of SSR motifs on the genome-wide level for pepper, tomato and potato. Here, the overall content of SSR motifs (1–6 bp) wit ≥4 repeats and a minimum of 10 bp length in all of the above genomic sequences was recorded (Table 1). Based on our search criteria, a total of 876,580 and 859,515 SSR loci with 136,857 and 123,281 presented in compound formation were identified in the whole genomes of Zunla-1 (N1) and Chiltepin (N2), respectively. The holistic view of their distribution patterns in the Zunla-1 and Chiltepin nuclear genomes is shown in Fig. 1 and Supplementary Figure S1, respectively. The number of SSRs identified in pepper nuclear genomes (average is 868,047.50) was approximately four times higher than the numbers found for other two Solanaceae species, tomato (235,398) and potato (241,570). If we consider into account all six species, the number of identified SSR motifs is significantly associated with genome size (R2 = 0.996, P < 0.01). However, despite possessing the largest genome size, pepper values for SSR density (SSRs/Mb) were the lowest (260.58 and 243.62 for N1 and N2, respectively) when compared to the other five species, although the two Solanaceae species, tomato and potato, showed comparable values (285.70 and 312.50 SSRs/Mb, respectively) (Table 1). In contrast, the highest density was found in cucumber (654.55 SSRs/Mb), followed by Arabidopsis (529.15 SSRs/Mb) and rice (527.62 SSRs/Mb). A similarly high SSR density was reported for another cucumber (Gy14) genome27. Nevertheless, the results from this study agreed with the trend that high SSR density is often observed in small genomes19. In addition, the cumulative sequence length of pepper SSR loci was approximately 16.81 Mbp and 15.90 Mbp, accounting for 0.50% and 0.45% of the assembled genomes of Zunla-1 and Chiltepin, respectively. This ratio was similar in tomato (0.59%) and potato (0.61%) but was higher than 1% in both cucumber (1.32%) and rice (1.12%) (Table 1).
Table 1. Frequency of SSR motifs (1–6 bp) in all genomes investigated in the present study.
| SSR type | Genome |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | M1 | M2 | C1 | C2 | Tomato | Potato | Cucumber | Arabidopsis | Rice | |
| Mono- | 377,376 | 375,358 | 22 | 23 | 25 | 27 | 86,922 | 121,911 | 69,085 | 34,751 | 64,831 |
| Di- | 198,033 | 185,688 | 7 | 8 | — | — | 62,486 | 41,875 | 25,117 | 9,375 | 37,370 |
| Tri- | 254,288 | 251,631 | 15 | 15 | 3 | 3 | 71,356 | 65,020 | 27,799 | 17,469 | 82,171 |
| Tetra- | 33,639 | 33,645 | — | 1 | 1 | 1 | 11,291 | 7,595 | 5,176 | 922 | 8,210 |
| Penta- | 6,663 | 6,507 | — | — | — | — | 1,761 | 3,122 | 1,665 | 351 | 2,958 |
| Hexa- | 6,581 | 6,686 | — | — | — | — | 1,582 | 2,047 | 1,105 | 178 | 1,393 |
| Total number | 876,580 | 859,515 | 44 | 47 | 29 | 31 | 235,398 | 241,570 | 129,947 | 63,046 | 196,933 |
| Compounda | 136,857 | 123,281 | 2 | 2 | 1 | 1 | 51,415 | 31,706 | 16,716 | 6,929 | 35,197 |
| Cumulative (%)b | 0.50 | 0.45 | 0.11 | 0.12 | 0.23 | 0.26 | 0.59 | 0.61 | 1.32 | 0.96 | 1.12 |
| Densityc | 260.58 | 243.62 | 86.71 | 91.88 | 184.97 | 197.90 | 285.70 | 312.50 | 654.55 | 529.15 | 527.62 |
aNumber of SSRs present in compound formation.
bThe ratio of cumulative sequence length of all SSR motifs to genome size.
cNumber of SSRs present in one million bases (SSRs/Mbp).
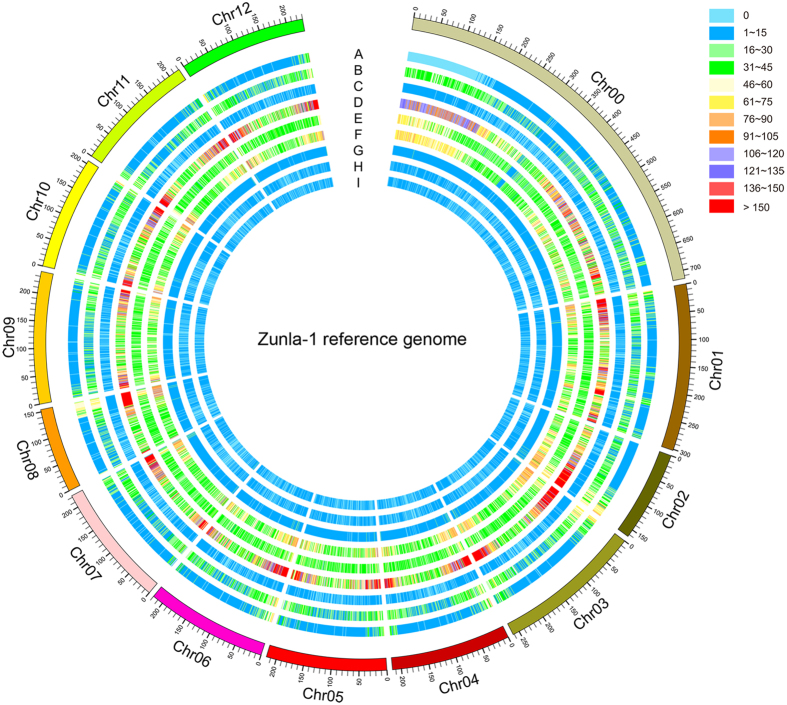
Figure 1. Overview of SSR distribution in the Zunla-1 reference genome.
A total of 876,580 SSR loci with 136,857 present in compound formation (C and C*) that form into 739,723 SSR units were identified in the Zunla-1 reference genome. The various numbers of SSR units and protein coding genes in each window size of 1000 kb were used for drawing this picture and are shown with different colours. Track A shows the gene density; tracks B to I refer to the C, C*, Mono-, Di-, Tri-, Tetra-, Penta-, Hexa- types, respectively.
Distribution of SSR motifs in pepper organelle genomes
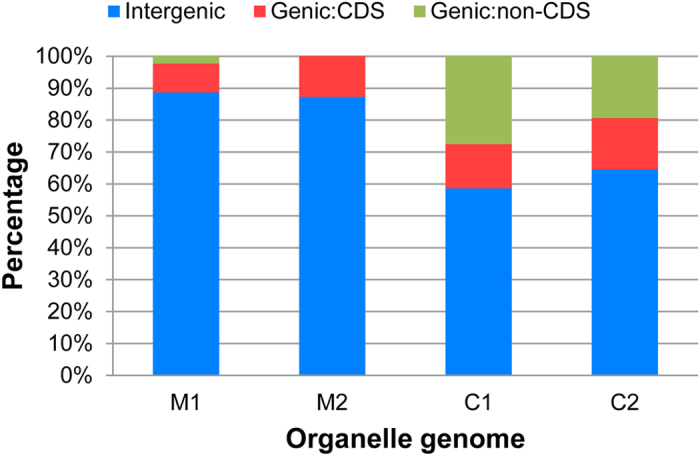
Similar to the results for the nuclear genomes, SSR motifs also have a wide range of distribution in the organelle genomes of various species, and they have become valuable resources for monitoring genetic flow and genetic diversity assessment at the cytoplasmic level31,32. Thus, to characterize the SSR distribution in pepper non-nuclear genomes, a total of four pepper organelle genomes were downloaded and analysed for SSR motifs. The analysis identified a total of 91 and 60 SSRs for pepper mitochondrial and chloroplast genomes, respectively (Table 1). The number of SSR loci found in each type of organelle genome was similar: 44 (M1) and 47 (M2) for the mitochondrial genomes (average 45.50) and 29 (C1) and 31 (C2) for chloroplast genomes (average 30.00) However, the overall density of SSRs in mitochondrial genomes was significantly lower than that of the pepper chloroplast (P = 0.000) and all the nuclear genomes (P = 0.001). In addition, the distribution of the mitochondrial SSRs (mtSSRs) across the genome was more even than what was observed for the chloroplast (hereafter called cpSSRs). Nevertheless, clustering of SSRs was also observed (Figs 2 and 3, Supplementary Figures S2 and S3). Notably, SSRs predominate in the long single copy (LSC) region of the chloroplast (Fig. 3 and Supplementary Figure S3). In addition, the pattern of SSR distribution in the C1 genome showed very high comparability with its wild type relative (C2) with the exception for the lack of two SSRs (*23 and #26 in Supplementary Figure S3). Furthermore, based on the current annotations, over 85% of the total mtSSRs were located in the intergenic region, whereas this value declined to no more than 65% for the cpSSRs (Fig. 4). Nevertheless, the number of SSRs located in the coding sequence (CDS) region was similar (P > 0.05). A significant discrepancy in the proportion of non-CDS (mainly intron)-located cpSSRs was still highlighted (Fig. 4). Intriguingly, all of the SSRs (#25 and #26 in Fig. 3 and #26, #27 and #28 in Supplementary Figure S3) from the short single copy (SSC) region were located in the genic region. In summary, the results obtained here will also provide basic resources for the next-step applications in the evaluation of cytoplasmic genetic differences in the pepper68.
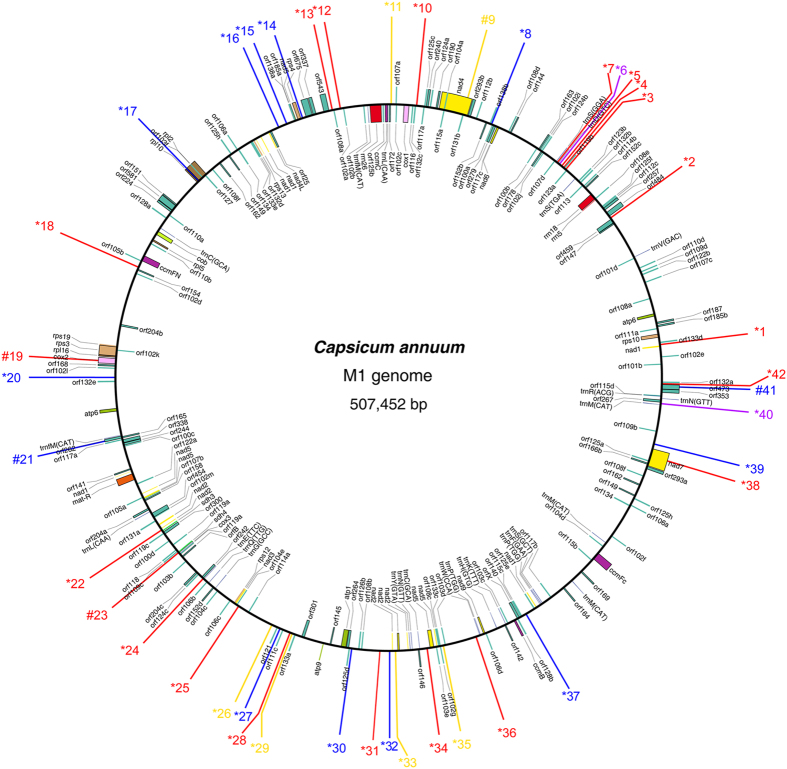
Figure 2. Overview of SSR distribution in pepper M1 mitochondrial genome.
The M1 genome refers to the mitochondrial genome of pepper line ‘FS4401’ (Capsicum annuum). Perfect SSRs with 1, 2, 3 and 4 bp length of motif are represented by red, yellow, blue and green lines, respectively. Compound SSRs are shown with purple lines. Numbers with * and # in the front means that the corresponding SSRs were located in the intergenic and genic region, respectively.
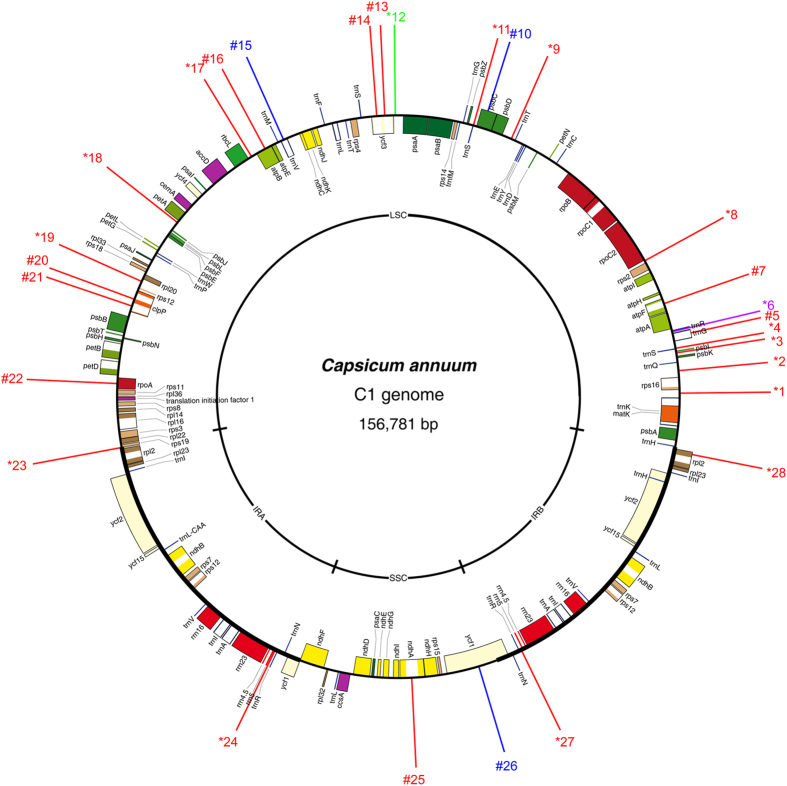
Figure 3. Overview of SSR distribution in pepper C1 chloroplast genome.
The C1 genome refers to the chloroplast genome of the pepper line ‘FS4401’ (Capsicum annuum). Perfect SSRs with 1, 3 and 4 bp length of motif are represented by red, blue and green lines, respectively. Compound SSRs are shown with purple lines. Numbers with * and # in the front means that the corresponding SSRs were located in the intergenic and genic region, respectively.
Figure 4. The relative proportion of SSR motifs in organelle genomes with different locations.
Characterization of SSR motifs by different length and repeat
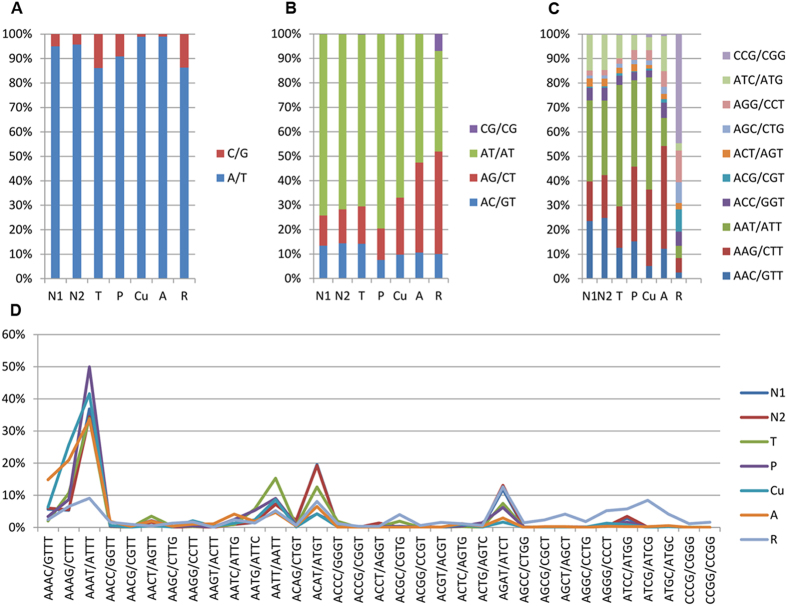
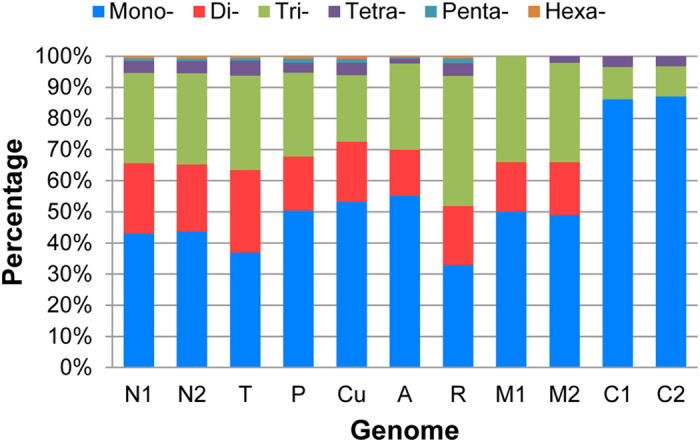
Data from many organisms indicated that SSR distribution across the genome is non-random19,69. Based on our search criteria, the sum of Mononucleotides (Mono-), Dinucleotides (Di-) and Trinucleotides (Tri-) accounted for the clear majority (>90%) of total SSRs for all genomes investigated in this study (Fig. 5). Of these, the Mono- was the most popular type for pepper nuclear genomes, followed by Tri-, Di-, Tetranucleotide (Tetra-), Pentanucleotide (Penta-) and Hexanucleotide (Hexa-) types. This pattern of distribution was in accordance with that found in tomato, potato, cucumber and Arabidopsis but not with the monocot plant (rice), in which Tri- was the main type (Fig. 5). With regard to the pepper organelle genomes, the frequency of Mono- reached higher than 80% in the pepper chloroplast whereas Tetra- was the least frequent type with percentages of 3.57% and 3.33% for C1 and C2, respectively. Furthermore, another signature of the chloroplast was the absence of Di- compared to the mitochondrial genomes. In addition, statistical results indicated an obvious trend in which the frequency of SSRs decreased with increasing repeat number regardless of species and motif length (Supplementary Table S3, Supplementary Figures S4 and S5). For example, the number of SSRs with repeat number ≤10 accounted for more than half of the total, and this rate dramatically declines to less than 4% when the repeat number is more than 20.
Figure 5. The relative proportion of SSR motifs with different lengths (1–6 bp) in eleven investigated genomes.
Characterization of SSR motifs by classified type
If the complementary sequence is taken into consideration, a total of 456 kinds of classified SSR motifs were detected in all genomes investigated in the present study (Table 2). The detailed frequency of classified SSR motif (1–6 bp) in different genomes is shown in the Supplementary Table S4. Out of the total of 456 kinds, 369 and 363 different motifs were identified in Zunla-1(N1) and Chiltepin (N2), respectively. Overall, we found 387 kinds of classified SSR motifs in the pepper genome. Additionally, all of the possible base combinations of Mono- (the number is 2), Di- (4) and Tri- (10) were detected in both pepper nuclear genomes as well as in all other species. However, there were inter-specific differences in both number and motif type with increasing motif length, such as Tetra-, Penta- and Hexa-. For example, 2 (CCCG/CGGG and CCGG/CCGG), 1 (CCGG/CCGG) and 6 (ACCT/AGGT, ACGC/CGTG, AGCC/CTGG, AGGC/CCTG, CCCG/CGGG and CCGG/CCGG) quadruplet motifs were not represented in potato, cucumber and Arabidopsis, respectively. The difference was more apparent for Penta- and Hexa-. As a result, we isolated a set of species-specific SSR motifs through integrative comparison, of which 15 motifs were found to be specific for pepper (Fig. 6, Supplementary Table S4). It is also worth mentioning that 17 species-specific SSR motifs were identified in rice (Fig. 6, Supplementary Table S4). Whether these SSR motifs play roles in the evolution of plants is unclear but we at least suggest the idea that SSR distribution and frequency are unequal27.
Table 2. Number of classified SSR motif types in different genomes.
| Genome | Mono- | Di- | Tri- | Tetra- | Penta- | Hexa- | Total |
|---|---|---|---|---|---|---|---|
| N1 | 2 | 4 | 10 | 33 | 92 | 228 | 369 |
| N2 | 2 | 4 | 10 | 33 | 89 | 225 | 363 |
| Tomato | 2 | 4 | 10 | 33 | 69 | 160 | 278 |
| Potato | 2 | 4 | 10 | 31 | 82 | 190 | 319 |
| Cucumber | 2 | 4 | 10 | 32 | 67 | 193 | 308 |
| Arabidopsis | 2 | 4 | 10 | 27 | 47 | 83 | 173 |
| Rice | 2 | 4 | 10 | 33 | 95 | 214 | 358 |
| M1 | 1 | 3 | 4 | 0 | — | — | — |
| M2 | 1 | 3 | 4 | 1 | — | — | — |
| C1 | 1 | — | 2 | 1 | — | — | — |
| C2 | 1 | — | 2 | 1 | — | — | — |
| Total | 2 | 4 | 10 | 33 | 101 | 306 | 456 |
“–” means not detected.
Figure 6. Identification of species-specific SSR motifs for the six species investigated in the present study.
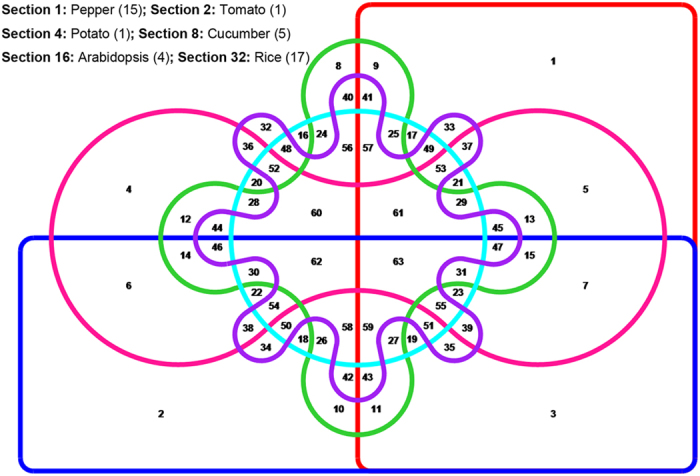
A set of 387 kinds of classified SSR motifs that were identified from the combination of Zunla-1(N1) and Chiltepin (N2) was used for the present analysis. Sets with red, blue, pink, green, light blue and purple colours represents pepper, tomato, potato, cucumber, Arabidopsis and rice, respectively. Intersection numbers are shown in the disjoint sets, and the total number of specific motifs for each species is shown with brackets in the upper left corner of the picture.
In terms of the distribution of different motifs, briefly, the A/T motif not only accounted for approximately 95% of the total Mono- type in the pepper (Fig. 7A), but was also found to be the most frequent motif across all the genomes examined. In addition, the AT/AT motif, as the most predominant duplets, accounted for over 70% of the total Di- motif in pepper, tomato and potato. However, this pattern did not apply for the monocot plant rice, in which the most frequent Di- motif was AG/CT. Moreover, compared to other species such as pepper, a significantly higher percentage of CG/CG was observed in rice (Fig. 7B). Then, of the 10 different triplets, AAT/ATT was overrepresented in pepper, tomato and potato as well as in cucumber, whereas the major Tri- motif in Arabidopsis was AAG/CTT. Similar to the Di- motif, the GC-rich motif CCG/CGG was dramatically predominant in rice, indicating a significant difference from the dicot plant species (Fig. 7C), which was also revealed in a previous study27. With regard to the Tetra- motif, AAAT/ATTT was the most frequent motif in all of the genomes with the exception of rice, in which AGAT/ATCT accounted for the highest percentage (12.62%) (Fig. 7D). Additionally, AAAAT/ATTTT and AGATAT/ATATCT were the major Penta- and Hexa- motifs in pepper, respectively. The former also prevailed among Penta- motifs in tomato, potato and Arabidopsis but not in cucumber and rice, where AAAAG/CTTTT was overrepresented. Lastly, AACAAT/ATTGTT, AAGAGG/CCTCTT, AAAAAG/CTTTTT, AAAAAT/ATTTTT (equal in number to ACCACG/CGTGGT), and ACATAT/ATATGT predominated in the Hexa- motif of tomato, potato, cucumber, Arabidopsis and rice, respectively.
Figure 7. The percentage of classified SSR motifs within the corresponding type with different lengths.
(A) Mono-; (B) Di-; (C) Tri-; (D) Tetra-.
Identification of unique SSR primer pairs in the Zunla-1 reference genome
In addition to understanding the characteristics of distribution, the development of molecular markers for pepper based on these SSR loci was one of the most important objectives in this study. For the purpose of convenient and efficient primer pair isolation, the compound SSRs are handled as a ‘unit’ in this section unless otherwise stated. As a result, in the Zunla-1 genome, a total of 739,723 SSR units were identified on all 13 chromosomes, including the pseudo-chromosome P0 (Table 3). The number of SSR units on each chromosome (P0-P12) ranged from 34,410 (chromosome P8) to 147,775 (chromosome P0), with an average number of 56,901.77, and the highest average density was observed on chromosome P2 (Supplementary Figure S6). Relatively, the SSR density was higher on both ends of all 12 chromosomes, which is in accordance with the distribution of protein coding genes (Fig. 1).
Table 3. Development and utility assessment of unique SSR primer pairs in the Zunla-1 genome.
| Total | Size filtering (%b) | Primer pairs isolation |
Utility assessment |
Average PIC | |||||
|---|---|---|---|---|---|---|---|---|---|
| SSR type | Total (%c) | Unique (%d) | Selected | Specific | Polymorphic (%e) | ||||
| Perfect | Mono- | 302,209 | 139,351 (46.11%) | 132,118 (94.81%) | 32,804 (23.54%) | 20 | 11 | 7 (35.00%) | 0.21 |
| Di- | 132,938 | 132,802 (99.90%) | 123,621 (93.09%) | 26,967 (20.31%) | 20 | 8 | 5 (25.00%) | 0.22 | |
| Tri- | 182,359 | 181,411 (99.48%) | 175,945 (96.99%) | 38,195 (21.05%) | 20 | 15 | 4 (20.00%) | 0.16 | |
| Tetra- | 18,958 | 18,910 (99.75%) | 17,935 (94.84%) | 4,192 (22.17%) | 20 | 12 | 8 (40.00%) | 0.21 | |
| Penta- | 5,065 | 5,018 (99.07%) | 4,839 (96.43%) | 1,125 (22.42%) | 20 | 11 | 11 (55.00%) | 0.23 | |
| Hexa- | 4,180 | 4,122 (98.61%) | 3,793 (92.02%) | 836 (20.28%) | 20 | 12 | 12 (60.00%) | 0.30 | |
| C | 88,729 | 40,559 (45.71%) | 36,789 (90.70%) | 8,647 (21.32%) | 20 | 10 | 10 (50.00%) | 0.27 | |
| C*a | 5,285 | 3,390 (64.14%) | 3,119 (92.01%) | 734 (21.65%) | 20 | 9 | 8 (40.00%) | 0.30 | |
| Total | 739,723 | 525,563 (71.05%) | 498,159 (94.79%) | 113,500 (21.60%) | 160 | 88 | 65 (40.63%) | 0.25 | |
aC with an asterisk means that the number of bases interrupting two adjacent SSR motifs within a compound microsatellite is 0.
bThe ratio of size filtering to the total number of SSR units.
cThe ratio of successful isolation to the remainder after size filtering.
dThe ratio of unique primer pairs to total primer pairs.
eThe ratio of polymorphic number to number selected.
However, similar to the maize genome24,70, the pepper genome (Zunla-1) consists of as high as 80.90% of repetitive sequences35, therefore, it is better to identify in silico the unique SSR loci set before preparing the primer pairs for practical evaluation. First, after the size filtering standard described in the Methods section, a total of 525,563 (71.05%) SSR units, including 481,614 perfect SSRs and 43,949 compound SSRs were selected for primer pair isolation (Table 3). Of these, except for the Mono- type, the vast majority (over 98%) of perfect SSRs were kept. In contrast, more than half of the compound SSRs were filtered out because they were too long for primer design (Table 3). Then, using the Primer3 software, a total of 498,159 (94.79%) primer pairs were successfully isolated and subsequently used to align back to the Zunla-1 reference genome. Finally, a large set of 113,500 (21.60%) in silico unique primer pairs were obtained (Supplementary Tables S5–S9). This local database of SSR primers will undoubtedly serve as an abundant mine for molecular marker development in the pepper.
Experimental validation of the SSR primers with a collection of pepper genotypes
To preliminarily test the usability of these SSR primers, a random set of 160 primer pairs (20 for each type) was intentionally selected from chromosome P0 to be synthesized and used for screening polymorphisms among a wide collection of 21 pepper genotypes (Table 3). These marker candidates were specially selected from chromosome P0 because they can not only to be used for possible versatile applications such as genetic diversity analysis, gene tagging and so on, but they also may be useful for anchoring some of the scaffolds that have not yet been assigned to current pepper chromosome buildings46. Of the initial 160 candidates, 88 (55.00%) primer pairs can specifically direct the amplification of one or two main bands (Supplementary Figure S7). In addition, out of those 88 primer pairs, 65 (73.86%) exhibited polymorphisms among the 21 pepper genotypes with the alleles per SSR marker ranging from 2 to 6 and the polymorphic information content (PIC) value for each SSR marker varying from 0.05 to 0.64 (Supplementary Table S10). Specifically, the results showed that the polymorphic rate was highest in Hexa-, followed by Penta- and C type, whereas the Tri- type exhibited the lowest polymorphic rate (Table 3). In addition, the PIC value was not significantly different between various SSR types (F = 0.59, P = 0.77). This information will provide a useful reference for the selection and design of further primer pairs to be tested.
A UPGMA-phylogenetic tree (Fig. 8) was constructed based on the 65 polymorphic SSR markers. The results showed that the SSR markers developed in this study were sufficient to classify the 21 pepper lines into 3 major groups that corresponded to the domesticated species taxonomy of the Capsicum genus. For example, the YNXML line (C. frutescens) showed a closer relationship to C. annuum than to C. baccatum and C. chinense Jacq. This result is consistent with previous studies71,72 . In addition, the semi-wild type Chiltepin (C. annuum var. glabriusculum) was distinguishable from the other C. annuum lines. Finally, it is worth noting that the genetic relationship of 11 of the 21 pepper lines revealed by these newly developed SSR markers was comparable to our previous results inferred from the genome-wide SNPs35. In summary, all experimental results indicated that the SSR information obtained in this study can be useful for SSR markers development in Capsicum.
Figure 8. Phylogenetic tree of 21 pepper lines based on 65 polymorphic SSR markers.
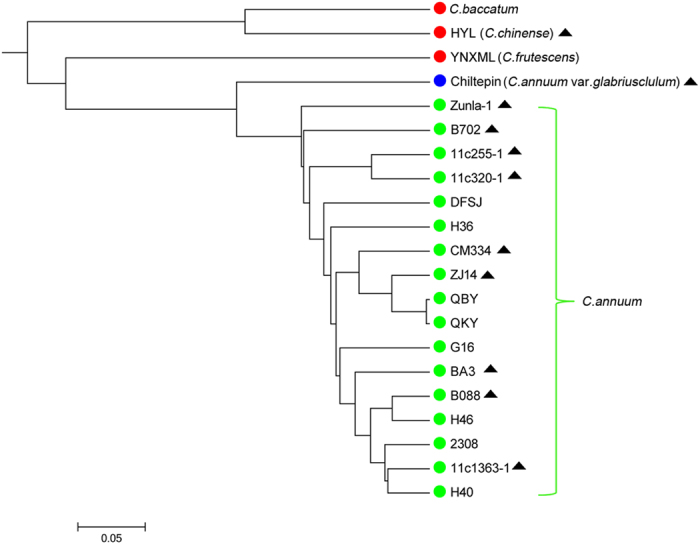
Genetic relationships of 11 pepper lines that were previously studied based on genome-wide SNP markers are marked with a solid black triangle suffix.
Materials and Methods
Plant materials and DNA extraction
A panel of 21 pepper genotypes (Supplementary Table S1) representing four cultivated species of the Capsicum genus, i.e., C. annuum, C. chinense Jacq., C. baccatum, C. frutescens and the semi-wild type, Chiltepin (C. annuum var. glabriusculum), were used to test potential application of the identified SSRs in this study. Of these, 18 genotypes were collected from a total of seven provinces/regions in China. The semi-wild type Chiltepin was collected from Queretaro in Mexico. The seeds of CM334 (Criollo de Morelos 334) and the local landrace C. baccatum were kindly provided by Dr. Paul W. Bosland from the Chile Pepper Institute, New Mexico State University, USA and Dr. Salvador Montes-Hernández from INIFAP-Mexico, respectively. This panel of genotypes also shows variability in fruit size, orientation, colour, pungency, and other characters. Genomic DNA was extracted from young leaves of each genotype according to the modified CTAB method73.
Collection of genomic sequences from different sources
Detailed information on the sources of different genomic sequences is summarized in Supplementary Table S2. Briefly, the complete genome sequences of Zunla-1 and its wild relative Chiltepin were downloaded from the Pepper Genome Database Release 2.0 (http://peppersequence.genomics.cn/page/species/index.jsp). Recently, two complete mitochondrial genomes of pepper from the respective cytoplasmic male sterility (CMS) line ‘FS4401’ and fertile line ‘Jeju’ were published59. Furthermore, the complete chloroplast genome sequence of ‘FS4401’ was previously reported61. With the addition of another recently published chloroplast genome60, a total of six different genomes, including two from each nucleus, mitochondria and chloroplast, were collected for further analysis. With the aim of comparison, whole genomic sequences of an additional five plant species, including tomato (Solanum lycopersicum), potato (Solanum tuberosum), cucumber (Cucumis sativus), Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), were also collected from the related public database (Supplementary Table S2).
Identification and characterization of SSR loci in different genomes
The software MISA (http://pgrc.ipk-gatersleben.de/misa/) was used for SSR identification, and both perfect and compound SSRs were recorded. The detailed search criteria were as follows: (1) ten repeat units for mononucleotide (Mono-) repeats, six for dinucleotide (Di-) repeats, four for trinucleotide (Tri-), tetranucleotide (Tetra-), pentanucleotide (Penta-) and hexanucleotide (Hexa-) repeats; (2) a compound microsatellite was defined if the number of bases between two adjacent microsatellites was ≤100. The circular framework maps for four organelle genomes were drawn with the online tool, OrganellarGenomeDRAW74. The distribution of SSRs in genic and intergenic regions was determined based on the information of genome sequence annotations available from NCBI GenBank. The Venn diagram investigational tool VENNTURE75 was adopted to identify the species-specific SSR motifs.
Identification of unique SSR primer pairs in the Zunla-1 reference genome
A homemade workflow was applied to identify the unique SSR loci in Zunla-1 genome due to the high percentage (80.90%) of repetitive sequences in the pepper genome revealed by a previous study35. At the same time, for the purpose of convenient and efficient primer pair isolation, the compound SSRs were handled as a ‘unit’ in the subsequent identification process unless otherwise stated. The primer modelling software Primer3 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html) accompanied by Perl scripts p3_in.pl and p3_out.pl (http://pgrc.ipk-gatersleben.de/misa/primer3.html) was used for the design of primers with SSR searching results as an input.
First, because the size of some SSR units was too large to obtain the ideal size (100–500 bp) of amplification products, they were filtered by 1) Mono-unit with repeats ≤ 10; 2) other types with unit size ≤60. Then, we constructed new sequence files by expanding 500 bp based on the Zunla-1 genome on both sides of the SSR (central SSR). This process greatly decreased the reference length in primer design process. The arguments used for primer design were as follow: 1) product size: 100–500 bp; 2) primer length: 18–25 bp with optimum: 20 bp; 3) annealing temperature: 57–62 °C; 4) number of returns: 2. Second, only primers with the central SSR as a target were reserved in the process. Finally, blast was used to align the forward primer and reverse primer to the Zunla-1 reference. Unique primer pairs were defined as cases in which both forward primers and reverse primers were uniquely aligned to the genome, or in which the e-value of primary alignment was 5 times larger than that of the secondary alignment.
Genotyping of pepper lines with a random set of selective unique SSR primers
To evaluate the utility of the above unique SSR primer pairs in Capsicum, a randomly selected set of 160 primer pairs was chosen to genotype the pepper lines with different sources. PCR amplification was conducted as follows: a final volume of 20 μL PCR mixture including 10 ng template DNA, 100 μM of each dNTP, 1.5 μM of each primer, 1 × reaction buffer (including Mg2+) and 1.0 unit of Taq DNA polymerase (Takara) was used for PCR reaction by an initial 3 min at 94 °C; 34 cycles of 45 s at 94 °C, 30 s at 55–58 °C, and 30 s at 72 °C, and a final 5 min at 72 °C. Then, the PCR products were electrophoresed on 6% polyacrylamide gels. A silver staining method was used to visualize bands in the gels.
Construction of a phylogenetic tree based on polymorphic SSR markers
Based on the genotypic data, the marker analysis software PowerMarker v3.2576 was used to calculate the allele number and polymorphism information content (PIC) value for each polymorphic marker. Then, a phylogenetic tree was constructed based on the Nei1983’s genetic distance by the unweighted pair group method with arithmetic averages (UPGMA). Finally, the dendrogram was viewed and plotted in MEGA 6.0 software77.
Additional Information
How to cite this article: Cheng, J. et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 6, 18919; doi: 10.1038/srep18919 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Guangdong Natural Science Foundation of China (S2011030001410, 2014A030313593), the National Natural Science Foundation of China (31372076), the National High Technology Research and Development Program (“863” Program) of China (2012AA100103), the Zunyi City Natural Science Foundation of China (No. 201201), the Guizhou Province and Zunyi City Science and Technology Cooperation Project of China (No. 201307) and the Zunyi County Technology Cooperation Project (SSX201407).
Footnotes
Author Contributions J.C., Z.Z. and K.H. conceived and designed the experiments; J.C., B.L. and J.C. performed the experiments; J.C., Z.Z., C.Q., Z.W., D.L., T.-S. and X.L. analysed the data; J.C., R.F., R.-B., S.L. and K.H. wrote the manuscript. All authors reviewed the manuscript.
References
- Tautz D. & Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res (12)10, 4127–38 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J., Guo X. & Shah A. Simple sequence repeats in prokaryotic genomes. Proc Natl Acad Sci USA (104)20, 8472–7 (2007). doi: 10.1073/pnas.0702412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L.M. Bouncing off microsatellites. Nat Genet (2)1, 8–9 (1992). doi: 10.1038/ng0992-8. [DOI] [PubMed] [Google Scholar]
- Zalapa J.E. et al. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am J Bot (99)2, 193–208 (2012). doi: 10.3732/ajb.1100394. [DOI] [PubMed] [Google Scholar]
- Powell W., Machray G.C. & Provan J. Polymorphism revealed by simple sequence repeats. Trends in Plant Science (1)7, 215–222 (1996). doi: 10.1016/1360-1385(96)86898-1. [DOI] [Google Scholar]
- Rongwen J., Akkaya M.S., Bhagwat A.A., Lavi U. & Cregan P.B. The use of microsatellite DNA markers for soybean genotype identification. Theor Appl Genet (90)1, 43–48 (1995). [DOI] [PubMed] [Google Scholar]
- Provan J., Kumar A., Shepherd L., Powell W. & Waugh R. Analysis of intra-specific somatic hybrids of potato (Solanum tuberosum) using simple sequence repeats. Plant Cell Rep (16)3–4, 196–9 (1996). doi: 10.1007/BF01890866. [DOI] [PubMed] [Google Scholar]
- Goldstein D.B., Linares A.R., Cavalli-Sforza L.L. & Feldman M.W. An evaluation of genetic distances for use with microsatellite loci. Genetics (139)1, 463–471 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Construction of a high-density simple sequence repeat consensus genetic map for Pear (Pyrus spp.). Plant Mol Biol Rep (33)2, 316–325 (2014). doi: 10.1007/s11105-014-0745-x. [DOI] [Google Scholar]
- Yu Y., Saghai Maroof M., Buss G., Maughan P. & Tolin S. RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology (84)1, 60–64 (1994). [Google Scholar]
- Moe A.M. & Weiblen G.D. Development and characterization of microsatellite loci in dioecious figs (Ficus, Moraceae). Am J Bot (98)2, e25–7 (2011). doi: 10.3732/ajb.1000412. [DOI] [PubMed] [Google Scholar]
- Wang H.L. et al. Developing conversed microsatellite markers and their implications in evolutionary analysis of the Bemisia tabaci complex. Sci. Rep. (4) (2014). doi: 10.1038/srep06351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell M.C., Frere C.H., Prentis P.J., Lambrides C.J. & Godwin I.D. Characterization and multiplexing of EST-SSR primers in Cynodon (Poaceae) species. Am J Bot (97)10, e99–e101 (2010). doi: 10.3732/ajb.1000254. [DOI] [PubMed] [Google Scholar]
- Lioi L., Galasso I. & Havey M. Development of genomic simple sequence repeat markers from an enriched genomic library of grass pea (Lathyrus sativus L.). Plant Breeding (132)6, 649–653 (2013). doi: 10.1111/pbr.12093. [DOI] [Google Scholar]
- Rico C. et al. Combining next-generation sequencing and online databases for microsatellite development in non-model organisms. Sci Rep (3), 3376 (2013). doi: 10.1038/srep03376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J.W. et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet (12)7, 499–510 (2011). doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Kumar S., Shah N., Garg V. & Bhatia S. Large scale in-silico identification and characterization of simple sequence repeats (SSRs) from de novo assembled transcriptome of Catharanthus roseus (L.) G. Don. Plant Cell Rep (33)6, 905–18 (2014). doi: 10.1007/s00299-014-1569-8. [DOI] [PubMed] [Google Scholar]
- Li Y.C., Korol A.B., Fahima T., Beiles A. & Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol (11)12, 2453–65 (2002). [DOI] [PubMed] [Google Scholar]
- Morgante M., Hanafey M. & Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet (30)2, 194–200 (2002). doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Kelkar Y.D., Tyekucheva S., Chiaromonte F. & Makova K.D. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res (18)1, 30–8 (2008). doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Li Q., Yu H. & Kong L. Genome-wide analysis of simple sequence repeats in marine animals-a comparative approach. Mar Biotechnol (NY) (16)5, 604–19 (2014). doi: 10.1007/s10126-014-9580-1. [DOI] [PubMed] [Google Scholar]
- Behura S.K. & Severson D.W. Motif mismatches in microsatellites: insights from genome-wide investigation among 20 insect species. DNA Res (22)1, 29–38 (2015). doi: 10.1093/dnares/dsu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J. et al. Genome-wide analysis of simple sequence repeats in the model medicinal mushroom Ganoderma lucidum. Gene (512)2, 331–6 (2013). doi: 10.1016/j.gene.2012.09.127. [DOI] [PubMed] [Google Scholar]
- Qu J. & Liu J. A genome-wide analysis of simple sequence repeats in maize and the development of polymorphism markers from next-generation sequence data. BMC Res Notes (6), 403 (2013). doi: 10.1186/1756-0500-6-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.S. et al. Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. (5) (2015). doi: 10.1038/srep08018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G. et al. Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res (20)2, 197–207 (2013). doi: 10.1093/dnares/dst002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavagnaro P.F. et al. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics (11)1, 569 (2010). doi: 10.1186/1471-2164-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Ge T., Li Y. & Hou X. Genome-wide identification of SSR and SNP markers from the non-heading Chinese cabbage for comparative genomic analyses. BMC Genomics (16)1, 328 (2015). doi: 10.1186/s12864-015-1534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W. et al. Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr Biol (5)9, 1023–9 (1995). [DOI] [PubMed] [Google Scholar]
- Delplancke M. et al. Gene flow among wild and domesticated almond species: insights from chloroplast and nuclear markers. Evol Appl (5)4, 317–329 (2012). doi: 10.1111/j.1752-4571.2011.00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.L. et al. Development of chloroplast simple sequence repeats (cpSSRs) for the intraspecific study of Gracilaria tenuistipitata (Gracilariales, Rhodophyta) from different populations. BMC Res Notes (7), 77 (2014). doi: 10.1186/1756-0500-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W., Morgante M., McDevitt R., Vendramin G.G. & Rafalski J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc Natl Acad Sci USA (92)17, 7759–63 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.X., Zhu R.L. & Liu Y. Simple sequence repeats in bryophyte mitochondrial genomes. Mitochondrial DNA (Early Online), 1–7 (2014). doi: 10.3109/19401736.2014.880889. [DOI] [PubMed] [Google Scholar]
- Rajendrakumar P., Biswal A.K., Balachandran S.M., Srinivasarao K. & Sundaram R.M. Simple sequence repeats in organellar genomes of rice: frequency and distribution in genic and intergenic regions. Bioinformatics (23)1, 1–4 (2007). doi: 10.1093/bioinformatics/btl547. [DOI] [PubMed] [Google Scholar]
- Qin C. et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci USA (111)14, 5135–40 (2014). doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnes S.C. et al. Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population. Genome (56)1, 61–74 (2013). doi: 10.1139/gen-2012-0083. [DOI] [PubMed] [Google Scholar]
- Holdsworth W.L. & Mazourek M. Development of user-friendly markers for the pvr1 and Bs3 disease resistance genes in pepper. Mol Breed (35)1, 1–5 (2015). doi: 10.1007/s11032-015-0260-2. [DOI] [Google Scholar]
- Devran Z., Kahveci E., Ozkaynak E., Studholme D.J. & Tor M. Development of molecular markers tightly linked to gene in pepper using next-generation sequencing. Mol Breed (35)4, 101 (2015). doi: 10.1007/s11032-015-0294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Nahm S.H., Kim Y.M. & Kim B.D. Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet (108)4, 619–27 (2004). doi: 10.1007/s00122-003-1467-x. [DOI] [PubMed] [Google Scholar]
- Hill T.A. et al. Characterization of Capsicum annuum genetic diversity and population structure based on parallel polymorphism discovery with a 30K unigene Pepper GeneChip. PLoS One (8)2, e56200 (2013). doi: 10.1371/journal.pone.0056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.K., Park S.W., Liu W.Y. & Kang B.C. Discovery of single nucleotide polymorphism in Capsicum and SNP markers for cultivar identification. Euphytica (175)1, 91–107 (2010). doi: 10.1007/s10681-010-0191-2. [DOI] [Google Scholar]
- Rodriguez J.M., Berke T., Engle L. & Nienhuis J. Variation among and within Capsicum species revealed by RAPD markers. Theor Appl Genet (99)1-2, 147–156 (1999). doi: 10.1007/s001220051219. [DOI] [Google Scholar]
- Paran I., Aftergoot E. & Shifriss C. Variation in Capsicum annuum revealed by RAPD and AFLP markers. Euphytica (99)3, 167–173 (1998). [Google Scholar]
- Tanksley S.D., Bernatzky R., Lapitan N.L. & Prince J.P. Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci USA (85)17, 6419–6423 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. et al. Construction of an interspecific genetic map based on InDel and SSR for mapping the QTLs affecting the initiation of flower primordia in pepper (Capsicum spp.). PLoS One (10)3, e0119389 (2015). doi: 10.1371/journal.pone.0119389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. An InDel-based linkage map of hot pepper (Capsicum annuum). Mol Breed (35)1, 1–10 (2015). doi: 10.1007/s11032-015-0219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T. et al. Development of simple sequence repeat markers and construction of a high-density linkage map of Capsicum annuum. Mol Breed (31)4, 909–920 (2013). doi: 10.1007/s11032-013-9844-x. [DOI] [Google Scholar]
- Nagy I., Stagel A., Sasvari Z., Roder M. & Ganal M. Development, characterization, and transferability to other Solanaceae of microsatellite markers in pepper (Capsicum annuum L.). Genome (50)7, 668–88 (2007). doi: 10.1139/g07-047. [DOI] [PubMed] [Google Scholar]
- Minamiyama Y., Tsuro M. & Hirai M. An SSR-based linkage map of Capsicum annuum. Mol Breed (18)2, 157–169 (2006). doi: 10.1007/s11032-006-9024-3. [DOI] [Google Scholar]
- Sanwen H. et al. Development of pepper SSR markers from sequence databases. Euphytica (117)2, 163–167 (2001). doi: 10.1023/a:1004059722512. [DOI] [Google Scholar]
- Portis E. et al. The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci (172)3, 640–648 (2007). doi: 10.1016/j.plantsci.2006.11.016. [DOI] [Google Scholar]
- Yi G., Lee J.M., Lee S., Choi D. & Kim B.D. Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet (114)1, 113–30 (2006). doi: 10.1007/s00122-006-0415-y. [DOI] [PubMed] [Google Scholar]
- Ahn Y.K. et al. Microsatellite marker information from high-throughput next-generation sequence data of Capsicum annuum varieties Mandarin and Blackcluster. Sci Hortic-Amsterdam (170)0, 123–130 (2014). doi: 10.1016/j.scienta.2014.03.007. [DOI] [Google Scholar]
- Ahn Y.K. et al. De novo transcriptome assembly and novel microsatellite marker information in Capsicum annuum varieties Saengryeg 211 and Saengryeg 213. Bot Stud (54)1, 58 (2013). doi: 10.1186/1999-3110-54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolai M., Pisani C., Bouchet J.P., Vuylsteke M. & Palloix A. Discovery of a large set of SNP and SSR genetic markers by high-throughput sequencing of pepper (Capsicum annuum). Genet Mol Res (11)3, 2295–300 (2012). doi: 10.4238/2012.August.13.3. [DOI] [PubMed] [Google Scholar]
- Lu F.H., Cho M.C. & Park Y.J. Transcriptome profiling and molecular marker discovery in red pepper, Capsicum annuum L. TF68. Mol Biol Rep (39)3, 3327–35 (2012). doi: 10.1007/s11033-011-1102-x. [DOI] [PubMed] [Google Scholar]
- Ashrafi H. et al. De novo assembly of the pepper transcriptome (Capsicum annuum): a benchmark for in silico discovery of SNPs, SSRs and candidate genes. BMC Genomics (13)1, 571 (2012). doi: 10.1186/1471-2164-13-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F.H. et al. Transcriptome analysis and SNP/SSR marker information of red pepper variety YCM334 and Taean. Sci Hortic-Amsterdam (129)1, 38–45 (2011). doi: 10.1016/j.scienta.2011.03.003. [DOI] [Google Scholar]
- Jo Y.D., Choi Y., Kim D.H., Kim B.D. & Kang B.C. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genomics (15)1, 561 (2014). doi: 10.1186/1471-2164-15-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F.C., Gao C.W. & Gao L.Z. The complete chloroplast genome sequence of American bird pepper (Capsicum annuum var. glabriusculum). Mitochondrial DNA (Early Online) 1–3 (2014). doi: 10.3109/19401736.2014.913160. [DOI] [PubMed] [Google Scholar]
- Jo Y.D. et al. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep (30)2, 217–29 (2011). doi: 10.1007/s00299-010-0929-2. [DOI] [PubMed] [Google Scholar]
- Kim S. et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet (46)3, 270–278 (2014). doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- Shirasawa K. et al. Development of Capsicum EST-SSR markers for species identification and in silico mapping onto the tomato genome sequence. Mol Breed (31)1, 101–110 (2013). doi: 10.1007/s11032-012-9774-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Zhang G., Chen W., Zhang Z. & Zou X. Identification and development of polymorphic EST-SSR markers by sequence alignment in pepper, Capsicum annuum (Solanaceae). Am J Bot (99)2, e59–61 (2012). doi: 10.3732/ajb.1100347. [DOI] [PubMed] [Google Scholar]
- Huang H.H. et al. Analysis of SSRs Information in Capsicum spp. from EST Database. Agr Sci China (10)10, 1532–1536 (2011). doi: 10.1016/S1671-2927(11)60148-X. [DOI] [Google Scholar]
- Lawson M.J. & Zhang L. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol (7)2, R14 (2006). doi: 10.1186/gb-2006-7-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temnykh S. et al. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res (11)8, 1441–52 (2001). doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann K., Hodkinson T.R. & Barth S. New chloroplast microsatellite markers suitable for assessing genetic diversity of Lolium perenne and other related grass species. Ann Bot (110)6, 1327–39 (2012). doi: 10.1093/aob/mcs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Mishra R.K. & Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol (4)2, R13 (2003). doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S. et al. The B73 maize genome: complexity, diversity, and dynamics. Science (326)5956, 1112–5 (2009). doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Nicolaï M., Cantet M., Lefebvre V., Sage-Palloix A.-M. & Palloix A. Genotyping a large collection of pepper (Capsicum spp.) with SSR loci brings new evidence for the wild origin of cultivated C. annuum and the structuring of genetic diversity by human selection of cultivar types. Genet Resour Crop Evol (60)8, 2375–2390 (2013). doi: 10.1007/s10722-013-0006-0. [DOI] [Google Scholar]
- Ibiza V.P., Blanca J., Canizares J. & Nuez F. Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genet Resour Crop Evol (59)6, 1077–1088 (2012). [Google Scholar]
- Murray M.G. & Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res (8)19, 4321–5 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Kahlau S. & Bock R. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res (41) Web Server issue, W575–81 (2013). doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. et al. VENNTURE–a novel Venn diagram investigational tool for multiple pharmacological dataset analysis. PLoS One (7)5, e36911 (2012). doi: 10.1371/journal.pone.0036911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui K. & Muse S. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics (21)2, 128–2 (2005). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol (30)12, 2725–9 (2013). doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.