Abstract
Intrinsically disordered proteins (IDPs), which lack persistent structure, are a challenge to structural biology due to the inapplicability of standard methods for characterization of folded proteins as well as their deviation from the dominant structure/function paradigm. Their widespread presence and involvement in biological function, however, has spurred the growing acceptance of the importance of IDPs and the development of new tools for studying their structure, dynamics and function. The interplay of folded and disordered domains or regions for function and the existence of a continuum of protein states with respect to conformational energetics, motional timescales and compactness is shaping a unified understanding of structure-dynamics-disorder/function relationships. On the 20th anniversary of this journal, Structure, we provide a historical perspective on the investigation of IDPs and summarize the sequence features and physical forces that underlie their unique structural, functional and evolutionary properties.
Keywords: Conformational ensemble, continuum, energy landscape, dynamics, responsiveness
One view of the overall goal of structural biology is to provide insights into the physical chemistry of biological function. This entails descriptions of the free energy landscapes of all relevant macromolecules in isolation and in their interactions so as to be able to predict the kinetics and thermodynamics underlying conformational states, catalytic reactions and binding properties. Such a goal is extremely challenging and many structural biologists have been satisfied by characterizations of the low energy wells on the energy landscapes of macromolecules and some limited numbers of combinations of macromolecules. This less ambitious scope has provided powerful correlations between structural properties of folded macromolecules and complexes and their biological functions. Thus, the broader goal has been thought by many to be superseded. However, it is becoming increasingly apparent that biological function utilizes higher energy states of folded macromolecules (Baldwin and Kay, 2009; Clore, 2011; Osawa et al., 2012) as well as macromolecules that do not predominantly sample low energy states (Dyson and Wright, 2005; Fink, 2005; Tompa, 2005; Meszaros et al., 2011). Proteins with this latter nature have been called intrinsically disordered as they do not adopt unique, well-folded three-dimensional structures but rather populate ensembles of diverse, interconverting conformations. Note that these ensembles, while lacking persistent structure, are “stable” in the thermodynamic sense. This means that the dynamic equilibrium of distinct conformations (i.e. disorder), with its significant conformational entropy, is lower in free energy under the prevailing conditions than a single conformation or narrow distribution of conformations. Intrinsically disordered proteins represent a significant challenge to the previously described body of structure/function correlations. However, from the standpoint of physical chemistry, it should not be surprising that all available energetic possibilities and all accessible conformations can be exploited for various functional purposes. In this regard, a powerful view that includes the entire continuum of dynamic and energetic properties of proteins as well as monomer, oligomer and higher order assemblies is emerging.
Interestingly, prior to the 1957 pioneering work on the binding of the S-protein and S-peptide of cleaved RNase A (Richards, 1997), and shortly thereafter the first crystal structures of ordered proteins (Kendrew et al., 1960), both demonstrating that proteins can be ordered in three dimensions with tight intra-molecular interactions, all proteins were thought of as highly malleable. The notion of proteins having similar structural and dynamic properties as small molecule crystals took some time to be established, with the application of the newly developed tool of protein crystallography having a significant role. To better understand this transition and the more recent change in view regarding the continuum of relevant protein states including disordered proteins, it is worth considering the nature of scientific paradigms and their roles in guiding scientific progress (Kuhn, 1962). In this conception, scientific progress is seen not as a linear enhancement of knowledge but as steady growth of understanding within certain defining and limiting theories or paradigms interrupted by dramatic changes in these paradigms to enable rapid development of knowledge. Thus, the paradigm of structure/function correlations, while facilitating significant biological insight, slowed the acceptance of the biological role of highly dynamic and disordered protein states. Dramatic shifts are often enabled by the application of new methods, with NMR and computational approaches driving the more recent appreciation for the full dynamic continuum of relevant protein states. Regardless of these philosophical considerations, the increasing body of evidence for the presence and functional importance of dynamic and disordered protein states has now led to a general acknowledgment of the structural biological significance of characterizing these states. After all, if these proteins have biological functions, such functions must be enabled by their conformations and dynamics, and structural biology should be able to enlighten us with respect to their structure-dynamics-disorder/function relationships.
Reviews of the history of the field of disordered proteins have been published with references to disorder from at least as far back as 1986 (Hernandez et al., 1986; Sigler, 1988; Spolar and Record, 1994). Within the past 20 years, significant milestones include the description as intrinsically disordered proteins of p21, which is required for the biologically critical function of inhibition of cyclin dependent kinases (Kriwacki et al., 1996), and of alpha-synuclein, which is involved in synaptic activity and a fragment of which is found in Alzheimer’s amyloid deposits (Weinreb et al., 1996). The observation that FlgM is required to be disordered for export but then folds upon binding to sigma factor (Daughdrill et al., 1997) highlighted the potential for disorder-to-order transitions and the advantages of being able to interconvert between states. The developing sparse data on protein disorder was used to train computational predictors, leading to the realization that disorder is not a property of a small subset of proteins but that it is an extremely common feature (Romero et al., 1998). Current predictions on multiple sequenced genomes suggest that up to 50% of amino acids in eukaryotic proteins are disordered, with about 35% of proteins containing stretches of greater than 30 residues of intrinsic disorder and potentially 15% being fully disordered, a not insignificant fraction of the proteome (Ward et al., 2004; Oates et al., 2013; Peng et al., 2013).
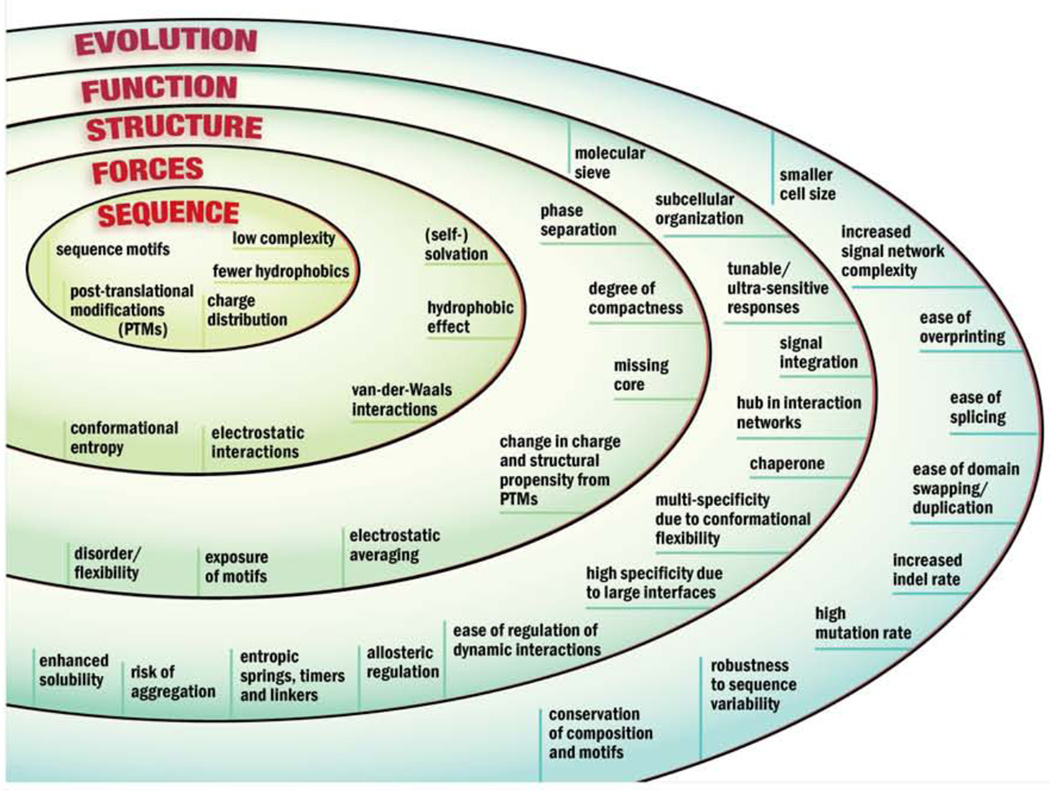
Based on these and many other important contributions over the past 20 years, a set of characteristics of intrinsically disordered proteins and regions (IDPs/IDRs) has been established that provide a context for functional correlations to augment those described for folded protein structures. (Note that for the remainder of the article, IDP will be used for both IDP and IDR.) These characteristics may be considered as an inter-related set beginning with their sequence features and physical forces dominating their interactions through their structural, functional and evolutionary properties (Figure 1).
Figure 1.
Summary of characteristics of intrinsically disordered proteins and regions (IDPs/IDRs) as an inter-related set rooted in their sequence features and physical forces dominating their interactions and continuing through their structural, functional and evolutionary properties. Posttranslational modifications; PTMs.
Sequence
As has been exploited within computational algorithms for predicting protein disorder, IDPs are highly enriched in charged and polar residues, as well as glycine and proline, with fewer hydrophobic residues (Uversky et al., 2000; Williams et al., 2001). IDPs have an overall tendency towards lower complexity sequences including the presence of sequence repeats (Romero et al., 2001; Jorda et al., 2010). Some IDPs can have high net charge, either positive or negative, with others having a mixture of charges with “blocks” or clusters of the same charge. These sequence features do not favor hydrophobic burial in the context of persistent secondary structural elements, precluding formation of folded protein structure. The few instances of large hydrophobic residues, such as tryptophan, tyrosine, phenylalanine and leucine, found in IDPs are almost always part of motifs that recognize binding partners (Fuxreiter et al., 2007; Brown et al., 2010). IDPs also contain sequence motifs for recognition by enzymes carrying out posttranslational modification (PTM) (Iakoucheva et al., 2004). The accessibility of disordered chains facilitates both PTM and binding, leading to enrichment of PTM sites in IDPs and their preponderance as hub proteins (those having >10 binding partners) (Dunker et al., 2005; Higurashi et al., 2008).
Forces
Obviously, the same physical forces apply to a disordered as to an ordered protein, but their relative importance in shaping structure and dynamics is different. The large degree of conformational sampling for IDPs gives them significant conformational entropy, which can be restricted by intra- and intermolecular interactions that lead to structure and limit sampling. It has been suggested that loss of conformational entropy upon binding results in significantly weaker binding for IDPs undergoing dramatic disorder-to-order transitions upon binding, facilitating high specificity but fast off-rates for regulatory interactions (Dunker et al., 1998). However, fine-tuning of enthalpically favorable interactions has been shown to lead to high affinities in some interactions involving significant disorder-to-order transitions (Kriwacki et al., 1996; Lacy et al., 2004; Ferreon et al., 2013; Follis et al., 2013). Given the significant numbers of charged residues and the presence of specific charge distributions in IDPs, electrostatic interactions, including cation-pi (Gallivan and Dougherty, 1999), must play prominent roles. Attractive or repulsive electrostatic interactions have thus been suggested to be critical for hydrodynamic properties (Mao et al., 2010; Marsh and Forman-Kay, 2010; Muller-Spath et al., 2010), as elaborated in the following section. The other forces, including van der Waals interactions and the hydrophobic effect, which are dominant for folded proteins, appear to be less important in general but specific cases of hydrophobic interactions and clustering in disordered proteins have been reported (Klein-Seetharaman et al., 2002; Mittag et al., 2008; Meng et al., 2013). Disordered protein energetics may be characterized by multiple weak interactions showing minimal cooperativity. The transient sampling of a large number of relatively weak intra-molecular interactions within an IDP can enable the protein to act as its own solvent, leading to self-association as intramolecular contacts become indistinguishable from inter-molecular contacts involving the same residues (Rauscher and Pomès, 2012).
Structure
The impact of these physical forces on IDPs leads to structural properties highly distinct from those of folded proteins. The dominant feature is lack of persistent secondary and tertiary structure, with a highly flexible chain transiently sampling fractional secondary structure and tertiary contacts (Daughdrill et al., 1998; Choy and Forman-Kay, 2001; Dunker et al., 2001). This flexibility enables motifs for PTM and binding to be accessible. Dynamic sampling of conformations can also facilitate averaging of electrostatic fields, leading to a dependence of structural (such as Rh) and binding properties on net charge or charge distributions (Borg et al., 2007; Serber and Ferrell, 2007). The fraction and linear patterning of charged residues significantly impacts IDP conformations (Das and Pappu, 2013). PTMs can in many cases change the net charge and charge distribution (Seet et al., 2006; Arif et al., 2010; Deribe et al., 2010; Sasaki, 2012; Bicker and Thompson, 2013), including phosphorylation of Ser, Thr and Tyr (changing charge from 0 to −2), sulfation of Tyr (0 to −1), deamination of Arg (+1 to 0) and acetylation of Lys (+1 to 0). These changes can dramatically affect structural and binding properties of IDPs, such as interactions of a phosphate with the helix dipole of a fluctuating helical element either stabilizing or destroying helical propensity depending on the phosphate position relative to the helix termini (Andrew et al., 2002). While the lack of persistent folded structure and fewer hydrophobic residues mean that there is no hydrophobic core, transient secondary structure and tertiary contacts, electrostatic interactions and backbone torsion angle propensities, particularly for proline, translate into different degrees of compactness for IDPs. These range from quite compact and only slightly expanded relative to a folded domain to extended beyond that expected for a fully denatured domain of the same number of residues (Mao et al., 2013).
One way of understanding the structural properties of disordered proteins is viewing them as the “polymer” state of proteins, capable of occupying many different parts of the phase diagram of accessible protein states (Mao et al., 2010). Many IDPs are monomeric or participate in defined oligomers upon binding protein partners, either folded or disordered. IDPs can undergo disorder-to-order transitions upon binding, stabilizing isolated helical or extended regions or even a small ordered domain (Wright and Dyson, 2009); other interactions result in only transient local ordering within a dynamic or “fuzzy” complex (Tompa and Fuxreiter, 2008; Mittag et al., 2010). The self-association of IDPs or dynamic multi-valent interaction of modular binding domains with motifs within IDPs can drive phase separation under certain conditions (Tompa, 2013), facilitating formation of micron-sized liquid protein-dense droplets (Li et al., 2012) that can function as non-membrane-bound organelles (Kato et al., 2012; Weber and Brangwynne, 2012; Malinovska et al., 2013) or precede gelation or fiber formation (see below). Liquid droplet formation is arguably one of the most important new roles for IDPs identified in the last decade. While cases of folding upon binding result in a straightforward extension of the structure/function paradigm, the retention of significant disorder in dynamic complexes and liquid droplets underscores the functional relevance of the full continuum of protein states.
Due to the dynamic nature of isolated IDPs and many of their complexes, standard biophysical tools are not easily applied for characterization of specific structural properties beyond polymer chain descriptors such as hydrodynamic radius (Rh), radius of gyration (Rg) and end-to-end distance distributions. NMR and SAXS data have been exploited within computational approaches for describing disordered state ensembles (Fisher and Stultz, 2011; Bernado and Svergun, 2012; Marsh and Forman-Kay, 2012; Schneider et al., 2012). Single molecule fluorescence has also been applied to obtain information on these states (Ferreon et al., 2010; Schuler et al., 2012). While for folded proteins, significant homology to an available structure (or sequence similarity for multiple shorter segments as for ROSETTA) (Das and Baker, 2008) is often required for successful structural prediction, recent success in predicting hydrodynamic properties from sequence composition and charge distribution (Hofmann et al., 2012; Das and Pappu, 2013) suggests that we may be able to reasonably predict the gross structural properties of ensembles of disordered states utilizing polymer models without the need to characterize many examples of IDP ensembles. However, combining experimental and computational methods will be required in order to describe more detailed structural properties of the many IDPs and dynamic complexes involving IDPs that underpin biological function.
Function
IDPs have a very large diversity in functional, as well as pathological, roles, in part due to their ability to sample various structural and oligomeric states in the continuum of accessible protein states. One perhaps surprising function involves the modulation of protein solubility. The high proportion of charged and polar residues often enhances solubility of IDPs and proteins containing IDRs (Santner et al., 2012) and, in fact, some IDPs can act as proteinaceous detergents due to unique sequence distributions (Bailey et al., 2001). In contrast, the exposure of hydrophobic residues and possible propensity for extended and turn structures can lead to risk for aggregation and formation of amyloid-type β-structure, with a number of IDPs being involved in aggregation-based diseases (Huang and Stultz, 2009; Uversky, 2010). The levels of IDPs in cells are therefore tightly controlled (Gsponer et al., 2008). IDPs can function to bridge distances, such as between enzyme active sites and substrate binding sites within large catalytic complexes (Mittag et al., 2010). The presence of IDRs as linkers between ordered domains enables control of orientation of and distances between the domains(Chong et al., 2010), as well as more complex mechanisms including roles as entropic springs (Lange et al., 2005; Smagghe et al., 2010) and timers (Wissmann et al., 1999; Bentrop et al., 2001). The length of linkers and their flexibility may, for example, determine how long it takes for two domains to encounter each other stochastically and may therefore time a subsequent signaling event.
A major function of IDPs is in mediating protein recognition due to the many favorable binding properties of IDPs. The complex energy landscape of IDPs and their potentially dynamic complexes facilitates allostery and allosteric regulation (Hilser and Thompson, 2007; Ferreon et al., 2013). The flexibility within dynamic bound states enables ease of regulation by PTM due to accessibility to modifying enzymes. PTM can have dramatic effects on binding due to structural, steric and/or electrostatic effects. Importantly, interactions can be dynamically turned on or off by PTM (Mittag et al., 2010). Many IDPs interact with extended structure across a folded partner with a large interface, providing high specificity and the potential for significant affinity; this can be modulated, however, by the loss of conformational entropy upon binding enabling tuning of binding affinity (Dunker et al., 1998). The larger hydrodynamic radii of some IDPs, but not of collapsed states, lead to higher capture radii (Shoemaker et al., 2000) but may also slow down diffusion (Huang and Liu, 2009), modulating the kinetics of binding and release, effects which may be more significant in the context of proteins containing folded domains and IDRs. Even if bound states of IDPs are ordered, the kinetics and thermodynamics of binding can hence be significantly different compared to folded proteins. An interesting observation is that the same region of an IDP can sometimes interact with different target proteins in distinct conformations, enabling multi-specificity due to the conformational plasticity of the chain (Oldfield et al., 2008; Hsu et al., 2013). Disordered regions of certain chaperones have been demonstrated to mediate the direct interactions with client proteins, highlighting another important function of disorder (Kovacs and Tompa, 2012; Foit et al., 2013).
Here the focus has been on the molecular functions of IDPs, but it is important to note the unique contributions of disorder to particular biological processes, based on these molecular functions. Many IDPs contain multiple binding motifs for interactions with target proteins. In cases where these motifs are distinct, IDPs can act as organizing scaffolds or as hubs in interaction networks facilitating signal integration and localization. Multi-valent interactions involving similar motifs, in contrast, can lead to dynamic exchange of these sites on and off of a single target (Mittag et al., 2008). Such dynamic complexes can generate ultra-sensitive, switch-like (Tang et al., 2012) or graded, rheostat-like (Lee et al., 2010; Mittag et al., 2010) responses, depending on the nature of the interactions. Under certain conditions, multi-valent interactions lead to phase separation behavior, as noted above (Li et al., 2012). This phase separation can generate non-membrane bound organelles involved in subcellular organization or elastic materials, gels and fibers performing various biological roles (Kato et al., 2012; Malinovska et al., 2013). For example, elastin and other elastic proteins are produced from association of disordered monomers in a dynamic, elastic matrix with covalent or β-sheet crosslinking (Muiznieks and Keeley, 2013), and the molecular sieve function of the nuclear pore is due to self-association of disordered nucleoporins (Frey and Gorlich, 2007; Milles and Lemke, 2011). Overall, disorder is significantly enriched in proteins associated with transcription, signaling, phosphorylation, RNA processing, ubiquitination, ion transport, cytoskeletal organization, cell cycle and other highly regulated biological processes (Ward et al., 2004; Lobley et al., 2007).
The correlation of disorder with these critical processes underlies the involvement of disordered proteins in a large number of diseases, including those characterized by loss of biological regulation such as cancer. Examples are the master tumor regulator proteins p53 and c-Myc, which are both transcription factors with folded DNA-binding domains and long disordered regions that mediate complex regulatory interactions (Oldfield et al., 2008; Andresen et al., 2012). The pathological association of disordered proteins into toxic aggregates contributes to another class of diseases, of which neurodegenerative diseases are representative. Alpha-synuclein and tau are examples of IDPs found in cellular aggregates in Parkinson’s and Alzheimer’s diseases (Huang and Stultz, 2009). Pathological effects can arise as consequences of different perturbations. Missense mutations may lead to disorder-to-order transitions (Vacic et al., 2012), changes in post-translational modifications and differences in binding affinities and competition among targets. Perhaps more importantly, changes in expression levels or availability of protein recognition elements can re-wire signaling networks (Gsponer et al., 2008; Babu et al., 2011) and differences in oligomerization or physical state can have dramatic loss- or gain-of-function consequences. The important role of IDPs in disease has lead to recent drug development efforts targeting disordered proteins directly or their interactions with folded partners (Metallo, 2010; Uversky, 2010; Rezaei-Ghaleh et al., 2012).
Evolution
Understanding the evolutionary correlations of IDPs has been challenging due to the use of tools developed for folded protein domains. Thus, conflicting conclusions have been drawn, at once suggesting that disordered regions are conserved as befits their functional significance and that they are much less conserved than folded regions. In different ways, both appear to be true. While folded proteins are usually more strictly positionally conserved than disordered proteins (Xia et al., 2009; Brown et al., 2010), IDPs can demonstrate high conservation of overall composition (Moesa et al., 2012) and of specific motifs for PTM and binding (Nguyen Ba et al., 2012). Importantly, the presence of specific segments of disorder of particular lengths is often highly conserved (Schlessinger et al., 2011). The looser positional conservation of IDPs explains why driver mutations in cancer, while they often involve signaling proteins with long disordered regions, occur more frequently in their folded than in their disordered domains because single amino acid substitutions are more likely to disrupt function in folded domains (Pajkos et al., 2012). This behavior allows high mutation rates in IDPs, increased rates of insertions and deletions (Nido et al., 2012), ease of domain swapping and duplication (Buljan et al., 2010) as well as ease of splicing (Buljan et al., 2013). In addition, the lack of strict constraints for maintaining folded structure facilitates overprinting, which is the use of multiple reading frames for protein translation, observed in a number of viruses (Rancurel et al., 2009). Since binding motifs within IDPs are usually relatively short primary sequence elements rather than full folded domains, protein interactions can be driven with significantly smaller protein lengths (Gunasekaran et al., 2003). This mechanism, together with the potential for overprinting, keeps genome sizes and cell sizes to a minimum.
Outside of constraints on composition, length and specific motifs, the robustness of disordered proteins to sequence variability has been suggested to facilitate rapid evolution of regulatory complexity, making disorder a major evolutionary tool for progressing from simple prokaryotes to complex multi-cellular eukaryotes (Schlessinger et al., 2011). Together with the role of IDPs in mediating regulatory interactions, this helps explain the preponderance of disorder in signaling networks within higher eukaryotes relative to their lower presence in constitutive metabolic proteins and in many bacteria (Xue et al., 2012). Interestingly, selective prokaryotes found in unusual environments (i.e. low temperature, acid, high radiation) and viruses known to undergo rapid selection (Tokuriki et al., 2009) also contain significant disorder. These observations argue for the utility of disorder for evolution.
Concluding remarks
These layers of IDP characteristics shown in Figure 1 demonstrate the large number of unique properties and advantages of IDPs. In assessing the relative roles of ordered and disordered proteins in function and pathology, it is interesting to consider the overall preponderance and expectation of order and disorder in biology, given the requirement for dynamic responsiveness as the primary feature of life. Cells are organized by membrane boundaries, both external and internal, with organelles and other localized features, yet there is considerable dynamics with fusion and fission events at these boundaries. The lipid bilayers that define these boundaries perform phase transitions in order to facilitate fusion and fission, as well as changes in composition and thickness (Furt and Moreau, 2009). While the core DNA structure is primarily ordered helix, the super-structure of DNA is in a state of flux depending on protein interactions and overall elastic properties of the long chain (Benham and Mielke, 2005; Vafabakhsh and Ha, 2012). RNA can have significant although not usually fully populated secondary structure as well as dynamic fluctuating tertiary contacts (Li et al., 2008; Lee et al., 2010). Sugars that are covalently linked to lipids and proteins are often highly dynamic, providing a disordered solubilizing coating (Hricovini, 2004). The primary solvent for all biological macromolecules, water, is significantly disordered at physiological temperatures yet can become partially ordered around various solute molecules (Yamaguchi et al., 2004). Thus, it should not be surprising that proteins occupy a continuum from folded to highly disordered, including protein interactions that may lead to ordered structure or retain significant dynamics.
Knowledge of the variety of conformational and dynamic states exploited by biology is currently developing within the structural biology community, particularly as more proteins containing significant disordered regions are being crystallized (Marsh et al., 2010). This paradigm shift is underway, with increasing appreciation for the extent and importance of disorder and dynamics in biology and the full continuum of protein states and their roles. Future progress towards understanding the complex relationship between energetic and structural features of biomolecules and biological function will require that all of these states be studied. Importantly, the more dynamic end of the continuum of protein states, including isolated intrinsically disordered proteins and their dynamic complexes and large-scale associated states, needs to be a focus. New methods for better characterizing IDPs and their dynamic complexes are required and more studies to demonstrate the functional link to advantages of disorder. Given the organization of many proteins with combinations of folded and disordered regions, as well as intra- and intermolecular interactions of folded and disordered regions (Babu et al., 2012; Tompa, 2012), the complex interplay between structure and disorder must also be understood to augment isolated structure/function and disorder/function relationships. Due to IDPs having multiple sites of PTMs, conformational states, functions, and binding partners, they are likely to be exploited to provide dynamic responsiveness and complex regulation in biology. Thus, future studies of IDPs and correlations between their biophysical properties and function are sure to provide critical insights into the structural basis of biological regulation, cellular organization and disease.
Acknowledgements
We thank A.K. Dunker and C. Morales for help with the figure and P.J. Farber, R.V. Pappu and R.W. Kriwacki for useful discussions. This work was supported by grants to J.D.F.-K. from the Canadian Institutes for Health Research, the Canadian Cancer Society Research Institute, Cystic Fibrosis Canada, the Cystic Fibrosis Foundation Therapeutics, and the Natural Science and Engineering Research Council of Canada and to T.M. by a V. Foundation Scholar Grant, a National Cancer Institute Cancer Center Support Grant P30CA21765 (at St. Jude Children's Research Hospital) and the American Lebanese Syrian Associated Charities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andresen C, Helander S, Lemak A, Fares C, Csizmok V, Carlsson J, Penn LZ, Forman-Kay JD, Arrowsmith CH, Lundstrom P, et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012;40(13):6353–6366. doi: 10.1093/nar/gks263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew CD, Warwicker J, Jones GR, Doig AJ. Effect of phosphorylation on alpha-helix stability as a function of position. Biochemistry. 2002;41(6):1897–1905. doi: 10.1021/bi0113216. [DOI] [PubMed] [Google Scholar]
- Arif M, Senapati P, Shandilya J, Kundu TK. Protein lysine acetylation in cellular function and its role in cancer manifestation. Biochim Biophys Acta. 2010;1799(10–12):702–716. doi: 10.1016/j.bbagrm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337(6101):1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21(3):432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Bailey RW, Dunker AK, Brown CJ, Garner EC, Griswold MD. Clusterin, a binding protein with a molten globule-like region. Biochemistry. 2001;40(39):11828–11840. doi: 10.1021/bi010135x. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5(11):808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- Benham CJ, Mielke SP. DNA mechanics. Annu Rev Biomed Eng. 2005;7:21–53. doi: 10.1146/annurev.bioeng.6.062403.132016. [DOI] [PubMed] [Google Scholar]
- Bentrop D, Beyermann M, Wissmann R, Fakler B. NMR structure of the "ball-and-chain" domain of KCNMB2, the beta 2-subunit of large conductance Ca2+ and voltage-activated potassium channels. J Biol Chem. 2001;276(45):42116–42121. doi: 10.1074/jbc.M107118200. [DOI] [PubMed] [Google Scholar]
- Bernado P, Svergun DI. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol Biosyst. 2012;8(1):151–167. doi: 10.1039/c1mb05275f. [DOI] [PubMed] [Google Scholar]
- Bicker KL, Thompson PR. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers. 2013;99(2):155–163. doi: 10.1002/bip.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A. 2007;104(23):9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27(3):609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Dunker AK, Bateman A, Balaji S, Fuxreiter M, Babu MM. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23(3):443–450. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Buljan M, Frankish A, Bateman A. Quantifying the mechanisms of domain gain in animal proteins. Genome Biol. 2010;11(7):R74. doi: 10.1186/gb-2010-11-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PA, Lin H, Wrana JL, Forman-Kay JD. Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity. Proc Natl Acad Sci U S A. 2010;107(43):18404–18409. doi: 10.1073/pnas.1003023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol. 2001;308(5):1011–1032. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- Clore GM. Exploring sparsely populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Science. 2011;20(2):229–246. doi: 10.1002/pro.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Das R, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1304749110. epub before print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4(4):285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Hanely LJ, Dahlquist FW. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry. 1998;37(4):1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 1998:473–484. [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498(7454):390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon AC, Moran CR, Gambin Y, Deniz AA. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15(1):35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Fisher CK, Stultz CM. Constructing ensembles for intrinsically disordered proteins. Curr Opin Struct Biol. 2011;21(3):426–431. doi: 10.1016/j.sbi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foit L, George JS, Zhang BW, Brooks CL, 3rd, Bardwell JC. Chaperone activation by unfolding. Proc Natl Acad Sci U S A. 2013;110(14):E1254–E1262. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis AV, Chipuk JE, Fisher JC, Yun MK, Grace CR, Nourse A, Baran K, Ou L, Min L, White SW, et al. PUMA binding induces partial unfolding within BCL-xL to disrupt p53 binding and promote apoptosis. Nat Chem Biol. 2013;9(3):163–168. doi: 10.1038/nchembio.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130(3):512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Furt F, Moreau P. Importance of lipid metabolism for intracellular and mitochondrial membrane fusion/fission processes. Int J Biochem Cell Biol. 2009;41(10):1828–1836. doi: 10.1016/j.biocel.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23(8):950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- Gallivan, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999;96(17):9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322(5906):1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci. 2003;28(2):81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- Hernandez MA, Avila J, Andreu JM. Physicochemical characterization of the heatstable microtubule-associated protein MAP2. Eur J Biochem. 1986;154(1):41–48. doi: 10.1111/j.1432-1033.1986.tb09356.x. [DOI] [PubMed] [Google Scholar]
- Higurashi M, Ishida T, Kinoshita K. Identification of transient hub proteins and the possible structural basis for their multiple interactions. Protein Science. 2008;17(1):72–78. doi: 10.1110/ps.073196308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104(20):8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci U S A. 2012;109(40):16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricovini M. Structural aspects of carbohydrates and the relation with their biological properties. Curr Med Chem. 2004;11(19):2565–2583. doi: 10.2174/0929867043364414. [DOI] [PubMed] [Google Scholar]
- Hsu WL, Oldfield CJ, Xue B, Meng J, Huang F, Romero P, Uversky VN, Dunker AK. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Science. 2013;22(3):258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Stultz CM. Finding order within disorder: elucidating the structure of proteins associated with neurodegenerative disease. Future Med Chem. 2009;1(3):467–482. doi: 10.4155/fmc.09.40. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu Z. Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the "fly-casting" mechanism. J Mol Biol. 2009;393(5):1143–1159. doi: 10.1016/j.jmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda J, Xue B, Uversky VN, Kajava AV. Protein tandem repeats - the more perfect, the less structured. FEBS J. 2010;277(12):2673–2682. doi: 10.1111/j.1742-464X.2010.07684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, Shore VC. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 A. resolution. Nature. 1960;185(4711):422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Long-range interactions within a nonnative protein. Science. 2002;295(5560):1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- Kovacs D, Tompa P. Diverse functional manifestations of intrinsic structural disorder in molecular chaperones. Biochem Soc Trans. 2012;40(5):963–968. doi: 10.1042/BST20120108. [DOI] [PubMed] [Google Scholar]
- Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93(21):11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TS. The Structure of Scientific Revolutions. Chicago: The University of Chicago Press; 1962. [Google Scholar]
- Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11(4):358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- Lange S, Agarkova I, Perriard JC, Ehler E. The sarcomeric M-band during development and in disease. J Muscle Res Cell Motil. 2005;26(6–8):375–379. doi: 10.1007/s10974-005-9019-4. [DOI] [PubMed] [Google Scholar]
- Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci U S A. 2010;107(45):19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PT, Vieregg J, Tinoco I., Jr How RNA unfolds and refolds. Annu Rev Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- Lobley A, Swindells MB, Orengo CA, Jones DT. Inferring function using patterns of native disorder in proteins. PLoS Comput Biol. 2007;3(8):e162. doi: 10.1371/journal.pcbi.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim Biophys Acta. 2013;1834(5):918–931. doi: 10.1016/j.bbapap.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci U S A. 2010;107(18):8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao AH, Lyle N, Pappu RV. Describing sequence-ensemble relationships for intrinsically disordered proteins. Biochem J. 2013;449(2):307–318. doi: 10.1042/BJ20121346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Dancheck B, Ragusa MJ, Allaire M, Forman-Kay JD, Peti W. Structural diversity in free and bound states of intrinsically disordered protein phosphatase 1 regulators. Structure. 2010;18(9):1094–1103. doi: 10.1016/j.str.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Forman-Kay JD. Sequence determinants of compaction in intrinsically disordered proteins. Biophys J. 2010;98(10):2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Forman-Kay JD. Ensemble modeling of protein disordered states: Experimental restraint contributions and validation. Proteins. 2012;80(2):556–572. doi: 10.1002/prot.23220. [DOI] [PubMed] [Google Scholar]
- Meng W, Lyle N, Luan B, Raleigh DP, Pappu RV. Experiments and simulations show how long-range contacts can form in expanded unfolded proteins with negligible secondary structure. Proc Natl Acad Sci U S A. 2013;110(6):2123–2128. doi: 10.1073/pnas.1216979110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B, Simon I, Dosztanyi Z. The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins. Phys Biol. 2011;8(3):035003. doi: 10.1088/1478-3975/8/3/035003. [DOI] [PubMed] [Google Scholar]
- Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14(4):481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles S, Lemke EA. Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153. Biophys J. 2011;101(7):1710–1719. doi: 10.1016/j.bpj.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23(2):105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18(4):494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci U S A. 2008;105(46):17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moesa HA, Wakabayashi S, Nakai K, Patil A. Chemical composition is maintained in poorly conserved intrinsically disordered regions and suggests a means for their classification. Mol Biosyst. 2012;8(12):3262–3273. doi: 10.1039/c2mb25202c. [DOI] [PubMed] [Google Scholar]
- Muiznieks LD, Keeley FW. Molecular assembly and mechanical properties of the extracellular matrix: A fibrous protein perspective. Biochim Biophys Acta. 2013;1832(7):866–875. doi: 10.1016/j.bbadis.2012.11.022. [DOI] [PubMed] [Google Scholar]
- Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci U S A. 2010;107(33):14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Yeh BJ, van Dyk D, Davidson AR, Andrews BJ, Weiss EL, Moses AM. Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Sci Signal. 2012;5(215):rs1. doi: 10.1126/scisignal.2002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nido GS, Mendez R, Pascual-Garcia A, Abia D, Bastolla U. Protein disorder in the centrosome correlates with complexity in cell types number. Mol Biosyst. 2012;8(1):353–367. doi: 10.1039/c1mb05199g. [DOI] [PubMed] [Google Scholar]
- Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztanyi Z, Uversky VN, Obradovic Z, Kurgan L, et al. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Takeuchi K, Ueda T, Nishida N, Shimada I. Functional dynamics of proteins revealed by solution NMR. Curr Opin Struct Biol. 2012;22(5):660–669. doi: 10.1016/j.sbi.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Pajkos M, Meszaros B, Simon I, Dosztanyi Z. Is there a biological cost of protein disorder? Analysis of cancer-associated mutations. Mol Biosyst. 2012;8(1):296–307. doi: 10.1039/c1mb05246b. [DOI] [PubMed] [Google Scholar]
- Peng Z, Mizianty MJ, Kurgan L. Genome-scale prediction of proteins with long intrinsically disordered regions. Proteins. 2013 doi: 10.1002/prot.24348. epub before print. [DOI] [PubMed] [Google Scholar]
- Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 2009;83(20):10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher S, Pomès R. Aggregated yet Disordered: A Molecular Simulation Study of the Self-Aggregation of Elastin. Biophys J. 2012;102(3) Supplement 1:40a. [Google Scholar]
- Rezaei-Ghaleh N, Blackledge M, Zweckstetter M. Intrinsically disordered proteins: from sequence and conformational properties toward drug discovery. Chembiochem. 2012;13(7):930–950. doi: 10.1002/cbic.201200093. [DOI] [PubMed] [Google Scholar]
- Richards FM. Whatever happened to the fun? An autobiographical investigation. Annu Rev Biophys Biomol Struct. 1997;26:1–25. doi: 10.1146/annurev.biophys.26.1.1. [DOI] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, Dunker AK. Thousands of proteins likely to have long disordered regions. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 1998:437–448. [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Santner AA, Croy CH, Vasanwala FH, Uversky VN, Van YY, Dunker AK. Sweeping away protein aggregation with entropic bristles: intrinsically disordered protein fusions enhance soluble expression. Biochemistry. 2012;51(37):7250–7262. doi: 10.1021/bi300653m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N. Current status and future prospects for research on tyrosine sulfation. Curr Pharm Biotechnol. 2012;13(14):2632–2641. doi: 10.2174/138920101314151120122922. [DOI] [PubMed] [Google Scholar]
- Schlessinger A, Schaefer C, Vicedo E, Schmidberger M, Punta M, Rost B. Protein disorder--a breakthrough invention of evolution? Curr Opin Struct Biol. 2011;21(3):412–418. doi: 10.1016/j.sbi.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Schneider R, Huang JR, Yao M, Communie G, Ozenne V, Mollica L, Salmon L, Jensen MR, Blackledge M. Towards a robust description of intrinsic protein disorder using nuclear magnetic resonance spectroscopy. Mol Biosyst. 2012;8(1):58–68. doi: 10.1039/c1mb05291h. [DOI] [PubMed] [Google Scholar]
- Schuler B, Muller-Spath S, Soranno A, Nettels D. Application of confocal single-molecule FRET to intrinsically disordered proteins. Methods Mol Biol. 2012;896:21–45. doi: 10.1007/978-1-4614-3704-8_2. [DOI] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7(7):473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Serber Z, Ferrell JE., Jr Tuning bulk electrostatics to regulate protein function. Cell. 2007;128(3):441–444. doi: 10.1016/j.cell.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A. 2000;97(16):8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler PB. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Smagghe BJ, Huang PS, Ban YE, Baker D, Springer TA. Modulation of integrin activation by an entropic spring in the {beta}-knee. J Biol Chem. 2010;285(43):32954–32966. doi: 10.1074/jbc.M110.145177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Mittag T, Csizmok V, Pawson T, Forman-Kay JD, Sicheri F, Tyers M. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc Natl Acad Sci U S A. 2012;109(9):3287–3292. doi: 10.1073/pnas.1116455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS. Do viral proteins possess unique biophysical features? Trends Biochem Sci. 2009;34(2):53–59. doi: 10.1016/j.tibs.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- Tompa P. On the supertertiary structure of proteins. Nat Chem Biol. 2012;8(7):597–600. doi: 10.1038/nchembio.1009. [DOI] [PubMed] [Google Scholar]
- Tompa P. Hydrogel formation by multivalent IDPs: A reincarnation of the microtrabecular lattice? Intrinsically Disordered Proteins. 2013;1(1):e24086. doi: 10.4161/idp.24068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: another illustration of the D(2) concept. Expert Rev Proteomics. 2010;7(4):543–564. doi: 10.1586/epr.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Vacic V, Markwick PR, Oldfield CJ, Zhao X, Haynes C, Uversky VN, Iakoucheva LM. Disease-associated mutations disrupt functionally important regions of intrinsic protein disorder. PLoS Comput Biol. 2012;8(10):e1002709. doi: 10.1371/journal.pcbi.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafabakhsh R, Ha T. Extreme bendability of DNA less than 100 base pairs long revealed by single-molecule cyclization. Science. 2012;337(6098):1097–1101. doi: 10.1126/science.1224139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149(6):1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35(43):13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- Williams RM, Obradovi Z, Mathura V, Braun W, Garner EC, Young J, Takayama S, Brown CJ, Dunker AK. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2001:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- Wissmann R, Baukrowitz T, Kalbacher H, Kalbitzer HR, Ruppersberg JP, Pongs O, Antz C, Fakler B. NMR structure and functional characteristics of the hydrophilic N terminus of the potassium channel beta-subunit Kvbeta1.1. J Biol Chem. 1999;274(50):35521–35525. doi: 10.1074/jbc.274.50.35521. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19(1):31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Franzosa EA, Gerstein MB. Integrated assessment of genomic correlates of protein evolutionary rate. PLoS Comput Biol. 2009;5(6):e1000413. doi: 10.1371/journal.pcbi.1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Matsuoka T, Koda S. Mode-coupling study on the dynamics of hydrophobic hydration. J Chem Phys. 2004;120(16):7590–7601. doi: 10.1063/1.1687319. [DOI] [PubMed] [Google Scholar]