Abstract
Attempts to isolate novel antimicrobial peptides from microbial sources have been on the rise recently, despite their low efficacy in therapeutic applications. Here, we report identification and characterization of a new efficient antimicrobial peptide from a bacterial strain designated A3 that exhibited highest identity with Paenibacillus ehimensis. Upon purification and subsequent molecular characterization of the antimicrobial peptide, referred to as penisin, we found the peptide to be a bacteriocin-like peptide. Consistent with these results, RAST analysis of the entire genome sequence revealed the presence of a lantibiotic gene cluster containing genes necessary for synthesis and maturation of a lantibiotic. While circular dichroism and one-dimension nuclear magnetic resonance experiments confirmed a random coil structure of the peptide, similar to other known lantibiotics, additional biochemical evidence suggests posttranslational modifications of the core peptide yield six thioether cross-links. The deduced amino acid sequence of the putative biosynthetic gene penA showed approximately 74% similarity with elgicin A and 50% similarity with the lantibiotic paenicidin A. Penisin effectively killed methicillin-resistant Staphylococcus aureus (MRSA) and did not exhibit hemolysis activity. Unlike other lantibiotics, it effectively inhibited the growth of Gram-negative bacteria. Furthermore, 80 mg/kg of body weight of penisin significantly reduced bacterial burden in a mouse thigh infection model and protected BALB/c mice in a bacteremia model entailing infection with Staphylococcus aureus MTCC 96, suggesting that it could be a promising new antimicrobial peptide.
INTRODUCTION
Rapid development of multiple-drug resistance in pathogenic and opportunistic pathogenic bacteria is posing a great threat to public health (1–3). As a consequence, research pertaining to antimicrobial peptides (AMPs) has increased recently as an alternative to antibiotics. AMPs produced by bacteria were recently classified as postribosomal peptide synthesis (PRPS) peptides and ribosomally synthesized and posttranslationally modified peptides (RiPPs), depending on their biosynthesis pathway (4, 5). RiPPs, also called bacteriocins, are encoded by a structural gene and are usually produced as a precursor peptide containing an N-terminal leader sequence involved in recognition of enzymes required for posttranslational modifications. Among the RiPPs, lanthipeptides undergo a two-step posttranslational modification process during their biosynthesis to yield meso-lanthionine (Lan) and 3-methyllanthionine (MeLan) residues. The first step involves dehydration of Ser and Thr residues in the precursor peptide to yield dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively. In a subsequent reaction, thioether bonds are formed by Cys residues with the dehydro amino acids. Based on enzymes involved in biosynthesis of lanthipeptides, they are grouped into four different classes (classes I to IV). Lanthipeptides exhibiting antimicrobial activity are called lantibiotics. Some of these lantibiotics are presently being tested as therapeutic agents in the treatment of drug-resistant bacterial infections (6). Similarly, PRPS peptides (previously known as nonribosomal peptide synthetases [NRPS]) represent a group of diverse lipopeptides containing unnatural amino acids in their composition. Additionally, they are an important class among the lipopeptide antibiotics used to inhibit the growth of Gram-negative bacteria and are being used for therapeutic applications (7). However, members of the Paenibacillus genus, like P. polymyxa, are known to produce both bacteriocins and lipopeptide antibiotics. In fact, P. polymyxa has been reported to produce polymyxins consisting of five chemically different compounds, polymyxins A to E (8–10). Apart from P. polymyxa, strains of a few other Paenibacillus species, like P. elgi, P. ehimensis, P. peoriae, and P. kobensis, have been reported to produce different antimicrobial agents, including lantibiotics and other non-lanthionine-containing bacteriocins (3, 11–18). However, a large number of other species of this genus have never been explored for their antimicrobial production and remain a potential source of novel antimicrobials. Therefore, in the present study we isolated, purified, and characterized an antimicrobial substance produced by Paenibacillus sp. strain A3.
MATERIALS AND METHODS
Strain isolation and characterization.
The bacterial strain A3 was isolated from a subsurface soil sample during a screening for bacteriocin-producing strains. All test strains used were procured from the Microbial Type Culture Collection (MTCC), Chandigarh, India. The morphological characteristics, such as Gram staining and endospore staining, and physiological tests, like growth at different temperatures, pH, and salt concentrations, were determined as per standard methods. Biochemical tests, including acid production from different carbohydrates, were performed as described by Smibert and Kreig (19). The genomic DNA was extracted using a DNA extraction kit (Zymo Research, USA) as per the manufacturer's instructions and used for 16S rRNA gene amplification, and subsequent sequence analysis was performed as described earlier (20).
Antimicrobial peptide production and activity assay.
A growth curve was established after up to 30 h of growth to test the antimicrobial peptide production in different growth phases. To test the effect of different carbon and nitrogen sources on antimicrobial production by strain A3, a 0.5% concentration of different substrates, like glucose, lactose, yeast extract, peptone, and beef extract, was added to minimal medium (composition: Na2HPO4 · 2H2O, 7.9 g/liter; KH2PO4, 3.0 g/liter; NaCl, 0.5 g/liter; NH4Cl, 1.0 g/liter; pH 7.2). The antimicrobial production ability of the strain was measured as the diameter of the inhibition zone in an antimicrobial bioassay using cell-free fermented broth (CFB). Strain A3 was grown in minimal medium containing different carbon and nitrogen sources for 48 h, and the CFB obtained was tested in a well diffusion assay. The CFB obtained by growing strain A3 in nutrient broth (NB; Himedia, India) was also used to test the activity. Test strains used in the present study included Listeria monocytogenes (MTCC 839), Staphylococcus aureus (MTCC 1430), Bacillus subtilis (MTCC 121), Pseudomonas aeruginosa (MTCC 1934), Vibrio cholerae (MTCC 3904), Escherichia coli (MTCC 1610), Candida albicans (MTCC 1637), Saccharomyces cerevisiae (MTCC 170), Fusarium oxysporum (MTCC 2773), and Asperigillus niger (MTCC 281). A methicillin-resistant S. aureus (MRSA) isolate was obtained from Guru Nanak Dev University.
Isolation and purification of bacteriocin-like peptide.
The antimicrobial substance was purified from CFB from 1,000 ml of nutrient broth in which strain A3 was grown for 48 h on a rotary shaker at 30°C (180 rpm). The crude extract was obtained by incubating CFB and 2% preactivated Diaion HP20 resin (Sigma, USA) as mentioned earlier (21). This crude extract was purified by gel filtration chromatography using a manually packed Sephadex G-50 column (50 mM NaCl with a flow rate of 1.0 ml/min), and fractions showing antimicrobial activity were collected and concentrated. Subsequently, salt was removed by dialysis using a Float-A-Lyzer G2 system (molecular mass cutoff, 0.5 to 1 kDa; Spectrum Laboratories, USA). Final purification of active compound was accomplished by reverse-phase high-pressure liquid chromatography (HPLC) on a 1260 Infinity instrument (Agilent Technologies, USA) using a semipreparative C18 column (Venusil, USA). The mobile phase consisted of HPLC-grade water containing 0.12% trifluoroacetic acid (TFA; mobile phase A) and HPLC-grade acetonitrile (ACN) with 0.1% TFA (mobile phase B), with monitoring via a UV detector set at 220 nm. A single peak was collected by using a fraction collector (DEABG01554; Agilent Technologies) coupled with the system and tested for antimicrobial activity. Purity of the peptide was tested by reinjecting into the HPLC column, and the pure peptide was used for circular dichroism (CD), NMR, PEGylation, MIC determinations, and an in-gel activity assay.
SDS-PAGE and in-gel activity bioassay.
The purified bacteriocin-like peptide was electrophoresed (Tricine–SDS-PAGE) with 16% polyacrylamide and used for an in-gel activity assay. The HPLC-purified peptide was loaded in duplicate lanes, and after electrophoresis it was divided into two vertical parts. The gel slice containing peptide along with marker was stained with Coomassie blue, and the other half containing only peptide was fixed in 20% isopropanol and 10% acetic acid solution for 2 h at room temperature (22). Subsequently, the gel slice was washed with sterile distilled water for 6 h. The gel was placed in a sterile petri dish, overlaid with 10 ml of soft agar (0.8%) containing an E. coli test strain (about 106 cells/ml), and checked for the zone of growth inhibition upon overnight incubation at 30°C.
Mass spectrometry analysis of antimicrobial peptide.
Matrix-assisted laser desorption ionization (MALDI) was used to determine the intact molecular mass of the HPLC-purified bacteriocin-like peptide. The lyophilized peptide was resuspended in methanol, 4 μl of peptide solution was mixed with 4 μl of matrix (α-cyano-4-hydroxycinnamic acid [CHCA]; 10 mg/ml), and 1.0 μl of this mixture was spotted onto the MALDI 100-well stainless steel sample plate and allowed to air dry prior to the MALDI analysis (23). MALDI mass spectra were obtained on an AB Sciex 5800 MALDI-time of flight (TOF-TOF) mass spectrometer. Tandem mass spectrometry (MS/MS) data were acquired at 1,000 Hz in 1-kV MS/MS mode with 2,000 laser shots/spectrum in CID (collision-induced dissociation) mode to obtain maximum resolution. The spectrum was recorded in positive ion linear mode. The reproducibility of the spectrum was checked several times from separately spotted samples.
N-terminal sequencing and analysis.
The HPLC-purified peptide was separated on a Tricine–SDS-PAGE (16%) gel, transferred onto a polyvinylidene difluoride membrane (Bio-Rad), rinsed with Milli-Q water, stained with amido black (Sigma, USA) for 2 to 3 min, and destained by placing in an aqueous solution containing 25% isopropanol and 10% acetic acid for 30 min. The membrane was finally rinsed multiple times in Milli-Q water. The excised peptide band was subjected to N-terminal sequencing as mentioned earlier (21). The partial sequence obtained was used to find the resembling sequences available in the database (http://aps.unmc.edu/AP/main.php).
Circular dichroism.
CD experiments were performed using a Jasco 815 spectropolarimeter (JASCO International Co. Ltd.), coupled to a Peltier Jasco PTC-423L system for temperature control. In experiments carried out in the presence of SDS and dodecyl phosphocholine (DPC) micelles, 50 μM peptide was added in solutions of 100 mM SDS or DPC in water. For experiments in the presence of 2,2,2-trifluoroethanol (TFE), 50 μM HPLC-purified peptide was added to solutions of 20, 60, and 100% TFE. For water experiments, a 50 μM solution of peptide was used. The pH variation was also studied, and samples were prepared to a final peptide concentration of 50 μM, 100 mM SDS, and all three TFE solutions, in phosphate buffer at pH 4.0, Tris-HCl buffer at pH 7.0, and ammonium chloride buffer at pH 11.0. Spectra were collected and averaged over six scans in the spectral range of 190 to 260 nm, with 0.1-mm-path length quartz cells at 37°C. A 0.2-nm step resolution, 50-nm/min speed, 1-s response time, and 1-nm bandwidth were used. Following baseline correction, the observed ellipticity, θ (measured in millidegrees) was converted to the molar mean residue ellipticity [θ] (units of degrees per square centimeter per decimole) (24).
Nuclear magnetic resonance analysis.
Nuclear magnetic resonance (NMR) data were acquired at 37°C with a Bruker Avance III 500 NMR spectrometer operated at 11.75 T (1H resonance frequency, 500.13 MHz), equipped with a 5-mm TBI probe (1H, 13C, and XBB). Water suppression was achieved using the presaturation technique. Sixty-four free induction decays (FIDs) were collected into 64,000 data points by using a 9.0-μs pulse width (90° pulse angle), a spectral width of 10,000.00 Hz, relaxation delay of 1.0 s, and an acquisition time of 3.30 s. All NMR spectra were processed using the program TopSpin version 3.2. The samples were prepared by dissolving the peptide in a solution of 90% TFE-deuterium (TFE-d3) and 10% heavy water–trimethylsilyl propionic acid-d4 (D2O/TMSP-d4; 95:5 [vol/vol]). Similarly, a solution of 90% water and 10% D2O/TMSP-d4 (95:5 [vol/vol]) was prepared. Both final solutions (TFE and H2O) resulted in a concentration of 1.0 mM peptide. The pH was adjusted to 7.0 with 10 mM aqueous Tris-HCl buffer.
PEGylation.
To determine the number of free cysteines in the AMP, maleimide polyethylene glycol (MalPEG; 5 kDa; Sigma-Aldrich) was reacted in different states (native, reduced, unfolded, and reduced unfolded) of the HPLC-purified peptide. TCEP (1 mM; Thermo Scientific, USA) at 25°C for 1 h was used as reducing agent and 6 M urea (Sigma-Aldrich) at 40°C for 1 h was used for unfolding of the native peptide (25).
Whole-genome sequencing, assembly, and annotation.
The genomic DNA for the whole genome sequence was isolated by using a ZR fungal/bacterial DNA isolation kit (Zymo Research). The quality of the isolated DNA was assessed using agarose gel electrophoresis, a Qubit fluorometer, and a Nanodrop spectrophotometer. An input of 1 μg of genomic DNA from each sample was used. The standard protocol for the Nextera XT DNA sample preparation kit was used for library construction. Purified fragmented DNA was used as a template for a limited cycle PCR with Nextera primers and index adaptors. Cluster generation and sequencing of libraries were performed on the Illumina MiSeq platform (Illumina, San Diego, CA) with a 2- by 250 paired-end run. The paired-end raw reads containing FASTQ files were assembled using CLC Genomics Workbench software version 7.0.3. The assembly was uploaded for annotation to the Rapid Annotation using Subsystems Technology (RAST) server (26).
Sensitivities of the antimicrobial peptide to temperature, pH, and proteolytic enzymes.
The sensitivities of the purified antimicrobial peptide toward pH, temperature, and hydrolytic enzymes were confirmed by performing bioassays. To determine the pH stability, aliquots of purified peptide were adjusted to pH 2.0 to 12.0 with an increment of 2 pH units (using 10 mM HCl or NaOH) followed by incubation at room temperature for 4 h, and the leftover activity was measured upon neutralizing the sample to pH 7.0. For the thermal stability assay, aliquots of purified peptide were exposed to 60, 80, and 100°C for 30 min and 121°C for 15 min and then antimicrobial activity was determined. Similarly, proteolytic enzymes such as trypsin and proteinase K were used at three different concentrations (0.1, 1.0, and 5.0 mg/ml) to ensure their effect on the peptide. The enzyme solutions were prepared in 50 mM phosphate buffer (pH 7.0). All reactions were performed at 37°C for 6 h followed by deactivation of enzyme by heating the solution in boiling water for 5 min before performing the activity assay.
Determination of MICs.
MICs of the purified AMP were determined by the microtiter plate dilution assay method. Initially, the peptide concentration was estimated using bovine serum albumin protein standards (Thermo Scientific, USA) according to the Bradford method (Bio-Rad) and a bicinchoninic acid assay (Thermo Scientific, USA) as described by the manufacturers. However, the concentration was confirmed based on the extinction coefficient, once we obtained the amino acid sequence of the peptide. Test strains were grown to logarithmic phase (between 0.3 to 0.4 optical density units) under optimal conditions and used for assays performed in triplicate, as described earlier (27).
Killing kinetics against MRSA.
To determine killing kinetics of the AMP, MRSA cells (∼2 × 104) were treated with the AMP at different concentrations (1×, 2×, and 5× the MIC) in a time-dependent manner. After incubation for different time intervals (30, 60, 90, and 120 min), the cells were removed from the reaction mixture by centrifugation (8,000 × g) and subsequently washed twice with phosphate-buffered saline (PBS; Gibco, USA). The CFU were counted after plating different dilutions of these cells, following vigorous vortexing to prevent clumping of cells. Untreated cells were used as a positive control and were processed along with treated cells. The experiment was performed in triplicate and repeated as three individual sets.
Mammalian cell toxicity.
Cultures of HeLa cells (human cervical cancer cells), RWPE1 cells (human prostate epithelial cells), and RAW cells (mouse macrophages) were subjected to an MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (Invitrogen, Life Technologies, USA) to determine the number of surviving cells after treatment with penisin. The cells were plated in 96-well BD Falcon vessels (5 × 103 to 10 × 103 cells/well) using Dulbecco's modified Eagle's medium (Gibco, USA) supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin-streptomycin cocktail (Sigma, USA). RWPE1 cells were grown in keratinocyte serum-free medium supplemented with recombinant human epidermal growth factor and bovine pituitary extract (Invitrogen, Life Technologies, USA). All cells were incubated under standard conditions (37°C, 5% CO2). After 24 h, the growth medium was replaced with fresh medium containing various concentrations of penisin (1 to 4.0 μM). Medium without peptide was used as a negative control, and 1% Triton X-100 was used as a positive control. The plates were incubated for an additional 24 h, 20 μl of MTT solution (5 mg/ml in PBS) was added to each well, and plates were incubated for 3 h at 37°C. Subsequently, after removal of MTT-containing medium, 50 μl of dimethyl sulfoxide was added to each well. To assess the percentage of live cells in samples, absorbance (590 nm) was assessed using an enzyme-linked immunosorbent assay plate reader (Thermo-Fisher Scientific, USA).
Hemolysis assay.
For the hemolysis assay, blood samples were collected from rabbits (New Zealand White) by using test tubes containing EDTA. The samples were centrifuged at 1,500 rpm for 5 min to remove morphotic elements and subsequently rinsed twice with PBS (28). The erythrocyte suspension (10%) in PBS was incubated along with different concentrations of the AMP in a 5% CO2 incubator (Thermo Fisher Scientific, USA) at 37°C. A 1% Triton X-100 (G-Biosciences, USA) solution in water was used as a positive control, and PBS was the negative control. The samples were centrifuged after 24 h of incubation, and supernatants were transferred into a 96-well plate to assay erythrocyte lysis at a wavelength of 405 nm (29).
TEM sample preparation protocol.
Since a high cell density of about 106 to 108 CFU/ml is required for observations under an electron microscope and the MIC for AMP was also determined with high inoculation doses, E. coli and MRSA cultures at mid-exponential growth phase were centrifuged at 8,000 rpm for 10 min and then cells were washed with PBS two times. Finally, the cell pellet was diluted with PBS to a cell density of 108 CFU/ml, subsequently treated with different concentrations of the AMP in a time-dependent manner, and incubated at 37°C (30). Untreated controls were prepared by diluting the cell pellet in PBS to a cell density of 108 CFU/ml. Treated cells were then subjected to negative stain with 0.1% (wt/vol) sodium phosphotungstate (PTA; Sigma, USA) in water for 1 min, on a carbon-coated copper grid (300 mesh; Polybioscience), and observed under a JEOL JEM 2100, 200-kV transmission electron microscope (TEM) at a resolution of 0.1 to 0.2 μm (31).
Fluorescence microscopy.
Fresh cells of E. coli prepared in PBS, as mentioned earlier, and 100-μl aliquots were treated with the AMP at 2× MIC for 30 min at 37°C. Cells were stained with fluorescent dyes according to the procedure described previously (32) with a few modifications. After treatment, cells were collected by centrifugation (6,000 × g, 10 min), resuspended in 100 μl of PBS containing 5 μl of propidium iodide (PI; 1 mg/ml; Invitrogen), and incubated for 30 min at room temperature or 37°C. Cells were washed three or four times with PBS to remove unbound dye. Slides were prepared by fixing cells on microscopic glass slides by air drying and then washing with PBS to remove any traces of the dye. Mowiol-DABCO antifade mounting medium (Sigma, USA) was used to mount the coverslip over the slide before visualization. An Evos fluorescence microscope (Life Technologies, USA) was used for imaging. Control slides of untreated cells were prepared and observed under the same conditions.
BALB/c thigh infection model.
Male BALB/c mice, 6 to 8 weeks old, weighing 18 to 24 g, were used in this study. All mice were rendered neutropenic by an intraperitoneal (i.p.) injection of 150 and 100 mg/kg of body weight cyclophosphamide (Sigma, USA) on day 0 and day 3, respectively, and infection was induced on the fourth day. The right thigh of mice was injected with S. aureus MTCC 96 (105, 106). Groups of mice (n = 6) were treated after 1 h of infection with a dose of 20, 40, 60, 80, or 100 mg/kg via single subcutaneous (s.c.) injection. While mice without infection and no AMP were considered negative controls, mice with infection but no AMP treatment served as positive controls to test the effect of the AMP. At 18 h postinoculation, mice were humanely sacrificed by cervical dislocation and thighs were harvested. The thighs were weighed and homogenized with a microelectric homogenizer (Thermo Fisher, USA) at 2,000 rpm for 5 min in 5 ml of PBS. To determine the bacterial load in the thighs, the homogenate was serially diluted in PBS and aliquots (50 μl) of each dilution were plated on Trypticase soy broth (TSB; Himedia, India) plates. The plates were incubated for 18 h at 37°C, and bacterial colonies were counted (33).
Survival studies.
Adult male BALB/c mice weighing 18 to 24 g were used for the survival studies. All mice were rendered neutropenic by i.p. injections of 150 and 100 mg/kg cyclophosphamide (Sigma, USA) on days 0 and 3, respectively, and infection was induced on the fourth day. Mice were grouped (n = 6) into negative controls (uninfected and no AMP but injected with PBS), positive controls (infected with S. aureus MTCC 96 but no AMP), and the test group. Bacteremia was produced in mice via an i.p. injection of 100 μl of bacterial suspension containing 105 to 106 CFU of S. aureus MTCC 96. After inoculation, the mice received peptide treatment (up to 100 mg/kg) i.p., once a day for the first day and twice daily for the next 3 days. Mice that survived 12, 24, or 72 h or 7 days after treatment were recorded, and the survival rate was calculated as described earlier (33). All experiments were carried out in a biosafety level 2 facility animal house at our institute. All animal protocols followed the National Regulatory Guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Ministry of Environment and Forests, Government of India.
Statistical analyses.
All comparisons were based on the mean ± standard deviation of the mean (SD). Parametric data were analyzed using two-way analysis of variance (ANOVA) with the Bonferroni posttest method for comparison between groups. Column statistics for nonparametric data were analyzed by using the D'Agostino and Pearson omnibus normality test along with a one-sample t test. Results were considered significant when P was <0.05 in experiments. All experiments were performed independently three times in triplicate.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence for the new AMP was submitted to the EMBL database and assigned accession number HG003584. The penisin biosynthetic cluster was assigned number KT934325, and the genome sequence was assigned number JTHN00000000.
RESULTS
Isolation, identification, and characterization of strain A3.
Strain A3 was isolated from a soil sample from the rhizosphere, and the antimicrobial substance produced inhibited growth of both Gram-positive and Gram-negative indicator strains. Phenotypic characterization of the strain revealed it to be a Gram-positive, aerobic, rod-shaped bacterium with the ability to form an endospore. It produced shiny and sticky colonies on nutrient agar and grew between pH 5 and 9, up to 2% NaCl, and 50°C. However, optimum growth was observed at pH 7.2 and a 30°C temperature. It showed a positive reaction for nitrate reduction, methyl red test, and esculin hydrolysis, but it could not hydrolyze starch, casein, or gelatin. A negative reaction was observed for Voges-Proskauer, indole, catalase, and oxidase tests, and no growth was observed on Simmon's citrate agar. The sugar utilization pattern revealed the ability of strain A3 to utilize sucrose or mannose as the sole carbon source but not arabinose or mannitol. Nevertheless, strain A3 was identified as a member of the genus Paenibacillus based on its 16S rRNA gene sequence and subsequent phylogenetic analysis. It displayed 99.7% identity with P. ehimensis KCTC 3748, followed by P. elgii SD17 (99.2%) and P. koreensis YC300 (98.8%); however, it formed a clade with P. elgii and a coherent cluster with the closest species in a neighbor-joining phylogenetic tree (see Fig. S1 in the supplemental material). The 16S rRNA gene sequence identity was much lower for other species of the genus Paenibacillus. Membrane fatty acid composition revealed the presence of C15:0 and aiC15:0 as major fatty acids (34). The polar lipid profile was predominated by phosphatidylglycerol (PG) and diphosphatidylethanolamine (DPG), and among the other lipids phosphatidylethanolamine (PE), unknown phospholipids, and unknown lipids were observed (see Fig. S2 in the supplemental material). The G+C content was estimated to be 53.6 mol%.
Production, purification, and characterization of the antimicrobial substance.
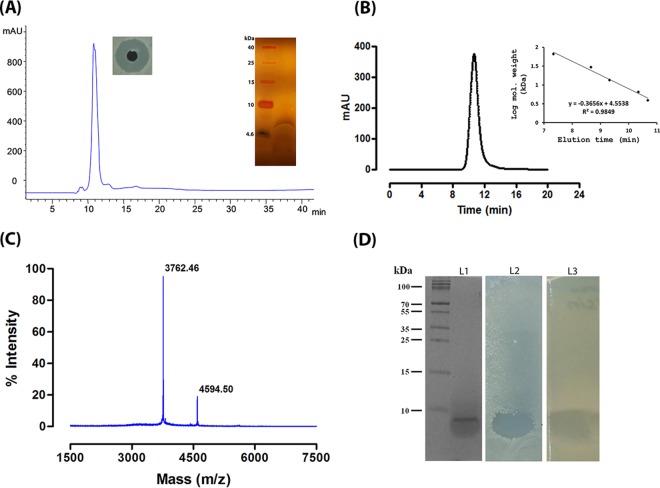
The antimicrobial production was observed at the late logarithmic phase of strain A3 growth and increased significantly thereafter. The CFB obtained after 24 h of growth inhibited all test strains of both Gram-positive and Gram-negative bacteria. No activity was observed against yeasts or fungus indicator strains. Though the antimicrobial substance was produced by strain A3 in minimal medium, nutrient broth was used as growth medium for optimal production. However, addition of different carbon and nitrogen sources to the growth media used for production of bacteriocin revealed that yeast extract and peptone increased the antimicrobial production. In contrast, a carbon source like glucose or lactose reduced the antimicrobial production. No change in pH was observed during or after the production of antimicrobial substances in the fermented broth. To obtain a larger quantity of antimicrobial substance, the CFB was collected after 48 h of incubation. The antimicrobial substance purified by Diaion HP20 affinity and size exclusion chromatography was found to be a bacteriocin-like peptide that was finally purified by reverse-phase HPLC. The purified peptide showed a single peak in the reverse-phase HPLC (Fig. 1A) and a sharp peak on a gel filtration column elution profile with an approximate molecular mass of 5 kDa (Fig. 1B). The bacteriocin-like peptide referred to as penisin displayed a molecular mass of 4,594.5 Da (Fig. 1C) based on MALDI-TOF analysis. The peptide yielded a single band with less than a 10-kDa molecular mass in gel electrophoresis, and a portion of the same gel showed antimicrobial activity in an in-gel activity assay (Fig. 1D) against E. coli MTCC 1610. The N-terminal sequencing of the peptide by Edman degradation yielded 4 amino acids in a continuous stretch. Antimicrobial activity of the purified peptide was not affected upon exposure to 60, 80, or 100°C, but a significant reduction was observed when incubated at 121°C for 15 min (see Fig. S3 in the supplemental material). Similarly, it was found to be stable between pH 2.0 and 12.0. Studies on tolerance of the peptide to proteolytic enzymes did not show any decrease in inhibition activity of penisin upon incubation for 6 h with trypsin and proteinase K.
FIG 1.
Purification, molecular mass determination, and in-gel activity assay of penisin. (A) Reverse-phase HPLC profile of penisin and silver stain gel of purified penisin after reverse-phase HPLC (inset shows the antimicrobial activity against S. aureus MTCC 1430). (B) Molecular mass determination on Shodex 802 HPLC column. The pensin elution profile suggested it is a monomer. (C) MALDI-TOF analysis of the purified peptide from Paenibacillus sp. strain A3 showed a mass of 4.5 kDa. (D) Tricine–SDS-PAGE and in-gel activity assay of purified penisin. Lanes: 1, molecular mass marker along with penisin; 2, portion of the SDS-PAGE gel overlaid with 0.8% soft agar containing E. coli MTCC 1610, showing zone of clearance; 3, portion of SDS-PAGE gel for nisin overlaid with 0.8% soft agar containing S. aureus MTCC 1430 showing zone of clearance. All electrophoresis and activity assays were performed with purified peptide dissolved in PBS.
Genetic characterization suggested penisin is a lantibiotic.
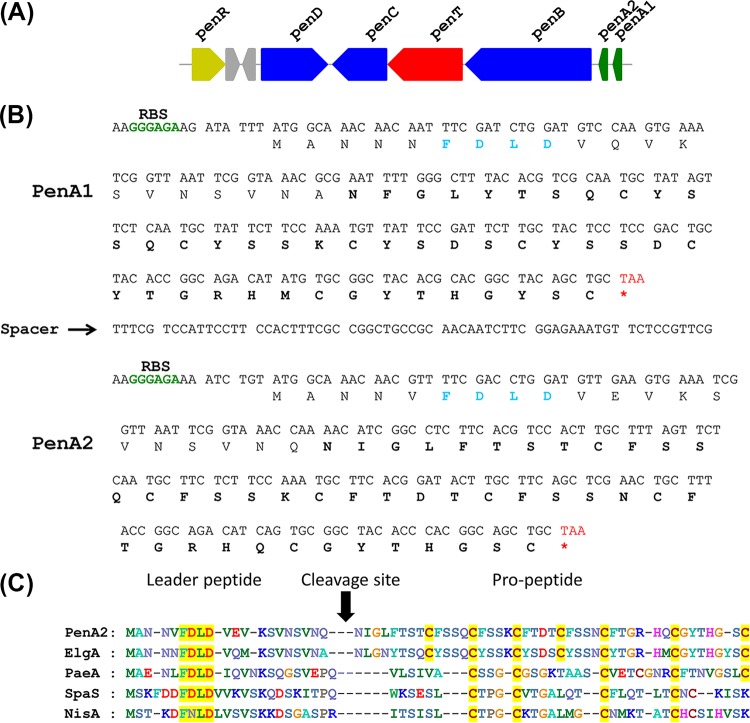
Aimed at identifying the antimicrobial peptide biosynthetic gene, we performed whole-genome sequencing of strain A3. The genome of A3 was checked for sequences of extrachromosomal (plasmid) origin by using Plasmid Finder and BLAST analysis, and the results showed no plasmid sequence in the genome. The reads assembled using a de novo approach produced 441 contigs. Analysis of the sequence using the RAST server predicted a total of 7,053 coding sequences (CDS) and 95 total RNAs (see Table S1 in the supplemental material). Since N-terminal sequencing of penisin yielded fewer amino acids, which also suggested ambiguities, we searched the genome sequence for the presence of bacteriocin-encoding genes. A putative lantibiotic biosynthetic gene cluster containing all genes involved in synthesis and maturation of a lantibiotic was identified. The annotated genome also resulted in the identification of a lantibiotic-like gene cluster containing six open reading frames (ORFs), designated penR, penD, penC, penT, penB, penA1, and penA2 (Fig. 2A). The total lantibiotic cluster in A3 is 9.8 kb (accession no. KT934325). Except for two genes, all other genes in the cassette are transcribed in the same orientation in the form of a cassette and are part of a single major contig. In silico analysis revealed the presence of two precursor lanthipeptide ORFs (structural genes penA1 and penA2). The deduced amino acid sequence of penA1 and penA2 displayed a similarity of 75% (Fig. 2B). However, the N-terminal sequence of the core peptide and molecular mass were identical with the deduced sequence of the penA2 gene. The translated sequence of structural gene penA1 (64 amino acids) showed high homology (94%) to elgicin (elgA), a predicted lantibiotic from Paenibacillus elgii B69. penA2 (63 amino acids) showed only 74% identity with elgicin, and it showed 49% identity to paenicidin, a highly cyclized lantibiotic produced by P. polymyxa (35). In addition to the structural gene (penA), the gene cluster encodes class Ia dehydratase (penB) and cyclase (penC) genes. While penB is responsible for dehydration of serine and threonine residues, the cyclase (penC) catalyzes the nucleophilic attack of a cysteine on the dehydrated residues. A dedicated lantibiotic ABC transporter gene (penT), which is required for the export of penisin, was also encoded with highly conserved domains such as Walker A (401-GRNGSGKTT-409), Q-loop (445-VVFQDFV-452), signature sequence (510-SGGQWQR-517), Walker B (529-LYVLDE-535), and H-loop (564-SHR-567), as described in the literature (36). Further, the genome also harbors an additional lantibiotic dehydratase gene similar to penD, which is unusual for the penisin gene cluster. Another gene, penR, was also annotated in the cluster, and this gene product might be involved in gene regulation by acting as a transcriptional regulator. The amino acid sequences of the four-gene product present in lantibiotic gene cluster penR-penD-penC-penT-penB revealed high levels of identity (92% to 99%) with homologous proteins from other reported lantibiotic gene clusters (see Table S1 in the supplemental material). The amino acid sequence of the penisin A2 precursor peptide was compared with that of other lantibiotics, including elgicin and nisin (Fig. 2C). This alignment revealed relatively higher homology in the leader peptides, with a conserved motif, FDLD, in all class Ia lantibiotics. The presence of genes encoding enzymes involved in posttranslational modifications also assigned penisin to the group class Ia lantibiotics.
FIG 2.
Penisin gene cluster, nucleotide sequence of the putative ORF with amino acid sequence, and sequence alignment with class Ia prelantibiotics. (A) The biosynthetic gene cluster of Paenibacillus sp. A3 consists ORFs penR, penD, penC, penT, penB, penA2, and penA1. The arrows indicate the relative directions of transcription. (B) Nucleotide sequence of the putative ORF encoding the penisin structural gene along with putative start and stop codons and the ribosome binding site (RBS). (C) Sequence alignment of the deduced prepenisin ORF (PenA2) with class Ia prelantibiotics of elgicin (ElgA), paenicidin (PaeA), subtilin (SpaS) and nisin (NisA). The conserved residues are shaded yellow, and the cleavage sites of the processing protease are indicated by the vertical solid arrow. The resulting propeptide of the cleaved PenA2 is shown. Penisin is a class Ia lantibiotic with the conserved FDLD motif in its leader peptide segment and the presence of the genes penB and penC.
Solution studies confirmed penisin as a random coil.
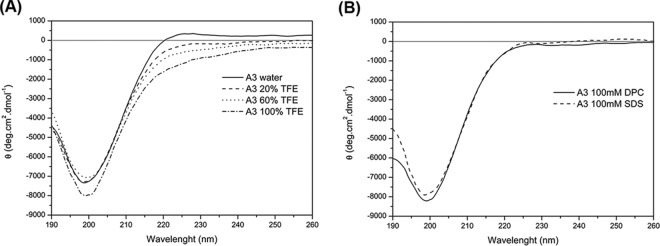
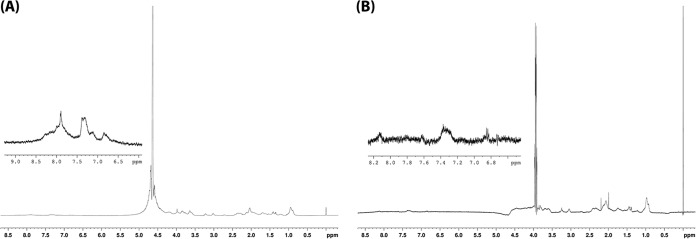
The CD spectrum of penisin showed a random structure in SDS, DPC, and TFE solutions at pH 4, 7 (Fig. 3A and B), and 11 that was characterized by negative values for the molar ellipticity (θ) for wavelengths shorter than 192 nm and a minimum close to 200 nm. Similar results was also observed for the water solution (Fig. 3A). The NMR analyses corroborated this random structure (Fig. 4A and B), based on the broadening of the signals and the loss of resolution mainly in the amide hydrogens region (δ 8.5 to 8.0).
FIG 3.
CD spectra of penisin under different conditions. (A) CD spectra of A3 peptide in water (solid line), 20% TFE (dashed line), 60% TFE (dotted line), and 100% TFE (dashed-dotted line). (B) CD spectra of 100 mM DPC (solid line) and 100 mM SDS (dashed line). All CD spectra were obtained at pH 7.0 in 10 mM Tris-HCl buffer.
FIG 4.
Penisin structure determination via NMR. (A) 1H NMR spectrum of the A3 peptide in water. (B) 1H NMR spectrum of the A3 peptide in TFE. The amide hydrogens region is highlighted (δ 8.5 to 8.0). All spectra were obtained at pH 7.0, in 10 mM Tris-HCl buffer at 37°C.
One cysteine is free in penisin.
The mass difference (108 Da) can be accounted for by a possible 6 dehydrations along with subsequent thioether bond formation in a characteristic posttranslational modification of lantibiotics. Moreover, we observed an additional strong signal at m/z 3,762 repeatedly during the MALDI analysis of penisin (Fig. 1C). The calculated mass of penisin with the additional 7-amino-acid residue deletion from the N teminus, past the leader peptide cleavage site, is 3,762, in agreement with the observed second peak in our MALDI data. Further, the involvement of six cysteine molecules in thioether bond formation was also confirmed in a PEGylation experiment; results for the electrophoretic mobility of native and PEGylated penisin molecules showed addition of a single PEG molecule (see Fig. S3 in the supplemental material). This suggests the presence of a free cysteine amino acid in the peptide and confirms the posttranslational modifications associated with the biosynthesis of penisin. However, dimerization of the penisin was not observed, based on the elution profile of the gel filtration column.
Penisin is a nonhemolytic antibacterial peptide.
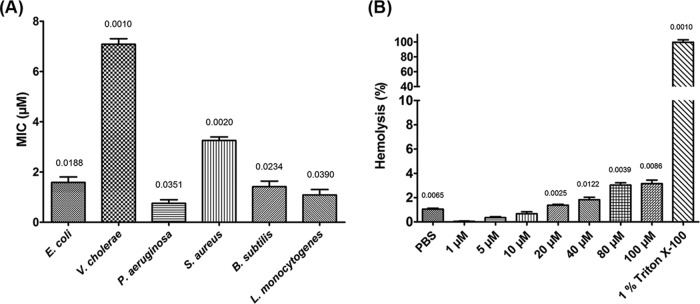
Penisin inhibited a range of Gram-positive and Gram-negative bacteria. The lowest MIC was observed against P. aeruginosa (Fig. 5A), followed by L. monocytogenes, a foodborne opportunistic pathogen. Among the Gram-negative strains tested, P. aeruginosa and E. coli were found to be highly sensitive and inhibited at 0.75 and 1.58 μM, respectively. No growth inhibition of yeast strains such as S. cerevisiae and C. albicans was observed. Results of hemolysis did not show any lysis of red blood cells (RBCs) (Fig. 5B), even at significantly high concentrations of the peptide.
FIG 5.
Determination of MICs and hemolysis activities of penisin in a microtiter plate assay performed in triplicate (n = 3). (A) MICs against Gram-positive and Gram-negative bacteria. (B) Hemolysis assay of penisin using rabbit RBCs. Purified peptide, bacterial cells, and RBC samples were prepared in PBS. Bars show SD. P values are indicated above each bar (significance was at a P level of <0.05).
Penisin causes alteration in cell membrane permeability.
In order to gain an insight into the inhibition activity of penisin, we have performed a series of microscopy experiments, including scanning electron microscopy, TEM, and fluorescence microscopy. The untreated E. coli cells for TEM images that were prepared in standard NB showed a smooth and intact shape (Fig. 6A); in contrast, indicator strains treated with a sublethal concentration of the lantibiotic penisin for 30 min showed alterations in morphological features. The changes included significant shrinkage of the cells, which seemed to be dehydration of the cell wall and cell membrane alterations, along with a disrupted periplasmic space. Cell surface roughening, along with multiple dents and blisters, was also observed after incubation for 60 min. Fluorescence microscopy was performed to get insights into the mechanism of action of penisin. Propidium iodide is a small (668.39-Da) red fluorescent nuclear and chromosome counterstain. Since propidium iodide is reported to be impermeant to live cells, it is also commonly used to detect dead cells in a population. Upon treatment with MIC multiples of penisin for 30 min, a uniform red fluorescence was observed in E. coli cells (Fig. 6B). However, no red fluorescence was observed in untreated cells, suggesting that the cell membrane was altered by penisin.
FIG 6.
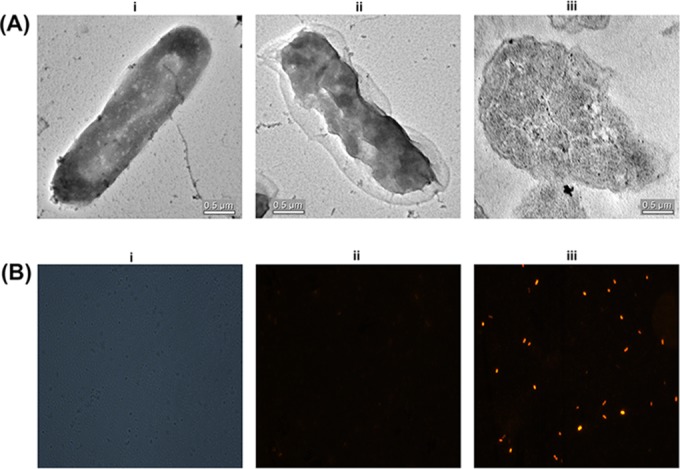
Determination of bactericidal effect of penisin on E. coli MTCC 1610. (A) Transmission electron microscopy without penisin treatment (i) or with penisin treatment (ii) in sections of an E. coli cell after penisin treatment. (B) Propidium iodide fluorescence microscopy phase-contrast micrograph of E. coli MTCC 1610 representing total number of bacteria (i), micrographs of E. coli MTCC 1610 cells without penisin treatment (ii) or after penisin treatment (4 μM) for 30 min. Bacterial samples for microscopy and purified peptide were prepared in PBS.
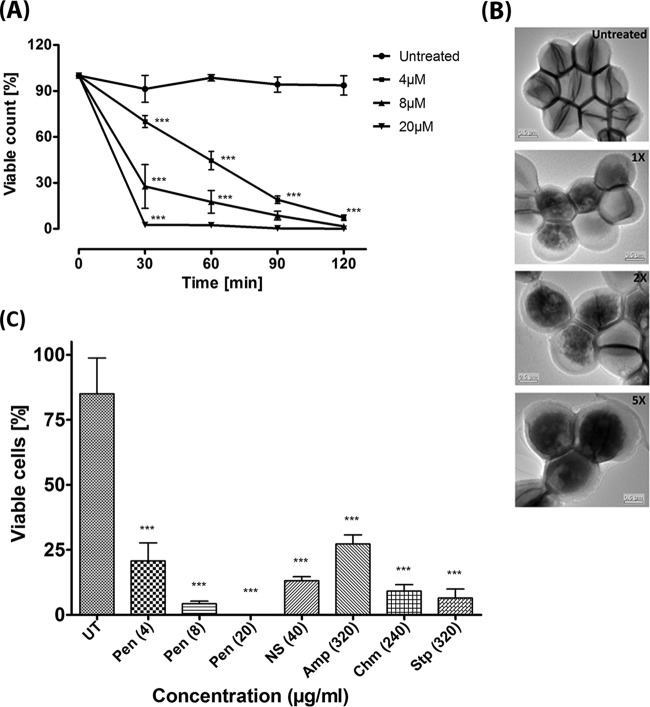
Penisin is effective against MRSA.
Penisin is effective against MRSA at 4 μM. Further, to determine the concentration and incubation time of penisin treatment, a killing kinetics experiment was performed. Less than 50% viability was observed after treatment with 1× MIC of penisin after incubation for 60 min, and only a 30-min incubation period was required for 100% killing of MRSA cells when penisin was used at 5× MIC (Fig. 7A). Results suggested that penisin is effective against MRSA in both a time-dependent as well as a concentration-dependent manner when used in the indicated range of multiples of the MIC. In fact, penisin is found to be much more effective than known antimicrobials like nisin, ampicillin, chloramphenicol, and streptomycin, as a low concentration (8 μg/ml) was found to be effective to reduce the number of viable cells of MRSA (Fig. 7B). TEM images confirmed the consistency of the killing kinetics assay results for a treatment time of 30 min (Fig. 7C). TEM result showed the shrinkage of cells upon treatment, most probably as a result of dehydration due to a compromised cell membrane. Interestingly, the absence of a division plate upon treatment with penisin suggested it is an inhibitor of cell division.
FIG 7.
Effect of penisin on MRSA and comparison with known antimicrobial agents. (A) Penisin-mediated killing kinetics of MRSA (n = 3). Symbols represent the viable cell counts of MRSA at different time intervals along with 1× (4 μM), 2× (8 μM), and 5× (20 μM) MIC of penisin. Error bars represent SD. All groups were compared with the untreated control for statistical significance. *, P < 0.05 in comparison to control. (B) TEM images of MRSA after treatment with penisin. (C) The viable cell percentage for penisin and other known antimicrobial agents. Error bars show SD (n = 3). *, P < 0.05 in comparison to the untreated control. NS, nisin; AMP, ampicillin; Chm, chloramphenicol; Stp, streptomycin. All bacterial samples and purified peptide were prepared in PBS.
In vivo properties of penisin.
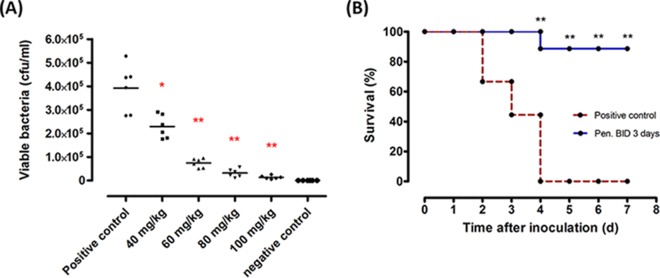
The in vivo efficacy of penisin was examined in a mouse model. A neutropenic thigh infection model was used to investigate the antimicrobial activity of penisin in vivo. Cyclophosphamide was used to render mice neutropenic, as this process decreases the influence of the host immune system on the initial inoculation of bacteria. The right thigh was inoculated with S. aureus (105 to 106 CFU). Treatments were initiated 1 h after inoculation via a single s.c. injection, and the bacterial load in the infected thighs was determined at 18 h postinoculation. The viable bacterial counts from infected thighs after treatment with penisin are shown in Fig. 8A; penisin was found significantly effective in reducing the bacterial load in a dose-dependent manner in treated mice compared to untreated mice (P < 0.05). Bacterial load was reduced more than 41% (P = 0.026) at a 40-mg/kg dose of penisin. Furthermore, at a 60-mg/kg dose, a more than 80% (P = 0.002) decrease was observed in bacterial load. Penisin was found to be significantly effective at the 80-mg/kg and 100-mg/kg doses, as bacterial load decreased up to 91% and 96%, respectively, compared to the negative control (P = 0.002 for both). Further, to confirm the protective efficacy of penisin against S. aureus MTCC 96, it was tested in vivo using a BALB/c bacteremia model. Bacteremia was produced in mice via an i.p. injection of bacterial suspension containing 105 to 106 CFU. This infection dose resulted in a gradual mortality increase over the first 3 days and 100% mortality rate by the fourth day after inoculation. The first penisin treatment was given 1 h after inoculation with bacteria, and further treatment was given two times a day until the fourth day of inoculation with bacteria. The mortality rate in penisin-treated animals was significantly reduced and no death was observed by the fourth day (Fig. 8B). Subsequently, the survival rate decreased to 88% and remained constant thereafter.
FIG 8.
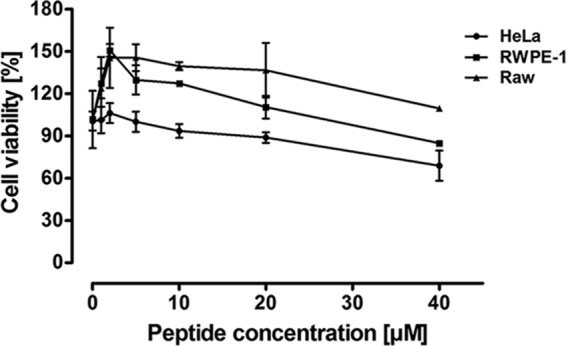
Cytotoxic effect of penisin on HeLa, RWPE-1, and RAW cell lines in the MTT assay. Cells were maintained in a humidified CO2 incubator at 37°C, and different concentrations of penisin were added after 24 h. A purified peptide sample was used for treatment in the growth medium used to grow cell lines. The percentage of cell viability was calculated as described in Materials and Methods. Results shown are means ± SD of three independent experiments performed in triplicate.
Mammalian cell toxicity.
In an attempt to test the toxicity of penisin against nucleated cells, we used mammalian cells of three different origins. Interestingly, penisin was found noncytotoxic up to 40 μM (Fig. 9), which is a significantly higher concentration than the MICs obtained for the different bacterial strains tested, including MRSA. More than 80% cell viability was observed in HeLa, RWPE1, and RAW cell lines in three independent experiments.
FIG 9.
Penisin reduces the bacterial load in a thigh infection model and confers protection against infection of mice with S. aureus MTCC 96 in a bacteremia model. (A) Viable bacterial counts in thigh infection model. (B) Survival of mice in a bacteremia model inoculated with S. aureus MTCC 96. Purified peptide samples for treatments were prepared in PBS. Results shown are means ±SD of three independent experiments performed in triplicate. *, P < 0.05 in comparison to the control.
DISCUSSION
Since bacterial resistance to antibiotics is increasing rapidly in pathogenic and opportunistic pathogenic bacteria, AMPs or bacteriocins are being studied as potential antimicrobials to replace antibiotics. As they involve a complicated mechanism of inhibitory action, bacteria have usually not shown resistance. Bacteriocins like nisin are approved by the World Health Organization for use as a food preservative (37). However, it is inactive against Gram-negative bacteria and does not inhibit the foodborne pathogens of the genera Salmonella and Yersinia (38). Antimicrobial peptides, including lipopeptides, lantibiotics, and other bacteriocins, are largely produced by species of the genera Bacillus and Paenibacillus. However, they have been explored in strains of only a few species. For example, P. polymyxa strain P13 produces polymyxin and a bacteriocin-like peptide (13) with activity against a wide range of bacteria. Another strain of P. polymyxa, NRRL-B-30509, produces a class IIa bacteriocin used to control Campylobacter spp. contamination in the poultry industry (39). Therefore, in the present study, an antimicrobial-producing strain designated A3 was isolated while screening for bacteriocin-producing strains from subsurface soil samples. Strain A3 was identified as a member of the genus Paenibacillus based on both 16S rRNA and rpoB gene sequences, and the major fatty acid composition was also in agreement with the genus Paenibacillus description (34, 40).The fermented broth of strain A3 inhibited the indicator strains of both Gram-positive and Gram-negative bacteria used in this study.
Different properties of the purified antimicrobial substance, including stability at higher temperatures, resistance to proteolytic enzymes, and low molecular mass, are in agreement with properties of bacteriocin-like peptides. Further, repeated N-terminal sequencing of this peptide yielded only a few amino acids, indicating the involvement of posttranslational modifications during its biosynthesis. Therefore, to identify the bacteriocin-like peptide, we used a whole-genome sequence analysis approach, because a similar method was used earlier by us as well as by others to characterize bacteriocins (12). Interestingly, RAST analysis revealed the presence of a lantibiotic biosynthetic gene that showed moderate similarity with a highly cyclized lantibiotic, paenicidin A (16). The theoretical molecular mass of the translated sequence of penisin (4,702.33 kDa) differed from the mass obtained by MALDI analysis (4,594.5 Da). Perhaps the posttranslational events involved in biosynthesis of penisin were supported by MALDI-MS of the peptide, as the difference in mass coincided with dehydration of six Ser or Thr molecules, which led to the formation of meso-lanthionine and methyllanthionine rings, respectively. Additionally, the presence of penB and penC genes that are involved in dehydration and cyclization, as well as the presence of cysteine, threonine, and serine residues at conserved positions that suggest they are substrates for the above-described gene products, confirmed the modifications involved in the synthesis of a mature active peptide, as observed for paenicidin A (38). In our annotation and analysis, we found a penC gene with 92% similarity to the LanC-like protein of P. elgii B69 with all conserved domains. Unlike paenicidin A, the ABC transporter present in the biosynthetic cluster of penisin is presumed to be involved in both transportation and immunity, as observed for other lantibiotics (36). To confirm the formation of the thioether bond between cysteine amino acids present in penisin, we performed PEGylation experiments with native and reduced forms of the peptide. Results showed that the addition of only single PEG molecules to both native and reduced forms of the peptide revealed the involvement of six cysteine residues in a thioether bond formation, and this was also supported by mass analysis.
The stability of the peptide at high temperatures and over a wide pH range starting from 2.0 to 12.0 in this study is also in agreement with other lantibiotics and posttranslational modifications (41). The results of CD and NMR analyses revealed it to be a random coil, which is in agreement with the other lantibiotics with multiple thioether bonds reported earlier (42–44). Thus, they do not exhibit any conformation-related obligations while present inside the host. The structures of several lantibiotics, like nisin, mutacin, epilancin, and leucocin A, have been elucidated using CD and NMR (45–51). Although antimicrobial peptides have attracted increasing attention over the last 2 decades, issues such as nonspecific cytotoxicity, hemolysis (7), and poor efficiency under in vivo conditions have limited their clinical utility (52). In contrast, penisin in this study showed neither hemolysis nor inhibition of growth of yeast or fungi, even at significantly higher concentrations, indicating its ability to discriminate cell membranes of prokaryotes and eukaryotes. Recently, it was shown that random coil peptides exhibit extremely low hemolytic activity compared to other peptides (53, 54). Primarily, most of the bacteriocins target the membrane and act by disrupting the membrane integrity (55), and also act as lytic agents (56). Similarly, electron and fluorescence microscopy experiments revealed penisin to be a membrane-active bacteriocin. Further, since AMPs with higher activity in vitro have been shown to be ineffective under in vivo conditions (57), we tested the efficacy of penisin against S. aureus, using thigh infection and bacteremia in a mouse model with BALB/c mice and an appropriate incubation period to allow the distribution of bacteria (58). Strikingly, results showed that penisin could efficiently protect 80% of the mice, which survived even after 8 days. It is pertinent to mention that treatment of mice with an 80-mg/kg dose of penisin demonstrated a significant reduction in the number of bacteria in thigh infection studies.
The MICs obtained for penisin were comparable with the MICs of other lantibiotics described in the literature (59). In accordance with several lantibiotics that have been shown to permeabilize the bacterial cytoplasmic membrane (60, 61), penisin in this study also showed disruption of membrane integrity in sensitive cells, and this was confirmed in electron and fluorescence microscopy experiments. Taken together, the results of our peptide characterization and animal studies lead us to conclude that penisin is a novel lantibiotic of class Ia with potential therapeutic applications.
In summary, synthetic peptide drugs produced by chemical synthesis are very expensive, so production of small AMPs naturally from bacteria is highly desirable and beneficial. Here we report the production of a lantibiotic by a rhizosphere soil isolate, identified as a new member of the genus Paenibacillus. The bacteriocin produced was found to be a novel lantibiotic with a broad antimicrobial spectrum and nonhemolytic properties. The antimicrobial activity of penisin was correlated with its ability to permeabilize bacterial membranes. Based on the striking physiochemical and biological findings, penisin has been found to be a potentially useful bacteriocin, and the antimicrobial-producing strain A3 exhibited applications in food preservation as a natural antimicrobial-producing strain that can preserve food by minimizing the metabolic capabilities of other bacteria.
Supplementary Material
ACKNOWLEDGMENTS
Financial assistance from the Department of Biotechnology (grant no. DBT/In-Bz/2013-16/16/SO-R1) is duly acknowledged and also the Council of Scientific and Industrial Research (CSIR).
We thank Sharanjeet Kaur for her help in MALDI analysis of the peptide, Paramjeet Kaur for her help in N-terminal sequencing of the peptide, and Subash Pawar for his help in electron microscopy. We also thank Rajesh Kumari, Department of Microbiology, Guru Nanak Dev University, for providing the methicillin-resistant S. aureus.
Funding Statement
Financial assistance from the Department of Biotechnology, Ministry of Science and Technology (DBT), India, and the Council of Scientific and Industrial Research (CSIR) is duly acknowledged.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01813-15.
REFERENCES
- 1.He Z, Kisla D, Zhang L, Yuan C, Green-Church KB, Yousef AE. 2007. Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl Environ Microbiol 73:168–178. doi: 10.1128/AEM.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh CT, Fischbach MA. 2009. New ways to squash superbugs. Sci Am 301:44–51. doi: 10.1038/scientificamerican0909-44. [DOI] [PubMed] [Google Scholar]
- 3.Huang Z, Hu Y, Shou L, Song M. 2013. Isolation and partial characterization of cyclic lipopeptide antibiotics produced by Paenibacillus ehimensis B7. BMC Microbiol 13:87. doi: 10.1186/1471-2180-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob Agents Chemother 43:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link A, Liu JW, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ökesli A, Cooper Fogle LE, van der Donk WA. 2011. Nine post-translational modifications during the biosynthesis of cinnamycin. J Am Chem Soc 133:13753–13560. doi: 10.1021/ja205783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock REW, Sahl H-G. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 8.Govaerts C, Orwa J, Van Schepdael A, Roets E, Hoogmartens J. 2002. Liquid chromatography-ion trap tandem mass spectrometry for the characterization of polypeptide antibiotics of the colistin series in commercial samples. J Chromatogr A 976:65–78. doi: 10.1016/S0021-9673(02)00375-8. [DOI] [PubMed] [Google Scholar]
- 9.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 10.Falagas ME, Kasiakou SK, Kofteridis DP, Roditakis G, Samonis G. 2006. Effectiveness and nephrotoxicity of intravenous colistin for treatment of patients with infections due to polymyxin-only-susceptible (POS) gram-negative bacteria. Eur J Clin Microbiol Infect Dis 25:596–599. doi: 10.1007/s10096-006-0191-2. [DOI] [PubMed] [Google Scholar]
- 11.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 12.Lohans CT, Vederas JC. 2014. Structural characterization of thioether-bridged bacteriocins. J Antibiot (Tokyo) 67:23–30. doi: 10.1038/ja.2013.81. [DOI] [PubMed] [Google Scholar]
- 13.Piuri M, Ruzal SM. 1998. A novel antimicrobial activity of a Paenibacillus polymyxa strain isolated from regional fermented sausages. Lett Appl Microbiol 27:9–13. doi: 10.1046/j.1472-765X.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 14.Stern NJ, Svetoch EA, Eruslanov BV, Kovalev YN, Volodina LI, Perelygin VV, Mitsevich EV, Mitsevich IP, Levchuk VP. 2005. Paenibacillus polymyxa purified bacteriocin to control Campylobacter jejuni in chickens. J Food Prot 68:1450–1453. [DOI] [PubMed] [Google Scholar]
- 15.Svetoch EA, Eruslanov BV, Perelygin VV, Levchuk VP, Seal BS, Stern NJ. 2010. Inducer bacteria, unique signal peptides, and low-nutrient media stimulate in vitro bacteriocin production by Lactobacillus spp. and Enterococcus spp. strains. J Agric Food Chem 58:6033–6038. doi: 10.1021/jf902802z. [DOI] [PubMed] [Google Scholar]
- 16.Wu X-C, Shen X-B, Ding R, Qian C-D, Fang H-H, Li O. 2010. Isolation and partial characterization of antibiotics produced by Paenibacillus elgii B69. FEMS Microbiol Lett 310:32–38. doi: 10.1111/j.1574-6968.2010.02040.x. [DOI] [PubMed] [Google Scholar]
- 17.von der Weid I, Alviano DS, Santos AL, Soares RM, Alviano CS, Seldin L. 2003. Antimicrobial activity of Paenibacillus peoriae strain NRRL BD-62 against a broad spectrum of phytopathogenic bacteria and fungi. J Appl Microbiol 95:1143–1151. doi: 10.1046/j.1365-2672.2003.02097.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin A., Camacho M, Portaels F, Palomino JC. 2003. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother 47:3616–3619. doi: 10.1128/AAC.47.11.3616-3619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smibert RM, Krieg NR. 1994. Phenotypic characterization, p 611–651. In Gerhardt P, Murray RGE, Wood WA, Krieg NR (ed), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 20.Suresh K, Mayilraj S, Chakrabarti T. 2006. Effluviibacter roseus gen. nov., sp. nov., isolated from muddy water, belonging to the family “Flexibacteraceae.” Int J Syst Evol Microbiol 56:1703–1707. [DOI] [PubMed] [Google Scholar]
- 21.Singh PK, Chittpurna Ashish Sharma V, Patil PB, Korpole S. 2012. Identification, purification and characterization of laterosporulin, a novel bacteriocin produced by Brevibacillus sp. strain GI-9. PLoS One 7:e31498. doi: 10.1371/journal.pone.0031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daba H, Pandian S, Gosselin JF, Simard RE, Huang J, Lacroix C. 1991. Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl Environ Microbiol 57:3450–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandal SM, Dey S, Mandal M, Sarkar S, Maria-Neto S, Franco OL. 2009. Identification and structural insights of three novel antimicrobial peptides isolated from green coconut water. Peptides 30:633–637. doi: 10.1016/j.peptides.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Kelly SM, Price NC. 2000. The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci 1:349–384. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- 25.Yoneyama F, Imura Y, Ohno K, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob Agents Chemother 53:3211–3217. doi: 10.1128/AAC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baindara P, Mandal SM, Chawla N, Singh PK, Pinnaka AK, Korpole S. 2013. Characterization of two antimicrobial peptides produced by a halotolerant Bacillus subtilis strain SK.DU.4 isolated from a rhizosphere soil sample. AMB Express 3:2. doi: 10.1186/2191-0855-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schibli DJ, Nguyen LT, Kernaghan SD, Rekdal Ø Vogel HJ. 2006. Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures. Biophys J 91:4413–4426. doi: 10.1529/biophysj.106.085837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smolarczyk R, Cichoń T, Kamysz W, Głowala-Kosińska M, Szydło A, Szultka L, Sieroń AL, Szala S. 2010. Anticancer effects of CAMEL peptide. Lab Invest 90:940–952. doi: 10.1038/labinvest.2010.58. [DOI] [PubMed] [Google Scholar]
- 30.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother 54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raje M, Dhiman R, Majumdar S, Dass T, Dikshit KL, Kaur R. 2006. Charged nylon membrane substrate for convenient and versatile high resolution microscopic analysis of Escherichia coli and mammalian cells in suspension culture. Cytotechnology 51:111–117. doi: 10.1007/s10616-006-9027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcellini L, Giammatteo M, Aimola P, Mangoni ML. 2010. Fluorescence and electron microscopy methods for exploring antimicrobial peptides mode(s) of action. Methods Mol Biol 618:249–266. doi: 10.1007/978-1-60761-594-1_16. [DOI] [PubMed] [Google Scholar]
- 33.Qian C-D, Wu X-C, Teng Y, Zhao W-P, Li O, Fang S-G, Huang Z-H, Gao H-C. 2012. Battacin (octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56:1458–1465. doi: 10.1128/AAC.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shida O, Takagi H, Knakamura LK, Komagata K. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298. doi: 10.1099/00207713-47-2-289. [DOI] [PubMed] [Google Scholar]
- 35.Lohans CT, Huang Z, van Belkum MJ, Giroud M, Sit CS, Steels EM, Zheng J, Whittal RM, McMullen LM, Vederas JC. 2012. Structural characterization of the highly cyclized lantibiotic paenicidin A via a partial desulfurization/reduction strategy. J Am Chem Soc 134:19540–19543. doi: 10.1021/ja3089229. [DOI] [PubMed] [Google Scholar]
- 36.Okuda K, Yanagihara S, Sugayama T, Zendo T, Nakayama J, Sonomoto K. 2010. Functional significance of the E loop, a novel motif conserved in the lantibiotic immunity ATP-binding cassette transport systems functional significance of the E loop, a novel motif conserved in the lantibiotic immunity ATP-binding cassette transport systems. J Bacteriol 192:2801–2808. doi: 10.1128/JB.00003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delves-Broughton J. 1990. Nisin and its application as a food preservative. Int J Dairy Technol 43:73–76. doi: 10.1111/j.1471-0307.1990.tb02449.x. [DOI] [Google Scholar]
- 38.Diep DB, Nes IF. 2002. Ribosomally synthesized antibacterial peptides in Gram positive bacteria. Curr Drug Targets 3:107–122. doi: 10.2174/1389450246055409. [DOI] [PubMed] [Google Scholar]
- 39.Svetoch E a. Stern NJ, Eruslanov BV, Kovalev YN, Volodina LI, Perelygin VV, Mitsevich EV, Mitsevich IP, Pokhilenko VD, Borzenkov VN, Levchuk VP, Svetoch OE, Kudriavtseva TY. 2005. Isolation of Bacillus circulans and Paenibacillus polymyxa strains inhibitory to Campylobacter jejuni and characterization of associated bacteriocins. J Food Prot 68:11–17. [DOI] [PubMed] [Google Scholar]
- 40.Ash C, Priest FG, Collins MD. 1993. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Van Leeuwenhoek 64:253–260. [DOI] [PubMed] [Google Scholar]
- 41.Georgalaki MD, Van den Berghe E, Kritikos D, Devreese B, Van Beeumen J, Kalantzopoulos G, De Vuyst L, Tsakalidou E. 2002. Macedocin, a food-grade lantibiotic produced by Streptococcus macedonicus ACA-DC 198. Appl Environ Microbiol 68:5891–5903. doi: 10.1128/AEM.68.12.5891-5903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Hooven HW, Rollema HS, Siezen RJ, Hilbers CW, Kuipers OP. 1997. Structural features of the final intermediate in the biosynthesis of the lantibiotic nisin. Influence of the leader peptide. Biochemistry 36:14137–14145. [DOI] [PubMed] [Google Scholar]
- 43.He Z, Yuan C, Zhang L, Yousef AE. 2008. N-terminal acetylation in paenibacillin, a novel lantibiotic. FEBS Lett 582:2787–2792. doi: 10.1016/j.febslet.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Escano J, Stauffer B, Brennan J, Bullock M, Smith L. 2014. The leader peptide of mutacin 1140 has distinct structural components compared to related class I lantibiotics. Microbiologyopen 3:961–972. doi: 10.1002/mbo3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Den Hooven HW, Doeland CC, Van De Kamp M, Konings RN, Hilbers CW, Van De Ven FJ. 1996. Three-dimensional structure of the lantibiotic nisin in the presence of membrane-mimetic micelles of dodecylphosphocholine and of sodium dodecylsulphate. Eur J Biochem 235:382–393. doi: 10.1111/j.1432-1033.1996.00382.x. [DOI] [PubMed] [Google Scholar]
- 46.Seco JM, Quiñoá E, Riguera R. 2004. The assignment of absolute configuration by NMR. Chem Rev 104:17–118. doi: 10.1021/cr000665j. [DOI] [PubMed] [Google Scholar]
- 47.Mota-Meira M, Lacroix C, LaPointe G, Lavoie MC. 1997. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett 410:275–279. doi: 10.1016/S0014-5793(97)00425-0. [DOI] [PubMed] [Google Scholar]
- 48.Krull RE, Chen P, Novak J, Kirk M, Barnes S, Baker J, Krishna NR, Caufield PW. 2000. Biochemical structural analysis of the lantibiotic mutacin II. J Biol Chem 275:15845–15850. doi: 10.1074/jbc.275.21.15845. [DOI] [PubMed] [Google Scholar]
- 49.Van de Kamp M, Horstink LM, van den Hooven HW, Konings RN, Hilbers CW, Frey A, Sahl HG, Metzger JW, van de Ven FJ. 1995. Sequence analysis by NMR spectroscopy of the peptide lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Eur J Biochem 227:757–771. doi: 10.1111/j.1432-1033.1995.tb20199.x. [DOI] [PubMed] [Google Scholar]
- 50.Fregeau Gallagher NL, Sailer M, Niemczura WP, Nakashima TT, Stiles ME, Vederas JC. 1997. Three-dimensional structure of leucocin a in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062–15072. doi: 10.1021/bi971263h. [DOI] [PubMed] [Google Scholar]
- 51.Freund S, Jung G, Gutbrod O, Folkers G, Gibbons WA, Allgaier H, Werner R. 1991. The solution structure of the lantibiotic gallidermin. Biopolymers 31:803–811. doi: 10.1002/bip.360310626. [DOI] [PubMed] [Google Scholar]
- 52.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. 1996. Membrane pores induced by magainin. Biochemistry 35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 53.Hong J, Oren Z, Shai Y. 1999. Structure and organization of hemolytic and nonhemolytic diastereomers of antimicrobial peptides in membranes. Biochemistry 38:16963–16973. doi: 10.1021/bi991850y. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Schlamadinger DE, Kim JE, McCammon JA. 2012. Comparative molecular dynamics simulations of the antimicrobial peptide CM15 in model lipid bilayers. Biochim Biophys Acta 1818:1402–1409. doi: 10.1016/j.bbamem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Héchard Y, Sahl HG. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545–557. doi: 10.1016/S0300-9084(02)01417-7. [DOI] [PubMed] [Google Scholar]
- 56.Martínez-Cuesta MC, Requena T, Peláez C. 2006. Permeabilization and lysis induced by bacteriocins and its effect on aldehyde formation by Lactococcus lactis. Biotechnol Lett 28:1573–1580. doi: 10.1007/s10529-006-9131-6. [DOI] [PubMed] [Google Scholar]
- 57.Hoffmann R, Bulet P, Urge L, Otvös L. 1999. Range of activity and metabolic stability of synthetic antibacterial glycopeptides from insects. Biochim Biophys Acta - Gen Subj 1426:459–467. doi: 10.1016/S0304-4165(98)00169-X. [DOI] [PubMed] [Google Scholar]
- 58.Noto PB, Abbadessa G, Cassone M, Mateo GD, Agelan A, Wade JD, Szabo D, Kocsis B, Nagy K, Rozgonyi F, Otvos L. 2008. Alternative stabilities of a proline-rich antibacterial peptide in vitro and in vivo. Protein Sci 17:1249–1255. doi: 10.1110/ps.034330.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Draper L a. Cotter PD, Hill C, Ross RP. 2013. The two peptide lantibiotic lacticin 3147 acts synergistically with polymyxin to inhibit Gram negative bacteria. BMC Microbiol 13:212. doi: 10.1186/1471-2180-13-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thennarasu S, Tan A, Penumatchu R, Shelburne CE, Heyl DL, Ramamoorthy A. 2010. Antimicrobial and membrane disrupting activities of a peptide derived from the human cathelicidin antimicrobial peptide ll37. Biophys J 98:248–257. doi: 10.1016/j.bpj.2009.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson RC, Hancock REW, Yu PL. 2004. Antimicrobial activity and bacterial-membrane interaction of ovine-derived cathelicidins. Antimicrob Agents Chemother 48:673–676. doi: 10.1128/AAC.48.2.673-676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.