Abstract
Isavuconazole is a novel, broad-spectrum, antifungal azole. In order to evaluate its interactions with known azole resistance mechanisms, isavuconazole susceptibility among different yeast models and clinical isolates expressing characterized azole resistance mechanisms was tested and compared to those of fluconazole, itraconazole, posaconazole, and voriconazole. Saccharomyces cerevisiae expressing the Candida albicans and C. glabrata ATP binding cassette (ABC) transporters (CDR1, CDR2, and CgCDR1), major facilitator (MDR1), and lanosterol 14-α-sterol-demethylase (ERG11) alleles with mutations were used. In addition, pairs of C. albicans and C. glabrata strains from matched clinical isolates with known azole resistance mechanisms were investigated. The expression of ABC transporters increased all azole MICs, suggesting that all azoles tested were substrates of ABC transporters. The expression of MDR1 did not increase posaconazole, itraconazole, and isavuconazole MICs. Relative increases of azole MICs (from 4- to 32-fold) were observed for fluconazole, voriconazole, and isavuconazole when at least two mutations were present in the same ERG11 allele. Upon MIC testing of azoles with clinical C. albicans and C. glabrata isolates with known resistance mechanisms, the MIC90s of C. albicans for fluconazole, voriconazole, itraconazole, posaconazole, and isavuconazole were 128, 2, 1, 0.5, and 2 μg/ml, respectively, while in C. glabrata they were 128, 2, 4, 4, and 16 μg/ml, respectively. In conclusion, the effects of azole resistance mechanisms on isavuconazole did not differ significantly from those of other azoles. Resistance mechanisms in yeasts involving ABC transporters and ERG11 decreased the activity of isavuconazole, while MDR1 had limited effect.
INTRODUCTION
Invasive fungal infections (IFIs) caused by yeasts are an important cause of morbidity and mortality. Candida spp. are opportunistic fungal pathogens and a particular cause for concern in immunocompromised individuals (1). Candida infections may account for more than 70% of all IFIs (2). Candidemia is a significant problem in the intensive care unit, as it is associated with a crude mortality rate of 42.6% and high resource use (3).
Azoles constitute a class of drug commonly used for treating infections caused by Candida spp., and the Infectious Diseases Society of America recommends them as a primary treatment for candidemia in nonneutropenic patients (1). They are fungistatic in most cases and work by binding and inhibiting the enzyme lanosterol 14-α-sterol-demethylase (encoded by ERG11), which is involved in converting lanosterol into ergosterol (4, 5). As ergosterol is an essential component of the fungal cell membrane, disruption of its biosynthesis inhibits fungal growth.
Repeated exposure to azoles and drug pressure in Candida spp. can lead to the emergence of azole-resistant strains, thereby increasing the risk of treatment failure and breakthrough infections (6, 7). Fluconazole resistance is common in many isolates of Candida spp., but they can remain susceptible to other azoles; for example, C. krusei has intrinsic resistance to fluconazole but is susceptible to voriconazole (8).
Different mechanisms of azole resistance exist, and more than one mechanism can be present in azole-resistant strains (9, 10). A common mechanism of resistance is the increased expression of multiple drug resistance genes, which result in the overexpression of efflux pumps, consequently decreasing the drug concentration within the fungal cell (11–13).
ATP binding cassette (ABC) transporter overexpression is caused by the presence of gain-of-function (GOF) mutations in the transcriptional activator of CDR genes, TAC1 (14), while the overexpression of the major facilitator superfamily (MFS) transporter MDR1 involves GOF mutations in the multidrug resistance regulator gene MRR1 (15). Point mutations in ERG11 are also an important resistance mechanism, reducing the ability of azoles to interact with or bind at the enzyme's target site, thereby reducing the effectiveness of the drug (9, 16). ERG11 upregulation also can occur via mechanisms that include GOF mutations in transcription factors such as UPC2 (17), thereby decreasing the ability of azoles to inhibit its action.
Isavuconazonium sulfate is a water-soluble antifungal prodrug that is hydrolyzed rapidly into isavuconazole, an active triazole. The oral and intravenous (i.v.) formulations of the prodrug have been approved by the U.S. Food and Drug Administration for treating adults with invasive aspergillosis and invasive mucormycosis (18). In phase 2 studies, isavuconazonium was observed to have efficacy and safety comparable to that of once-daily fluconazole in the primary treatment of uncomplicated esophageal candidiasis (19). In vitro, isavuconazole is highly active against bloodstream isolates of Candida spp. and has demonstrated good efficacy in animal models of candidiasis (20–22). However, it is important to understand how isavuconazole interacts with known mechanisms of azole resistance and what patterns of cross-resistance it shares with other azoles. In this study, we assessed the possible role of isavuconazole as a substrate for yeast multidrug efflux transporters. We also evaluated the susceptibility profile of isavuconazole when cytochrome P450 proteins encoded by different ERG11 alleles are expressed in a heterologous yeast host. In addition, isavuconazole susceptibility of clinical strains of Candida spp. with different azole susceptibility profiles and known azole resistance mechanisms were evaluated and compared to other azoles (fluconazole, itraconazole, voriconazole, and posaconazole).
MATERIALS AND METHODS
Strains.
Saccharomyces cerevisiae isolates expressing Candida albicans ATP binding cassette (ABC) transporter genes CDR1 and CDR2, the C. albicans major facilitator BENr (MDR1), FLU1, and ERG11 alleles, and C. glabrata CDR1 and CDR2 (CgCDR1 and CgCDR2) were used in this study; these resistance-conferring alleles have been described elsewhere (23–25). The S. cerevisiae strain YKKB13 (MATa ura3-53 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 pdr5Δ::TRP1) was used for all transporter and ERG11 allele expression experiments with YEp24- and YEp51-derived plasmids, respectively (23, 26, 27).
Antifungal drugs.
Azole antifungal drugs were obtained as pure substances from pharmaceutical companies (fluconazole and voriconazole, Pfizer Inc., Groton, CT; itraconazole, Janssen Pharmaceuticals, Inc., Titusville, NJ; posaconazole, Schering-Plough, Kenilworth, NJ; isavuconazole, Basilea Pharmaceutica International Ltd., Basel, Switzerland).
Isavuconazole susceptibility profile of S. cerevisiae strains expressing drug resistance genes.
Susceptibility testing with S. cerevisiae was performed with broth microdilutions based on protocol M27-A3 of the Clinical and Laboratory Standards Institute (CLSI) (28). This protocol uses selective yeast medium (yeast nitrogen base with 2% added glucose) containing doubling dilutions of azoles (fluconazole, 128 to 0.0625 μg/ml; itraconazole, voriconazole, posaconazole, and isavuconazole, 8 to 0.0039 μg/ml) at 30°C for 48 h. We grew S. cerevisiae isolates for C. albicans ERG11 alleles expressed from plasmid YEp51 in YKKB13 in medium containing 1% galactose and 1% raffinose to induce ERG11 expression as reported previously (23). MICs were obtained with a microplate reader at a wavelength of 540 nm. Normalization was performed by allowing growth to 100% in the absence of drugs. The MIC was defined as the concentration of the drug needed to decrease fungal growth by at least 50%.
Isavuconazole susceptibility of clinical Candida species isolates and C. albicans mutants.
Sequential C. albicans and C. glabrata clinical isolates with increasing MICs for several azoles were obtained from the University Hospital Centre (Lausanne, Switzerland) between 1993 and 2004; their azole resistance mechanisms have been described elsewhere (14, 15, 29–32), and their origins were summarized (14, 15, 25, 29, 31–36) (Table 1).
TABLE 1.
Clinical isolates used in the study
| Clinical strain |
Species | Parent strain | Resistance mechanism/genotypea | Reference or source | |
|---|---|---|---|---|---|
| DSY no. | Alternative no. | ||||
| DSY281 | C23 | C. albicans | Related to DSY284 | WT | 32 |
| DSY284 | C39 | C. albicans | ERG11 (S405F), TAC1 (G980E)b | 32 | |
| DSY347 | C33 | C. albicans | Related to DSY348 and DSY289 | WT | 25 |
| DSY288 | C34 | C. albicans | ERG11 (S405F) | 25 | |
| DSY289 | C26 | C. albicans | ERG11 (S405F, Y132H), TAC1 (A736V) | 25, 29 | |
| DSY348 | C82 | C. albicans | ERG11 (S405F), TAC1 (A736V) | 25, 29 | |
| DSY290 | C27 | C. albicans | Related to DSY291, DSY292 | WT | 25 |
| DSY291 | C37 | C. albicans | ERG11 (G464S) (R467K) | 15, 25 | |
| DSY292 | C40 | C. albicans | ERG11 (G464S) (R467K, Y132H),b MRR1 (P683H)b | 15, 25 | |
| DSY294 | C43 | C. albicans | Related to DSY296 | WT | 14 |
| DSY296 | C56 | C. albicans | ERG11 (G464S), TAC1 (N977D) | 14 | |
| DSY2321 | C. albicans | Related to DSY2322 and DSY2323 | ERG11 (S405F)b | 29 | |
| DSY2322 | C. albicans | ERG11 (S405F), TAC1 (G980E) | 29 | ||
| DSY2323 | C. albicans | ERG11 (S405F), TAC1 (G980E) | 29 | ||
| DSY731 | C. albicans | Related to DSY732 and DSY735 | WT | 29 | |
| DSY732 | C. albicans | TAC1 (ΔM677)c | 29 | ||
| DSY735 | C. albicans | TAC1 (ΔM677), i (5L) | 29 | ||
| DSY544 | C. albicans | Related to DSY775 | WT | 32 | |
| DSY775 | C. albicans | ERG11 (G464S), TAC1 (G980W) | 32 | ||
| DSY2309 | C. albicans | Related to DSY750 and DSY751 | WT | 33 | |
| DSY750 | C. albicans | MRR1 (N378D)b | 33 | ||
| DSY751 | C. albicans | ERG11 (S405F), MRR1 (N378D)b | 33 | ||
| DSY2243 | C. albicans | Related to DSY2242 | ERG11 (S442F, R467K) | 32 | |
| DSY2242 | C. albicans | ERG11 (S442F, R467K), TAC1 (G980E) | 32 | ||
| DSY2284 | C. albicans | Related to DSY2285 | WT | 15, 36 | |
| DSY2285 | C. albicans | MRR1 (T896I), i (5L) | 15, 36 | ||
| DSY550 | C. albicans | Related to DSY551 | WT | 15 | |
| DSY551 | C. albicans | ERG11 (G464S, Y132H) | 33 | ||
| DSY520 | C. albicans | Related to DSY522 | TAC1 (N972D) | 32 | |
| DSY522 | C. albicans | ERG11 (G464S), TAC1 (N972D) | 32 | ||
| DSY2250 | C. albicans | Related to DSY2250 | ERG11 (S442F, G465S) | 32 | |
| DSY2251 | C. albicans | ERG11 (S442F, G465S), TAC1 (N972D) | 32 | ||
| DSY741 | C. albicans | Related to DSY742 | WT | 33 | |
| DSY742 | C. albicans | MRR1 (T360I) | 33–35 | ||
| DSY562 | C. glabrata | WT | 33 | ||
| DSY565 | C. glabrata | DSY562 | CgPDR1 (I280F) | 33–35 | |
| DSY1041 | C. glabrata | DSY1029 | Cgcdr1Δ | 34 | |
| DSY1612 | C. glabrata | DSY1029 | Cgcdr2Δ | 34 | |
| DSY1613 | C. glabrata | DSY1056 | Cgcdr1Δ, Cgcdr2Δ | 34 | |
| DSY529 | C. glabrata | WT | 31 | ||
| DSY530 | C. glabrata | DSY529 | CgPDR1 (E1083Q) | 31 | |
| DSY2324 | C. glabrata | WT | 31 | ||
| DSY2325 | C. glabrata | DSY2324 | Mutation petite (loss of mitochondrial DNA) | 31 | |
| DSY724 | C. glabrata | WT | This study | ||
| DSY726 | C. glabrata | DSY724 | WT | 31 | |
| DSY727 | C. glabrata | DSY726 | CgPDR1 (D876Y) | 31 | |
| DSY2270 | C. glabrata | WT | 31 | ||
| DSY2271 | C. glabrata | DSY2270 | CgPDR1 (D261G) | 31 | |
| DSY759 | C. glabrata | WT | 31 | ||
| DSY2268 | C. glabrata | DSY759 | CgPDR1 (S316I) | 31 | |
Mutations are indicated in parentheses. Amino acid changes at given codon numbers of the corresponding gene are given. i (5L), isochromosome 5L.
Heterozygous state.
Deletion of methionine at position 677 in TAC1.
Several C. albicans mutants lacking the major multidrug transporters involved in azole resistance were used in this study (37). DSY1054 (flu1Δ/Δ), DSY1021 (cdr1Δ/Δ, cdr2Δ/Δ, flu1Δ/Δ), and DSY1055 (flu1Δ/Δ, mdr1Δ/Δ) are equivalent to DCY2, DCY20, and DCY7, respectively, and have been described by Calabrese et al. (38). DSY1050 (cdr1Δ/Δ, cdr2Δ/Δ, mdr1Δ/Δ) is the parent of DSY1024 (cdr1Δ/Δ, cdr2Δ/Δ, flu1Δ/Δ, mdr1Δ/Δ) and has been described previously (39).
C. albicans mutants with the inactivation of known resistance mechanisms also were used in this study; they originated from strain DSY296 (12). This strain originally had a GOF mutation in TAC1 and a G464S mutation in ERG11, and both are homozygous in DSY296. The MICs for Candida clinical isolates and mutants were determined using the standard protocol (CLSI M27-A3) (28). Isolates were grown at 35°C for 48 h in RPMI 1640 medium (Sigma-Aldrich, Switzerland) with azole doubling dilutions, as described previously in this report. Quality controls included C. albicans ATCC 928, C. krusei ATCC 6258, C. tropicalis ATCC 750, C. parapsilosis ATCC 22019, and C. glabrata ATCC 930 strains, and these were tested with all azoles used in the study.
RESULTS
Susceptibility testing of S. cerevisiae containing drug resistance genes.
In order to test whether isavuconazole is a potential substrate for ABC and MFS transporters, drug susceptibility testing was carried out with S. cerevisiae containing specific transporter genes associated with drug resistance. Changes to the MICs of isavuconazole upon expression of a given transporter indicated whether this azole was a substrate for the expressed transporter.
The expression of ABC transporters, including CDR1, CDR2, CgCDR1, and CgCDR2, elevated the MICs of isavuconazole from 2- to 32-fold compared with the MICs of the recipient S. cerevisiae strain (Table 2). These values depended on the expressed ABC transporters. CDR1 expression had the greatest impact on MICs (32-fold increase), while CgCDR2 expression had the least impact (2-fold increase). Fluconazole and voriconazole showed the largest increases in MICs, ranging from 16- to 128-fold and 8- to 56-fold, respectively.
TABLE 2.
Activity of azoles in S. cerevisiae containing several Candida multidrug transporters
| S. cerevisiae strain | Resistance mechanism/genotype | MIC (μg/ml [fold differencea]) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | ||
| DSY390 | YEp24b in YKKB13 | 1 (1) | 0.0625 (1) | 0.25 (1) | 0.0156 (1) | 0.25 (1) |
| DSY415 | CDR1 in YKKB13 | 128 (128) | 8 (128) | 2 (8) | 4 (256) | 8 (32) |
| DSY417 | CDR2 in YKKB13 | 16 (16) | 0.5 (8) | 1 (4) | 0.125 (8) | 1 (4) |
| DSY416 | MDR1 in YKKB13 | 128 (128) | 0.0625 (1) | 0.0625 (0) | 0.5 (32) | 0.25 (1) |
| DSY426 | FLU1 in YKKB13 | 16 (16) | 0.0625 (1) | 0.125 (1) | 0.125 (8) | 0.125 (1) |
| DSY691 | CgCDR1 in YKKB13 | 64 (64) | 1 (16) | 1 (4) | 0.5 (32) | 2 (8) |
| DSY687 | CgCDR2 in YKKB13 | 128 (128) | 0.0625 (1) | 0.125 (1) | 0.5 (32) | 0.5 (2) |
Fold difference is relative to the level for DSY390 for each azole.
Yeast replicating vector (parent vector) into which drug resistance genes were cloned.
Expression of MFS transporters MDR1 and FLU1 did not increase the MICs of isavuconazole, which was a characteristic shared with itraconazole and posaconazole (Table 2). In contrast, MDR1 and FLU1 expression increased the MICs of fluconazole by 128-fold and 16-fold, respectively, and those of voriconazole by 32-fold and 8-fold, respectively. These results indicated that, in contrast to isavuconazole, fluconazole and voriconazole were substrates for these MFS transporters.
Ten S. cerevisiae strains, each expressing an ERG11 allele from different C. albicans clinical isolates, were tested for their susceptibility to isavuconazole and other azoles and were compared to a strain carrying a wild-type (WT) allele. The ERG11 alleles used in this study carried nine distinct single or combined mutations that are known to be involved in azole resistance (23). Isavuconazole MICs ranged from 0.125 to 1 μg/ml. The highest increase for isavuconazole was between 4- and 8-fold and was associated with the substitution Y132H alone or combined with the substitution S405F or G464S.
When C. albicans ERG11 mutant alleles were expressed in S. cerevisiae, increases in azole MICs relative to those of the control strain were observed for fluconazole (4- to 32-fold) and voriconazole (1- to 16-fold) when at least two mutations were present in the same ERG11 allele (Table 3). Correlation coefficient analyses between the different azoles demonstrated that isavuconazole was more closely related to voriconazole and itraconazole than to fluconazole (see Fig. SA1 in the supplemental material). Interestingly, fluconazole MICs in S. cerevisiae showed the highest variations, followed by voriconazole MICs. No changes in posaconazole MICs were observed with any of the ERG11 mutant alleles expressed in S. cerevisiae.
TABLE 3.
Activity of azoles in S. cerevisiae containing C. albicans ERG11 alleles
| S. cerevisiae | Resistance mechanism/genotype | MIC (μg/ml [fold differencea]) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | ||
| DSY595b | YKKB13 and C. albicans ERG11 | 4 (1) | 0.25 (1) | 0.5 (1) | 0.125 (1) | 0.125 (1) |
| DSY1109 | YKKB13 and C. albicans ERG11 (G129A) | 4 (1) | 0.25 (1) | 0.5 (1) | 0.0156 (1) | 0.125 (1) |
| DSY949 | YKKB13 and C. albicans ERG11 (S405F) | 8 (2) | 0.25 (1) | 0.5 (1) | 0.125 (1) | 0.25 (2) |
| DSY952 | YKKB13 and C. albicans ERG11 (Y132H) | 8 (2) | 0.5 (2) | 0.25 (1) | 0.25 (2) | 0.5 (4) |
| DSY950 | YKKB13 and C. albicans ERG11 (G464S) | 8 (2) | 0.25 (1) | 0.5 (1) | 0.125 (1) | 0.125 (1) |
| DSY951 | YKKB13 and C. albicans ERG11 (R467K) | 8 (2) | 0.25 (1) | 0.5 (1) | 0.125 (1) | 0.25 (2) |
| DSY963 | YKKB13 and C. albicans ERG11 (S405F/Y132H) | 128 (32) | 1 (4) | 0.5 (1) | 2 (16) | 1 (8) |
| DSY965 | YKKB13 and C. albicans ERG11 (G464S/Y132H) | 32 (8) | 0.5 (2) | 0.5 (1) | 2 (16) | 1 (8) |
| DSY1107 | YKKB13 and C. albicans ERG11 (G464S/R467K) | 32 (8) | 0.25 (1) | 0.5 (1) | 0.25 (2) | 8 (4) |
| DSY1108 | YKKB13 and C. albicans ERG11 (G464S/G129A) | 15 (4) | 0.25 (1) | 0.5 (1) | 0.125 (1) | 0.25 (2) |
| DSY612 | YKKB13 and YEp51c | 1 (0.25) | 0.0625 (0.25) | 0.25 (0.5) | 0.0078 (0.0625) | 0.0625 (0.5) |
Fold difference is relative to the level for DSY595 for each azole.
Baseline strain.
Yeast replicating vector (parent vector) into which ERG11 alleles were cloned.
Susceptibility testing of Candida species isolates with known azole resistance mechanisms.
Our strain collection included a number of clinical isolates of C. albicans and C. glabrata with known and diverse resistance mechanisms. Paired isolates exhibited closely related genotypic patterns deduced from multilocus sequence typing (31, 40) (see Table SA1 in the supplemental material). Therefore, it was possible to compare azole MICs from the most susceptible isolates to those of the most resistant isolates. All quality controls were observed to have azole MICs within the accepted range (20, 41) (Table 4).
TABLE 4.
Azole MIC quality controls
| Reference strain | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | |
| C. albicans ATCC 928 | 0.125 | 0.0625 | 0.0078 | <0.0039 | 0.0078 |
| C. krusei ATCC 6258 | 32 | 0.125 | 0.125 | 0.125 | 0.5 |
| C. tropicalis ATCC 750 | 0.5 | 0.0078 | 0.0156 | 0.0078 | 0.0312 |
| C. parapsilosis ATCC 22019 | 2 | 0.0625 | 0.0625 | 0.0156 | 0.125 |
| C. glabrata ATCC 930 | 4 | 0.125 | 0.125 | 0.0312 | 0.125 |
Susceptibility testing of C. albicans clinical isolates.
The MIC range for isavuconazole (0.004 to 8 μg/ml) was similar to that for voriconazole (0.004 to 4 μg/ml) (Table 5). The MICs for fluconazole, itraconazole, and posaconazole ranged from 0.125 to 128, 0.016 to 2, and 0.016 to 4 μg/ml, respectively. The relative increase in MICs for each azole compared to that of a susceptible control ranged from 32- to 512-fold for fluconazole, voriconazole, and isavuconazole but from only 4- to 32-fold for itraconazole and 2- to 128-fold for posaconazole. The MIC90s for the matched clinical C. albicans isolates for fluconazole, voriconazole, itraconazole, posaconazole, and isavuconazole were 128, 2, 1, 0.5, and 2 μg/ml, respectively. This suggests that isavuconazole had activity similar to those of itraconazole, posaconazole, and voriconazole.
TABLE 5.
Activity of azoles in C. albicans clinical isolates with known resistance mechanisms
| C. albicans clinical strain | Resistance mechanism/genotype | MIC (μg/ml [fold differenceb]) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | ||
| DSY281 | WT | 0.250 (1) | 0.125 (1) | 0.031 (1) | 0.004 (1) | 0.016 (1) |
| DSY284 | ERG11 (S405F), TAC1 (G980E)a | 8 (32) | 0.25 (2) | 0.25 (8) | 0.063 (16) | 0.25 (16) |
| DSY347 | WT | 0.250 (1) | 0.125 (1) | 0.125 (1) | 0.004 (1) | 0.031 (1) |
| DSY288 | ERG11 (S405F) | 0.5 (2) | 0.125 (1) | 0.125 (1) | 0.031 (8) | 0.063 (2) |
| DSY289 | ERG11 (S405F, Y132H), TAC1 (A736V) | >128 (512) | 2 (16) | 0.5 (4) | 4 (1026) | 8 (256) |
| DSY348 | ERG11 (S405F), TAC1 (A736V) | 8 (32) | 1 (8) | 0.5 (4) | 0.063 (16) | 1 (32) |
| DSY290 | WT | 0.500(1) | 0.063 (1) | 0.063 (1) | 0.004 (1) | 0.008 (1) |
| DSY291 | ERG11 (G464S, R467K) | 2 (1) | 0.25 (4) | 0.063 (1) | 0.031 (8) | 0.031 (4) |
| DSY292 | ERG11 (G464S, R467K, Y132H),a MRR1 (P683H)a | 128 (256) | 1 (16) | 0.25 (4) | 0.5 (128) | 2 (256) |
| DSY294 | WT | 0.5 (1) | 0.016 (1) | 0.063 (1) | 0.004 (1) | 0.016 (1) |
| DSY296 | ERG11 (G464S), TAC1 (N977D) | 64 (128) | 0.125 (8) | 0.25 (4) | 0.5 (128) | 2 (128) |
| DSY2321 | ERG11 (S405F)a | <0.125 (1) | 0.031 (1) | 0.063 (1) | 0.004 (1) | <0.0039 (1) |
| DSY2322 | ERG11 (S405F), TAC1 (G980E) | 16 (128) | 1 (32) | 0.5 (8) | 0.125 (32) | 2 (513) |
| DSY2323 | ERG11 (S405F), TAC1 (G980E) | 32 (256) | 0.5 (16) | 0.5 (8) | 0.125 (32) | 2 (513) |
| DSY731 | WT | 0.250 (1) | 0.063 (1) | 0.031 (1) | 0.004 (1) | 0.008 (1) |
| DSY732 | TAC1 (ΔM677) | 16 (64) | 1 (16) | 0.5 (16) | 0.25 (64) | 4 (513) |
| DSY735 | TAC1 (ΔM677), i (5L) | 64 (256) | 0.5 (8) | 1 (32) | 0.25 (64) | 2 (256) |
| DSY544 | WT | 0.125 (1) | 0.063 (1) | 0.063 (1) | <0.0039 (1) | <0.0039 (1) |
| DSY775 | ERG11 (G464S), TAC1 (G980W) | 64 (512) | 1 (16) | 0.5 (8) | 0.5 (128) | 2 (513) |
| DSY2309 | WT | 2 (1) | 0.125 (1) | 0.125 (1) | 0.004 (1) | <0.0039 (1) |
| DSY750 | MMR1 (N378D)a | 16 (8) | 0.25 (2) | 0.125 (1) | 0.063 (16) | 0.063 (16) |
| DSY751 | ERG11 (S405F), MMR1 (N378D)a | 128 (64) | 0.5 (4) | 0.25 (2) | 0.125 (32) | 1 (256) |
| DSY2243 | ERG11 (S442F, R467K) | 0.25 (1) | 0.125 (1) | 0.063 (1) | 0.063 (1) | <0.0039 (1) |
| DSY2242 | ERG11 (S442F, R467K), TAC1 (G980E) | 8 (32) | 0.5 (4) | 0.5 (8) | 1 (16) | 1 (256) |
| DSY2284 | WT | 0.125 (1) | 0.063 (1) | 0.063 (1) | 0.004 (1) | 0.004 (1) |
| DSY2285 | MRR1 (T896I), i (5L) | 64 (512) | 0.25 (4) | 0.25 (4) | 0.125 (32) | 0.25 (64) |
| DSY550 | WT | 0.125 (1) | 0.016 (1) | 0.016 (1) | 0.004 (1) | 0.008 (1) |
| DSY551 | ERG11 (G464S, Y132H) | 64 (512) | 0.5 (32) | 0.5 (32) | 2 (513) | 2 (256) |
| DSY520 | TAC1 (N972D) | 16 (1) | 0.063 (1) | 0.5 (1) | 1 (1) | 0.250 (1) |
| DSY522 | ERG11 (G464S), TAC1 (N972D) | 128 (8) | 2 (32) | 1 (2) | 2 (2) | 2 (8) |
| DSY2250 | ERG11 (S442F, G465S) | 1 (1) | 0.125 (1) | 0.031 (1) | 0.004 (1) | 0.008 (1) |
| DSY2251 | ERG11 (S442F, G465S), TAC1 (N972D) | 32 (32) | 1 (8) | 4 (128) | 0.25 (64) | 4 (513) |
| DSY741 | WT | 0.125 (1) | 0.063 (1) | 0.125 (1) | 0.008 (1) | 0.008 (1) |
| DSY742 | MRR1 (T360I) | 16 (128) | 0.250 (4) | 0.125 (1) | 0.031 (4) | 0.125 (16) |
| DSY757 | WT | 0.5 (1) | 0.031 (1) | 0.063 (1) | <0.0039 (1) | <0.0039 (1) |
| DSY758 | ERG11 (G464S, F145L),a TAC1 (A736V) | 32 (64) | 0.5 (16) | 0.25 (4) | 0.125 (32) | 1 (256) |
Heterozygous state.
Fold difference is relative to the WT or to the most susceptible isolate for each related isolate group.
Isavuconazole MICs for each isolate were compared to those of other azoles, separating non-WT isolates from WT isolates, i.e., those with and without known azole resistance mechanisms (Fig. 1A). The upper isavuconazole MIC value for WT isolates was 0.031 μg/ml. This value is in agreement with another study which reported that 90% of C. albicans isolates exhibit isavuconazole MICs of ≤0.03 μg/ml (42). Correlation curves between isavuconazole MICs of WT and non-WT isolates and other azoles were established (Fig. 1). Isavuconazole MICs correlated with other azoles MICs in the following order of correlation strength: itraconazole < voriconazole < fluconazole < posaconazole. However, the analysis of relative MIC increases of matched isolates for each azole correlation coefficient demonstrated that the MIC profile of isavuconazole was related significantly to those of posaconazole and itraconazole, while profiles of fluconazole and voriconazole were significantly more similar to each other (see Fig. SA2 in the supplemental material).
FIG 1.
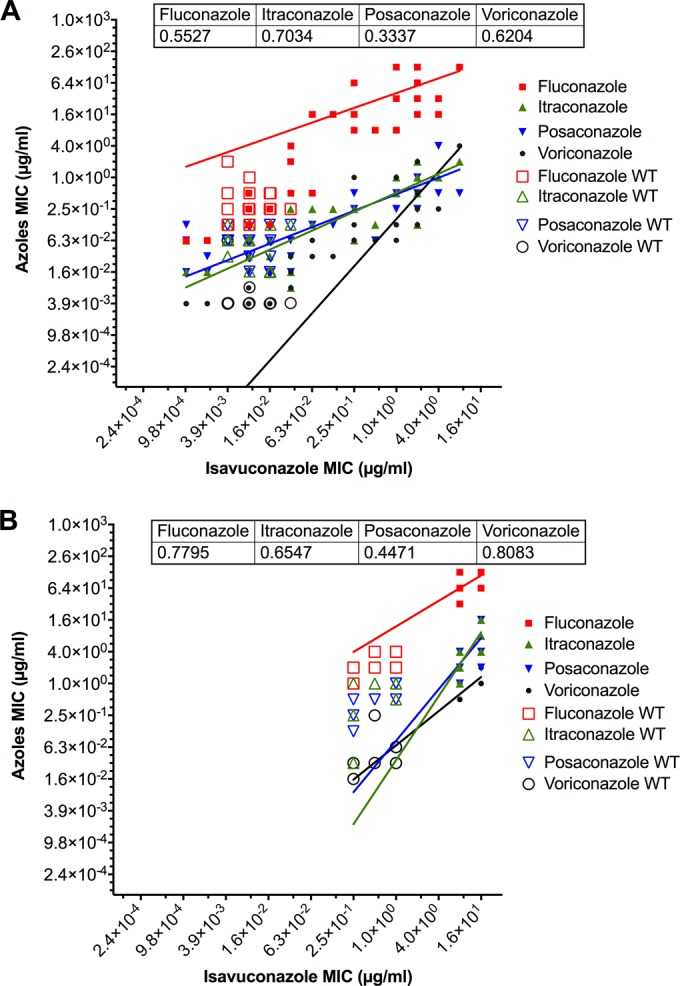
Relationship of isavuconazole MICs to those of other azoles for C. albicans isolates (MICs are from Tables 5, 7, and 8) (A) and C. glabrata isolates (MICs are from Table 6) (B). Correlations were calculated with nonlinear regression fits and log-log lines as an option using GraphPad Prism 6. The R2 values are indicated within figure captions. Wild-type (WT) isolates and non-WT isolates with resistance mechanisms are shown with empty and filled symbols, respectively.
Susceptibility testing of C. glabrata isolates.
The MIC range for isavuconazole (0.25 to 16 μg/ml) was similar to the range for posaconazole (0.125 to 16 μg/ml) (Table 6). The MIC ranges for fluconazole, itraconazole, and voriconazole were 1 to 128, 0.0312 to 16, and 0.0156 to 2 μg/ml, respectively. MIC90s of clinical isolates for fluconazole, voriconazole, itraconazole, posaconazole, and isavuconazole were 128, 2, 4, 4, and 16 μg/ml, respectively. These values were higher than those observed for the C. albicans isolates DSY565 and DSY2325 and were associated with the highest relative increase in MICs for isavuconazole (32-fold). Comparison of isavuconazole MICs from WT isolates and non-WT isolates revealed that the upper isavuconazole MIC value of WT isolates was 1.0 μg/ml (Fig. 1B). This value was equivalent to that observed in another study (42). Correlation curves between isavuconazole MICs of WT and non-WT isolates and other azoles highlighted that isavuconazole MICs were correlated with MICs of other azoles in the following order: voriconazole < fluconazole < itraconazole < posaconazole. When we analyzed correlation coefficients between relative MIC increases of matched isolates for each azole in C. glabrata, we observed that the profile of isavuconazole was related significantly only to that of fluconazole, in contrast to the case for C. albicans (see Fig. SA3 in the supplemental material).
TABLE 6.
Activity of azoles in C. glabrata clinical isolates with known resistance mechanisms
| C. glabrata clinical strain | Resistance mechanism/genotype | MIC (μg/ml [fold differencea]) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | ||
| DSY562 | WT | 2 (1) | 1 (1) | 0.5 (1) | 0.25 (1) | 0.5 (1) |
| DSY565 | CgPDR1 (I280F) | 64 (32) | 4 (4) | 2 (4) | 1 (4) | 16 (32) |
| DSY1041 | Cgcdr1Δ | 2 (1) | 1 (1) | 0.5 (1) | 0.0312 (0.12) | 1 (2) |
| DSY1612 | Cgcdr2Δ | 32 (16) | 1 (1) | 1 (2) | 0.5 (2) | 8 (16) |
| DSY1613 | Cgcdr1Δ, Cgcdr2Δ | 1 (0.5) | 0.0312 (0.03) | 0.125 (0.25) | 0.0156 (0.06) | 0.25 (0.5) |
| DSY529 | WT | 4 (1) | 1 (1) | 0.5 (1) | 0.0312 (1) | 0.5 (1) |
| DSY530 | CgPDR1 (E1083Q) | 64 (16) | 2 (2) | 2 (4) | 0.5 (32) | 8 (16) |
| DSY2324 | WT | 2 (1) | 1 (1) | 0.25 (1) | 0.0312 (1) | 0.25 (1) |
| DSY2325 | Mutation petite | 128 (64) | 2 (0.5) | 2 (8) | 1 (32) | 8 (32) |
| DSY724 | WT | 4 (1) | 0.5 (1) | 1 (1) | 0.0625 (1) | 1 (1) |
| DSY726 | WT | 2 (0.5) | 0.25 (0.5) | 0.5 (0.5) | 0.0312 (0.5) | 0.25 (0.25) |
| DSY727 | CgPDR1 (D876Y) | 128 (32) | 8 (16) | 4 (4) | 1 (16) | 16 (16) |
| DSY2270 | WT | 4 (1) | 1 (1) | 1 (1) | 0.0625 (1) | 1 (1) |
| DSY2271 | CgPDR1 (D261G) | 128 (32) | 16 (16) | 16 (16) | 2 (32) | 16 (16) |
| DSY759 | WT | 4 (1) | 1 (1) | 1 (1) | 0.0312 (1) | 1 (1) |
| DSY2268 | CgPDR1 (S316I) | 64 (16) | 4 (4) | 4 (4) | 0.5 (16) | 8 (8) |
Fold difference relative to the level for the WT for each related isolate group.
Susceptibility testing of C. albicans multidrug transporter mutants.
MICs for isavuconazole and other azoles were determined for C. albicans mutants lacking the major multidrug transporters involved in azole resistance in order to test their impact on drug resistance. The absence of CDR1 had the largest impact on azole MICs (Table 7). For isavuconazole, the MIC decreased from 0.0078 μg/ml in WT CAF2-1 to <0.001 μg/ml in the cdr1Δ/Δ mutant. The fluconazole MIC decreased from 0.25 to <0.0625 μg/ml in the absence of CDR1. MIC decreases in itraconazole and posaconazole were less pronounced, 0.0625 to 0.0156 μg/ml, when multiple transporters were deleted.
TABLE 7.
Activity of azoles in C. albicans multidrug transporter mutants
| C. albicans transporter mutant | Resistance mechanism/genotype | MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | ||
| CAF2-1 | URA3/ura3Δ | 0.25 | 0.0625 | 0.0625 | <0.0039 | 0.0078 |
| DSY448 | cdr1Δ/Δ | <0.0625 | 0.0625 | 0.0625 | <0.0039 | <0.001 |
| DSY465 | mdr1Δ/Δ | 0.25 | 0.0625 | 0.0625 | <0.0039 | 0.0078 |
| DSY468 | cdr1Δ/Δ mdr1Δ/Δ | <0.0625 | 0.0625 | 0.0156 | <0.0039 | <0.001 |
| DSY653 | cdr2Δ/Δ | 0.125 | 0.0625 | 0.0156 | <0.0039 | 0.0078 |
| DSY654 | cdr1Δ/Δ cdr2Δ/Δ | <0.0625 | 0.0156 | 0.0156 | <0.0039 | 0.002 |
| DSY1054 | flu1Δ/Δ | 0.25 | 0.0625 | 0.0625 | <0.0039 | 0.0156 |
| DSY1021 | cdr1Δ/Δ cdr2Δ/Δ flu1Δ/Δ | <0.0625 | 0.0625 | 0.0625 | <0.0039 | <0.001 |
| DSY1024 | cdr1Δ/Δ cdr2Δ/Δ flu1Δ/Δ mdr1Δ/Δ | <0.0625 | 0.0156 | 0.125 | <0.0039 | <0.001 |
| DSY1050 | cdr1Δ/Δ cdr2Δ/Δ mdr1Δ/Δ | <0.0625 | 0.0156 | 0.0312 | <0.0039 | 0.002 |
| DSY1055 | flu1Δ/Δ mdr1Δ/Δ | 0.25 | 0.0156 | 0.125 | <0.0039 | 0.0156 |
Susceptibility testing of C. albicans mutants with inactivation of known azole resistance mechanisms.
The DSY3706 derivative, which lacks TAC1 (the transcriptional activator of CDR1 and CDR2) and contains WT ERG11, had MICs for all azoles tested that were similar to those of DSY294, which is the susceptible parent strain (Table 8). Reintroduction of the TAC1 mutation in DSY3606-1 increased the MIC of fluconazole by approximately 32-fold. The largest increases in MICs for isavuconazole were observed in the DSY3606-1 (64-fold) and DSY296 (128-fold) strains. These strains contain TAC1 GOF mutations, suggesting that increases in CDR1 and CDR2 are associated with reduced susceptibility to isavuconazole. The G464S mutation in DSY296 resulted in the largest increases in MICs (128-fold) for fluconazole, voriconazole, and isavuconazole. The reintroduction of ERG11 WT alleles into the TAC1 deletion mutant DSY3083 resulted in MICs that were decreased by 16-fold (fluconazole) and 4-fold (voriconazole) in strain DSY3706.
TABLE 8.
Activity of azoles in C. albicans mutants reconstituting the azole-susceptible parent
| C. albicans ERG11/TAC1 mutant | Resistance mechanism/genotype | MIC (μg/ml [fold differencea]) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Itraconazole | Posaconazole | Voriconazole | Isavuconazole | ||
| DSY294 | WT | 0.5 (1) | 0.0156 (1) | 0.0156 (1) | <0.0039 (1) | 0.0156 (1) |
| DSY296 | ERG11 (G464S), TAC1 (N977D) | 64 (128) | 0.125 (8) | 0.25 (16) | 0.5 (128) | 2 (128) |
| DSY3082 | ERG11, tac1Δ/Δ | 0.5 (1) | 0.0156 (1) | 0.0156 (1) | 0.0078 (2) | 0.0312 (2) |
| DSY3083 | ERG11 (G464S), tac1Δ/Δ | 4 (8) | 0.0078 (0.5) | 0.0312 (2) | 0.0156 (4) | 0.0312 (2) |
| DSY3706 | tac1Δ/Δ, ERG11 | 0.25 (0.5) | 0.0156 (1) | 0.0156 (1) | <0.0039 (1) | 0.0156 (1) |
| DSY3606-1 | tac1Δ/Δ, ERG11, TAC1-5 (N977D) | 8 (32) | 0.125 (4) | 0.0625 (4) | 0.0625 (16) | 0.5 (64) |
| DSY3608-2 | tac1Δ/Δ, ERG11, TAC1-1 | 0.125 (0.5) | 0.0312 (1) | 0.0156 (1) | <0.0039 (1) | 0.0156 (2) |
| DSY3604 | tac1Δ/Δ, ERG11 (G464S)b | 0.5 (1) | 0.0312 (2) | 0.0312 (2) | 0.0078 (2) | 0.0078 (0.5) |
Fold difference relative to the level of the WT for each azole.
Heterozygous state.
DISCUSSION
This study assessed the mechanisms of isavuconazole resistance in Candida species using either S. cerevisiae as a vector for expressing specific azole resistance genes or various C. albicans and C. glabrata isolates with known resistance mechanisms to determine azole susceptibility. Isavuconazole had mechanisms of resistance similar to those of other azoles. Its range of activity was comparable to that of voriconazole in the Candida strains tested in this study.
The presence or absence of ABC transporters had the greatest effect on the MICs of isavuconazole as well as those of voriconazole, posaconazole, and itraconazole. The expression of CDR1, CDR2, CgCDR1, and CgCDR2 in S. cerevisiae resulted in elevated isavuconazole MICs compared to those of the controls, indicating that isavuconazole is a substrate for these transporters. This is particularly the case with the CDR1 and CgCDR1 transporters of C. albicans and C. glabrata, respectively. The expression of these transporters resulted in the largest increases in MICs for isavuconazole. Changes in MICs due to the expression of CDR genes were similar to those for isavuconazole, voriconazole, and itraconazole, although isavuconazole exhibited average MIC increases compared to those of other drugs.
MDR1 and FLU1 expression did not result in increases in MIC levels for isavuconazole, demonstrating that isavuconazole is not a substrate for MFS transporters. The opposite was observed for fluconazole and voriconazole, for which increases in MICs were linked to the expression of these transporters. This is supported in a study by Cheng et al. (43) in which MDR1 overexpression in a C. albicans petite mutant was associated with increased resistance to fluconazole and voriconazole, but the strain remained susceptible to itraconazole and ketoconazole.
The MIC increases for isavuconazole were moderate when associated with ERG11 mutations in the S. cerevisiae model. The MICs for fluconazole demonstrated the greatest variations, followed by those for voriconazole. A closer look at all azole susceptibility profiles indicated that the profile of fluconazole was distinct from those of isavuconazole and itraconazole. Interestingly, posaconazole MICs exhibited no changes with any of the mutant alleles expressed in S. cerevisiae. The largest effect on the MIC for isavuconazole was with S. cerevisiae strains expressing ERG11 alleles with the Y132H mutation either alone or combined with mutation S405F or G464S. These results are consistent with recent Erg11p crystallographic data published by Monk et al. (44), who posited that individual azole molecules interact with specific Erg11p residues depending on their structure. Therefore, we expect that ERG11 mutations do not have the same effect on the binding efficiency of specific azoles.
When testing C. albicans and C glabrata isolates, MIC changes for isavuconazole were similar to those observed for fluconazole, voriconazole, itraconazole, and posaconazole. In general, isavuconazole was more active in C. albicans than in C. glabrata (MIC ranges of 0.004 to 8 μg/ml and 0.25 to 16 μg/ml, respectively). This difference also has been observed in other studies (20, 45). C. albicans clinical isolate DSY289 had the highest isavuconazole MIC (8 μg/ml). This strain originally was isolated from an HIV-positive patient who had oropharyngeal candidiasis treated with fluconazole (25); this strain is associated with the ERG11 mutation S405F/Y132H and with the A736V mutation in TAC1. The TAC1 mutation is associated with increased CDR1 and CDR2 levels (29). This suggests that combined resistance mechanisms result in high resistance levels against isavuconazole and other azoles. This is consistent with data in the present study, in which the MICs of non-WT isolates were shifted to higher values than those of WT isolates. However, comparison of relative increases in MICs for fluconazole (512-fold) and voriconazole (1,026-fold) to that of isavuconazole (256-fold) indicates that the effect of a combined azole resistance mechanism was reduced on isavuconazole.
Increases in MICs for isavuconazole in C. glabrata clinical isolates were observed when the CgPDR1 GOF mutation was present in non-WT isolates. These mutations cause the upregulation of CgCDR1 and CgCDR2 (46, 47). Relative increases for isavuconazole ranged from 8- to 32-fold, similar to values reached by other azoles. However, the analysis of correlation coefficients between relative MIC increases by each azole in specific isolates highlighted that the susceptibility profile of isavuconazole was closely related to those of posaconazole and itraconazole in C. albicans but distinct from that of fluconazole. In C. glabrata this tendency was not verified, since the profile of isavuconazole was more closely related to that of fluconazole. The present study also reflects these features, as the MIC profile for isavuconazole was similar to the MIC profile for itraconazole in C. albicans and for the fluconazole profile in C. glabrata. Azole resistance mechanisms in C. albicans and C. glabrata are not equally distributed between the two species and therefore may have contributed to the observed susceptibility profile differences. On the other hand, isavuconazole exhibits a chemical structure that is different from those of the two structurally related groups of fluconazole/voriconazole and posaconazole/itraconazole (48). Therefore, isavuconazole might adopt susceptibility profiles that cannot be predicted simply from structural resemblance to the two known azole groups.
The absence of CDR1 from C. albicans mutants had the greatest effect on the MICs of azoles. The lowest MICs for both fluconazole and isavuconazole were associated with CDR1 knockout either on its own or in association with the deletion of other transporters. Isavuconazole and fluconazole had similar MIC profiles in these mutants, while decreases in MICs were less pronounced for itraconazole and posaconazole. The inactivation of known resistance mechanisms demonstrated that TAC1 had the greatest effect on isavuconazole, with its deletion resulting in decreases in MICs and its reintroduction increasing them.
This study suggests that isavuconazole follows the resistance patterns observed in other azoles, with slight variations depending on the investigated fungal species. Currently, there is a lack of data on isavuconazole susceptibility patterns among clinical Candida isolates. In an Egyptian epidemiology study, all strains of Candida isolated from 187 patients were susceptible to isavuconazole (range, < 0.016 to 1 μg/ml), while overall resistance to voriconazole was 2.5% in all strains (49). Another study demonstrated isavuconazole to be highly active against 296 Candida bloodstream isolates; the activity of isavuconazole was more potent than that of fluconazole against all organisms tested, and often it was more potent than that of itraconazole or voriconazole (20). It also was noted that only two isolates, both C. glabrata, had a MIC for isavuconazole of >0.5 μg/ml. In a study investigating approximately 1,400 Candida species, isavuconazole exhibited high activity. A few isolates with high isavuconazole MICs in this study (1 to 8 μg/ml) could not be analyzed for their cross-resistance to other azoles (42). C. albicans and C. glabrata isolates were directly compared by using isavuconazole MICs. Fluconazole- and voriconazole-resistant isolates corresponded to high isavuconazole MICs in C. albicans (0.12 to 1 μg/ml) and C. glabrata (1 to 8 μg/ml), which overlap values obtained in the study reported here (42).
Isavuconazole activity has been investigated in other fungal species with respect to a possible correlation between azole resistance and levels of isavuconazole MICs. One study on Aspergillus fumigatus highlighted that the acquisition of resistance to voriconazole and itraconazole due to Cyp51 mutations was followed by an increase of isavuconazole MICs compared to those of susceptible isolates (50). Thus, isavuconazole does not deviate from other azoles in the characteristic decrease of activity in the presence of azole resistance mechanisms.
In conclusion, isavuconazole, as a substrate of multidrug ABC transporters, has properties in common with other azoles. However, in contrast to fluconazole and voriconazole, isavuconazole is not a substrate for the MFS transporter MDR1 or FLU1. Isavuconazole was sensitive to mutations in ERG11, but these mutations had less impact on MIC increases in isavuconazole than they did on MIC increases for fluconazole and voriconazole. These mutations had minimal effect on MICs of posaconazole. Isavuconazole had an activity range similar to that of voriconazole in the Candida strains tested in this study.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by an investigation grant from Basilea Pharmaceutica AG, Basel, Switzerland, and partially by the Swiss National Research Foundation (31003A_146936/1).
Technical support was provided by Francoise Ischer. Editorial support was provided by Barrie J. Anthony (Envision Scientific Solutions), who is funded by Astellas Pharma Global Development, Inc.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02157-15.
REFERENCES
- 1.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. 2012. The PATH (Prospective Antifungal Therapy) Alliance registry and invasive fungal infections: update 2012. Diagn Microbiol Infect Dis 73:293–300. doi: 10.1016/j.diagmicrobio.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Kett DH, Azoulay E, Echeverria PM, Vincent JL. 2011. Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Care Med 39:665–670. doi: 10.1097/CCM.0b013e318206c1ca. [DOI] [PubMed] [Google Scholar]
- 4.Alcazar-Fuoli L, Mellado E. 2012. Ergosterol biosynthesis in Aspergillus fumigatus: its relevance as an antifungal target and role in antifungal drug resistance. Front Microbiol 3:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanafani ZA, Perfect JR. 2008. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis 46:120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 6.Borst A, Raimer MT, Warnock DW, Morrison CJ, Arthington-Skaggs BA. 2005. Rapid acquisition of stable azole resistance by Candida glabrata isolates obtained before the clinical introduction of fluconazole. Antimicrob Agents Chemother 49:783–787. doi: 10.1128/AAC.49.2.783-787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F. 2011. Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob Agents Chemother 55:532–538. doi: 10.1128/AAC.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuoka T, Johnston DA, Winslow CA, de Groot MJ, Burt C, Hitchcock CA, Filler SG. 2003. Genetic basis for differential activities of fluconazole and voriconazole against Candida krusei. Antimicrob Agents Chemother 47:1213–1219. doi: 10.1128/AAC.47.4.1213-1219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franz R, Kelly SL, Lamb DC, Kelly DE, Ruhnke M, Morschhauser J. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother 42:3065–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med 125:S3–S13. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. 2010. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res 38:536–545. doi: 10.1177/147323001003800216. [DOI] [PubMed] [Google Scholar]
- 12.MacCallum DM, Coste A, Ischer F, Jacobsen MD, Odds FC, Sanglard D. 2010. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob Agents Chemother 54:1476–1483. doi: 10.1128/AAC.01645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torelli R, Posteraro B, Ferrari S, La Sorda M, Fadda G, Sanglard D, Sanguinetti M. 2008. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol Microbiol 68:186–201. doi: 10.1111/j.1365-2958.2008.06143.x. [DOI] [PubMed] [Google Scholar]
- 14.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunkel N, Blass J, Rogers PD, Morschhauser J. 2008. Mutations in the multidrug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng LJ, Wan Z, Wang XH, Li RY, Liu W. 2010. Relationship between antifungal resistance of fluconazole resistant Candida albicans and mutations in ERG11 gene. Chin Med J 123:544–548. [PubMed] [Google Scholar]
- 17.Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhauser J, Rogers PD. 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 11:1289–1299. doi: 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astellas Pharma Inc. 2015. Cresemba (isavuconazonium sulfate) prescribing information. Astellas Pharma, Northbrook, IL: http://www.astellas.us/docs/cresemba.pdf. [Google Scholar]
- 19.Viljoen J, Azie N, Schmitt-Hoffmann AH, Ghannoum M. 2015. A phase 2, randomized, double-blind, multicenter trial to evaluate the safety and efficacy of three dosing regimens of isavuconazole compared with fluconazole in patients with uncomplicated esophageal candidiasis. Antimicrob Agents Chemother 59:1671–1679. doi: 10.1128/AAC.04586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seifert H, Aurbach U, Stefanik D, Cornely O. 2007. In vitro activities of isavuconazole and other antifungal agents against Candida bloodstream isolates. Antimicrob Agents Chemother 51:1818–1821. doi: 10.1128/AAC.01217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guinea J, Pelaez T, Recio S, Torres-Narbona M, Bouza E. 2008. In vitro antifungal activities of isavuconazole (BAL4815), voriconazole, and fluconazole against 1,007 isolates of zygomycete, Candida, Aspergillus, Fusarium, and Scedosporium species. Antimicrob Agents Chemother 52:1396–1400. doi: 10.1128/AAC.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majithiya J, Sharp A, Parmar A, Denning DW, Warn PA. 2009. Efficacy of isavuconazole, voriconazole and fluconazole in temporarily neutropenic murine models of disseminated Candida tropicalis and Candida krusei. J Antimicrob Chemother 63:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14alpha-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42:241–253. doi: 10.1093/jac/42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard D, Ischer F, Monod M, Bille J. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143(Part 2):405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 25.Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bille J. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother 39:2378–2386. doi: 10.1128/AAC.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altboum Z, Gottlieb S, Lebens GA, Polacheck I, Segal E. 1990. Isolation of the Candida albicans histidinol dehydrogenase (HIS4) gene and characterization of a histidine auxotroph. J Bacteriol 172:3898–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bissinger PH, Kuchler K. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem 269:4180–4186. [PubMed] [Google Scholar]
- 28.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, approved standard, 3rd ed Document M27-A3. CLSI, Wayne, PA. [Google Scholar]
- 29.Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d'Enfert C, Berman J, Sanglard D. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6:1889–1904. doi: 10.1128/EC.00151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coste A, Turner V, Ischer F, Morschhauser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, Rohde B, Bauser C, Bader O, Sanglard D. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 5:e1000268. doi: 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coste AT, Crittin J, Bauser C, Rohde B, Sanglard D. 2009. Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot Cell 8:1250–1267. doi: 10.1128/EC.00069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posteraro B, Martucci R, La Sorda M, Fiori B, Sanglard D, de Carolis E, Florio AR, Fadda G, Sanguinetti M. 2009. Reliability of the Vitek 2 yeast susceptibility test for detection of in vitro resistance to fluconazole and voriconazole in clinical isolates of Candida albicans and Candida glabrata. J Clin Microbiol 47:1927–1930. doi: 10.1128/JCM.02070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanglard D, Ischer F, Bille J. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob Agents Chemother 45:1174–1183. doi: 10.1128/AAC.45.4.1174-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob Agents Chemother 43:2753–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanglard D, Ischer F, Monod M, Bille J. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother 40:2300–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calabrese D, Bille J, Sanglard D. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146(Part 11):2743–2754. doi: 10.1099/00221287-146-11-2743. [DOI] [PubMed] [Google Scholar]
- 39.Marchetti O, Moreillon P, Entenza JM, Vouillamoz J, Glauser MP, Bille J, Sanglard D. 2003. Fungicidal synergism of fluconazole and cyclosporine in Candida albicans is not dependent on multidrug efflux transporters encoded by the CDR1, CDR2, CaMDR1, and FLU1 genes. Antimicrob Agents Chemother 47:1565–1570. doi: 10.1128/AAC.47.5.1565-1570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odds FC, Jacobsen MD. 2008. Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell 7:1075–1084. doi: 10.1128/EC.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barry AL, Pfaller MA, Brown SD, Espinel-Ingroff A, Ghannoum MA, Knapp C, Rennie RP, Rex JH, Rinaldi MG. 2000. Quality control limits for broth microdilution susceptibility tests of ten antifungal agents. J Clin Microbiol 38:3457–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castanheira M, Messer SA, Rhomberg PR, Dietrich RR, Jones RN, Pfaller MA. 2014. Isavuconazole and nine comparator antifungal susceptibility profiles for common and uncommon Candida species collected in 2012: application of new CLSI clinical breakpoints and epidemiological cutoff values. Mycopathologia 178:1–9. doi: 10.1007/s11046-014-9772-2. [DOI] [PubMed] [Google Scholar]
- 43.Cheng S, Clancy CJ, Nguyen KT, Clapp W, Nguyen MH. 2007. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob Agents Chemother 51:1855–1858. doi: 10.1128/AAC.00182-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JDA, Connell JD, Cannon RD, McDonald JG, Rodriguez A, Finer-Moore JS, Stroud RM. 2014. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc Natl Acad Sci U S A 111:3865–3870. doi: 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfaller MA, Castanheira M, Messer SA, Rhomberg PR, Jones RN. 2014. Comparison of EUCAST and CLSI broth microdilution methods for the susceptibility testing of 10 systemically active antifungal agents when tested against Candida spp. Diagn Microbiol Infect Dis 79:198–204. doi: 10.1016/j.diagmicrobio.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Vale-Silva L, Ischer F, Leibundgut-Landmann S, Sanglard D. 2013. Gain-of-function mutations in PDR1, a regulator of antifungal drug resistance in Candida glabrata, control adherence to host cells. Infect Immun 81:1709–1720. doi: 10.1128/IAI.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brun S, Berges T, Poupard P, Vauzelle-Moreau C, Renier G, Chabasse D, Bouchara JP. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob Agents Chemother48:1788–179610.1128/AAC.48.5.1788-1796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov 9:719–727. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- 49.Taj-Aldeen SJ, Kolecka A, Boesten R, Alolaqi A, Almaslamani M, Chandra P, Meis JF, Boekhout T. 2014. Epidemiology of candidemia in Qatar, the Middle East: performance of MALDI-TOF MS for the identification of Candida species, species distribution, outcome, and susceptibility pattern. Infection 42:393–404. doi: 10.1007/s15010-013-0570-4. [DOI] [PubMed] [Google Scholar]
- 50.Gregson L, Goodwin J, Johnson A, McEntee L, Moore CB, Richardson M, Hope WW, Howard SJ. 2013. In vitro susceptibility of Aspergillus fumigatus to isavuconazole: correlation with itraconazole, voriconazole, and posaconazole. Antimicrob Agents Chemother 57:5778–5780. doi: 10.1128/AAC.01141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


