Abstract
An interregional surveillance program was conducted in the northwestern part of France to determine the prevalence of carbapenem-nonsusceptible Enterobacteriaceae (CNSE) isolates and their susceptibility to ceftazidime-avibactam and aztreonam-avibactam combinations. Nonduplicate CNSE clinical isolates were prospectively collected from six hospitals between June 2012 and November 2013. MICs of ceftazidime and aztreonam, alone or combined with a fixed concentration of avibactam (4 μg/ml), and those of carbapenems (comparator agents) were determined. MICs of ertapenem in combination with phenylalanine arginine-naphthylamide dihydrochloride (PAβN) were also determined to assess active efflux. Genes encoding carbapenemases, plasmid-mediated AmpC enzymes, extended-spectrum β-lactamases (ESBLs), and major outer membrane proteins (OMPs) were amplified and sequenced. OMPs were also extracted for SDS-PAGE analysis. Among the 139 CNSE isolates, mainly Enterobacter spp. and Klebsiella pneumoniae, 123 (88.4%) were ertapenem nonsusceptible, 12 (8.6%) exhibited reduced susceptibility to all carbapenems, and 4 Proteeae isolates (2.9%) were resistant to imipenem. Carbapenemase production was detected in only two isolates (producing OXA-48 and IMI-3). In contrast, OMP deficiency, in association with AmpCs and/or ESBLs (mainly CTX-M-9, SHV-12, and CTX-M-15), was largely identified among CNSE isolates. The ceftazidime-avibactam and aztreonam-avibactam combinations exhibited potent activity against CNSE isolates (MIC50/MIC90, 1/1 μg/ml and 0.5/0.5 μg/ml, respectively) compared to that of ceftazidime and aztreonam alone (MIC50/MIC90, 512/512 μg/ml and 128/512 μg/ml, respectively). This study reveals the in vitro activity of ceftazidime-avibactam and aztreonam-avibactam combinations against a large collection of porin-deficient enterobacterial isolates that are representative of the CNSE recovered in the northern part of France.
INTRODUCTION
Carbapenems are broad-spectrum antibiotics usually reserved for severe life-threatening infections. Some isolates of Enterobacteriaceae have developed carbapenem resistance, which results in limited options for the treatment of infections caused by these organisms. Except for the members of the Proteeae tribe, carbapenem resistance in Enterobacteriaceae is almost always attributable to the production of β-lactamases, which can be distinguished according to their carbapenemase activity. “True” carbapenemases (e.g., KPC, OXA-48, and metallo-β-lactamases [MBLs]) confer resistance per se to carbapenems, whereas extended-spectrum β-lactamases (ESBLs) and AmpC-type enzymes require an additional mechanism of resistance, such as a decrease in the uptake of antibiotics by porin deficiency (1, 2) or efflux system overexpression (3), to be responsible for carbapenem resistance.
The rapid dissemination of carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CP-CRE) that account for worldwide outbreaks (4) could be attributable to the location of the carbapenemase-encoding genes on mobile elements that facilitate transmission within and between isolates of Enterobacteriaceae. CP-CRE infections are associated with high rates of morbidity and mortality and occur most frequently among inpatients with a prolonged hospitalization stay and those who are critically ill or exposed to invasive devices (4). The antibiotic agents for treating carbapenem-nonsusceptible Enterobacteriaceae (CNSE) infections, such as colistin, are extremely limited and are often associated with adverse reactions.
CP-CRE isolates emerged in France in the late 2000s, and their number has increased particularly in Paris and in the northern part of the country (5). The aim of this study was to conduct a prospective interregional surveillance program to assess the prevalence of CNSE clinical isolates at six health care facilities located in the North of France and to identify their mechanisms of resistance. We also tested the activity of ceftazidime-avibactam and aztreonam-avibactam against these strains to determine whether these novel combinations could constitute therapeutic alternatives in the future.
MATERIALS AND METHODS
Bacterial strains.
Enterobacterial clinical isolates that were recovered from clinical samples and presented reduced susceptibility to at least one carbapenem, according to the CLSI criteria (6), were collected from six health care facilities (the teaching hospitals of Amiens, Lille, and Caen and the general hospitals of Abbeville, Compiègne, and Beauvais). The isolates were collected during a 1-year period, between June 2012 and November 2013, according to the hospital centers. Only one isolate per patient was included in the study. All isolates were sent to the laboratory of bacteriology of the teaching hospital of Amiens for reidentification by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), antimicrobial susceptibility testing, and characterization of resistance mechanisms.
Antimicrobial susceptibility testing.
MIC measurements were performed by the reference broth microdilution method as described in CLSI document M07-A9 (6). MICs of ceftazidime and aztreonam alone or in combination with avibactam (at a fixed concentration of 4 μg/ml; AstraZeneca Pharmaceuticals) and MICs of ertapenem, imipenem, meropenem, and doripenem, which were used as comparators, were also measured. Moreover, MICs of carbapenems in combination with cloxacillin (at a fixed concentration of 250 μg/ml; bioMérieux, Marcy l'Etoile, France), which acts as an inhibitor of AmpC-type enzymes (7), were also determined. Interpretation was made according to the criteria in CLSI document M100-S23 (8). Quality control (QC) was performed using Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, and Pseudomonas aeruginosa ATCC 27853.
Efflux pump inhibitor tests.
MICs of ertapenem in combination with phenylalanine arginine-naphthylamide dihydrochloride (PAβN; Sigma-Aldrich) (26.3 μg/ml), an inhibitor of RND pumps of enterobacteria (9), were determined. A 2-fold decrease in MIC after addition of PAβN was considered significant (9). The laboratory mutant E. coli AG102, which overexpresses the AcrAB transporter, was used as a positive control (10).
β-Lactamase detection.
The double-disk synergy test was performed to screen for ESBLs as described by the CLSI (8). AmpC overproduction was investigated using agar plates containing cloxacillin (250 μg/ml) (7). Isolates were screened for class A, B, and D carbapenemases by using the modified Hodge test (MHT) as recommended by the CLSI (8) and the Carba NP test as described previously (11).
β-Lactamase identification.
PCR was used to amplify carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM, blaIMI, blaOXA-48-like, blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, and blaOXA-50-like), ESBL-encoding genes (blaCTX-M, blaTEM-like, blaSHV-like, blaPER, blaVEB, and blaGES), and plasmid-mediated AmpC-encoding genes (blaCMY-2, blaDHA, blaACC, blaECT, blaMOX, and blaFOX) according to previously described procedures (12–15). All PCR products were sequenced by use of an ABI7570 sequencer (Applied Biosystems, Foster City, CA).
Examination of porin genes and porin expression.
The sequences of the ompK35 and ompK36 genes from K. pneumoniae, the ompC and ompF genes from Enterobacter cloacae and E. coli, the omp-35 and omp-36 genes from Enterobacter aerogenes, and the ompC-like and ompF-like genes from Citrobacter freundii and Serratia marcescens were amplified and sequenced using the primers shown in Table S1 in the supplemental material. The amplification conditions used for the ompK35, ompK36, ompC, ompF, omp-35, and omp-36 genes were identical to those previously described (16, 17). The ompC-like and ompF-like genes from Citrobacter freundii and Serratia marcescens were amplified using the following program: 95°C for 5 min; 35 cycles of 94°C for 1 min, 51°C for 1 min, and 72°C for 1 min; and a final elongation step of 72°C for 7 min. Strains used for the comparison of porin gene sequences were as follows: K. pneumoniae NTUH-K2044, E. cloacae ATCC 13047, E. aerogenes ATCC 13048, Citrobacter freundii ATCC 8090, and Serratia marcescens ATCC 13880. Outer membrane proteins (OMPs) were isolated according to the rapid procedure of Carlone et al. (18) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (16).
Molecular typing.
Relatedness among isolates was established by random amplified polymorphic DNA (RAPD) analysis and by enterobacterial repetitive intergenic consensus PCR (ERIC-PCR), using Ap12h and ERIC-2 primers, respectively, as previously described (19).
RESULTS
Diversity of CNSE isolates.
A total of 139 CNSE isolates, including 104 E. cloacae (75%), 16 K. pneumoniae (11.5%), 5 E. aerogenes (3.5%), 5 C. freundii (3.5%), 3 Morganella morganii (2.1%), 3 E. coli (2.1%), 2 S. marcescens (1.6%), and 1 Proteus mirabilis (0.8%) isolate, were collected. Eighty-four (60.4%), 43 (31%), 9 (6.5%), 2 (1.4%), 1 (0.7%), and 0 (0%) isolates were collected from the hospitals of Amiens, Lille, Caen, Beauvais, Abbeville, and Compiègne, respectively. A total of 11,431, 12,338, 1,862, 2,186, 1,235, and 1,138 enterobacterial isolates were recovered in the respective hospital centers during the study period.
Genotyping analysis revealed that 36 E. cloacae and 4 K. pneumoniae isolates in the teaching hospital of Amiens displayed undistinguishable genotyping patterns (data not shown) (clones A and B, respectively). Moreover, 3 E. cloacae and 2 K. pneumoniae isolates were genotypically related in the teaching hospital of Lille (clones C and D, respectively). All other CNSE isolates were genotypically unrelated.
Characterization of carbapenem resistance.
Only two strains were detected as CP-CRE (1.4%). One K. pneumoniae isolate and one E. cloacae strain, recovered in the hospital of Caen and in the hospital of Amiens, respectively, tested positive for carbapenemase production. The K. pneumoniae isolate expressed an OXA-48 enzyme (in association with an OXA-1 penicillinase), whereas the E. cloacae isolate produced an IMI-3 β-lactamase (in combination with the chromosomally encoded cephalosporinase) (Table 1).
TABLE 1.
β-Lactamase production and OMP profiles for the 139 CNSE isolates
| OMP profile | Organism with β-lactamase production, β-lactamase(s) (no. of strains) |
||||
|---|---|---|---|---|---|
| Carbapenemase | ESBL | AmpC | ESBL + AmpC | No β-lactamase | |
| Wild-type outer membrane profile | E. cloacae, IMI-3 (1) | M. morganii (3) | P. mirabilis (1) | ||
| K. pneumoniae, OXA-48 (1) | |||||
| Loss of one major outer membrane porin | K. pneumoniae, CTX-M-15 (10) | E. aerogenes (4) | E. cloacae, CTX-M-9/SHV-12 (36) | ||
| E. coli, CTX-M-15 (2) | E. cloacae (39) | E. cloacae, CTX-M-9 (20) | |||
| C. freundii (4) | E. cloacae, SHV-12 (4) | ||||
| S. marcescens (1) | C. freundii, CTX-M-9 (1) | ||||
| E. coli, ACC-1 (1) | |||||
| Loss of OmpC-like and OmpF-like porins | K. pneumoniae, CTX-M-15 (1) | K. pneumoniae, CMY-2 (4) | E. cloacae, CTX-M-9/SHV-12 (4) | ||
| S. marcescens (1) | E. aerogenes, TEM-24 (1) | ||||
The other 137 CNSE isolates (98.6%) did not produce true carbapenemases, as deduced from the Hodge test, PCR screening, and the Carba NP test. One hundred twenty-four strains (90.5%) among these 137 strains produced chromosomally encoded or plasmid-mediated AmpC-type enzymes, as deduced from the significant decreases in the MICs of cephalosporins with the use of cloxacillin-containing plates (Table 1). One hundred nineteen strains expressed their chromosomally encoded AmpC enzymes, whereas 4 K. pneumoniae strains, which were clonally related (clone B), and one E. coli isolate turned out to be positive for blaCMY-2 and blaACC-1 genes, respectively.
Among the 139 CNSE isolates included in this study, 79 (57%) were detected as ESBL producers by using the CLSI screening criteria and the PCR screening method (Table 1) (8). They included 64 E. cloacae (62% of the overall samples for this species), 11 K. pneumoniae (69%), 2 E. coli (66%), 1 E. aerogenes (20%), and 1 C. freundii (20%) isolate. CTX-M-type β-lactamases were detected among 93% of ESBL-producing isolates. CTX-M-9 was the most prevalent ESBL detected in E. cloacae isolates (94% of ESBL-producing E. cloacae isolates), frequently in combination with the SHV-12 enzyme (66%). In contrast, CTX-M-15 was the most prevalent ESBL in E. coli and K. pneumoniae isolates (100% of ESBL producers).
PAβN failed to restore susceptibility to ertapenem in the 139 CNSE isolates included in this study, suggesting that efflux systems are unlikely to be involved. SDS-PAGE analysis of OMPs revealed a deficit of at least one major OMP for all CNSE isolates (Fig. 1), except the two CP-CRE strains and the four Proteeae strains (Table 1). Sequence analysis of the ompC-like and ompF-like genes revealed that the porin deficiency was attributable, in 48% of cases, to disruption by insertion sequence, nonsense mutations leading to a premature stop codon, amino acid insertion in the L3 loop (20). Some ompC-like or ompF-like genes were not amplified, in agreement with complete gene deletion or a large insertion preventing amplification. Moreover, 52% of CNSE isolates with an altered OMP profile did not present mutations in the coding sequences of the ompC-analogue and ompF-analogue genes. Among the 133 porin-deficient CNSE isolates, 122 lacked only one major porin, and the other 11 had a simultaneous loss of both OMPs.
FIG 1.
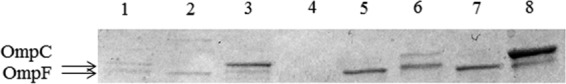
OMP profiles of representative CNSE isolates compared to wild-type reference strains. OMPs were profiled by SDS-PAGE. Lanes: 1, E. coli porin-deficient strain BH01043; 2, E. coli porin-deficient strain DP02127; 3, E. coli wild-type strain TOP10; 4, E. cloacae porin-deficient strain BH01124; 5, E. cloacae porin-deficient strain AJP01076; 6, E. cloacae wild-type strain ATCC 13047; 7, K. pneumoniae porin-deficient strain AG02123; 8, K. pneumoniae wild-type strain NTUH-K2044. The arrows indicate the positions of the OMPs OmpC and OmpF.
CNSE isolates fell into three groups according to their carbapenem MICs. The first one included 123 isolates (88% of the overall CNSE strains) that presented a reduced susceptibility to ertapenem, whereas they remained susceptible to imipenem, meropenem, and doripenem (Table 2). All these strains, except the OXA-48-producing isolate, were also resistant to extended-spectrum cephalosporins. The 12 isolates (9% of the overall CNSE strains) of the second group were resistant or had intermediate resistance to all carbapenems (Table 2). They included three clonally related SHV-12/CTX-M-9-producing E. cloacae strains, derived from clone A isolated in the city of Amiens, one clonally unrelated SHV-12/CTX-M-9-producing E. cloacae strain that was recovered in Caen, one IMI-3-producing E. cloacae isolate, four clonally related CMY-2-producing K. pneumoniae strains (clone B), one CTX-M-15-producing K. pneumoniae strain, one TEM-24-producing E. aerogenes strain, and one non-ESBL-producing S. marcescens strain. These strains were resistant to all β-lactams, except the IMI-producing E. cloacae isolate, which remained susceptible to cefepime. The four strains of the tribe Proteeae, which were susceptible to all carbapenems but imipenem, were integrated in the third group (3% of the overall CNSE strains). They did not produce ESBL and carbapenemase and were susceptible to cefepime.
TABLE 2.
MIC distributions for the 139 carbapenem-nonsusceptible Enterobacteriaceae clinical isolates, with cumulative percentages
| Organism (no. of strains) | Agent(s)a | MIC (μg/ml) (cumulative %) |
MIC50 | MIC90 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥32 | ||||
| E. cloacae (104) | CAZ | 2 (2) | 3 (5) | 99 (100) | ≥32 | ≥32 | |||||||
| CAZ-AVI | 9 (9) | 21 (29) | 58 (85) | 12 (96) | 4 (100) | 0.5 | 1 | ||||||
| ATM | 2 (2) | 3 (5) | 14 (18) | 85 (100) | ≥32 | ≥32 | |||||||
| ATM-AVI | 14 (13) | 23 (36) | 63 (96) | 4 (100) | 0.5 | 0.5 | |||||||
| ERTA | 82 (79) | 12 (90) | 5 (93) | 5 (100) | 1 | 2 | |||||||
| IMP | 5 (5) | 8 (13) | 76 (86) | 10 (95) | 4 (99)b | 1 (100)c | 0.5 | 1 | |||||
| MERO | 11 (11) | 79 (87) | 9 (95) | 4 (99)b | 1 (100)c | 0.5 | 1 | ||||||
| DORI | 18 (17) | 71 (86) | 9 (94) | 1 (95) | 4 (99)b | 1 (100)c | 0.25 | 1 | |||||
| K. pneumoniae (16) | CAZ | 1 (6) | 15 (100) | ≥32 | ≥32 | ||||||||
| CAZ-AVI | 3 (19) | 8 (69) | 1 (75) | 4 (100) | 0.25 | 4 | |||||||
| ATM | 1 (6) | 15 (100) | ≥32 | ≥32 | |||||||||
| ATM-AVI | 1 (6) | 9 (63) | 2 (75) | 4 (100) | 0.125 | 4 | |||||||
| ERTA | 7 (44) | 3 (63) | 2 (75) | 5 (100)b | 2 | ≥32 | |||||||
| IMP | 2 (13) | 8 (63) | 1 (69) | 1 (75)b | 4 (100)b | 0.5 | ≥32 | ||||||
| MERO | 2 (13) | 9 (69) | 1 (75)b | 4 (100)b | 1 | ≥32 | |||||||
| DORI | 2 (13) | 7 (56) | 2 (69) | 1 (75)b | 4 (100)b | 0.5 | 16 | ||||||
| E. aerogenes (5) | CAZ | 1 (20) | 4 (100) | ≥32 | ≥32 | ||||||||
| CAZ-AVI | 2 (40) | 2 (80) | 1 (100) | 0.5 | 2 | ||||||||
| ATM | 1 (20) | 1 (40) | 3 (100) | ≥32 | ≥32 | ||||||||
| ATM-AVI | 3 (60) | 1 (80) | 1 (100) | 0.25 | 1 | ||||||||
| ERTA | 4 (80) | 1 (100)b | 2 | ≥32 | |||||||||
| IMP | 4 (80) | 1 (100)b | 1 | 8 | |||||||||
| MERO | 4 (80%) | 1 (100)b | 1 | 8 | |||||||||
| DORI | 1 (20%) | 3 (80%) | 1 (100%)b | 0.5 | 4 | ||||||||
| E. coli (3) | CAZ | 2 (67) | 1 (100) | 2 | ≥32 | ||||||||
| CAZ-AVI | 1 (33) | 2 (100) | 0.25 | 0.25 | |||||||||
| ATM | 2 (66) | 1 (100) | 8 | ≥32 | |||||||||
| ATM-AVI | 1 (33) | 1 (67) | 1 (100) | 0.125 | 0.25 | ||||||||
| ERTA | 2 (67) | 1 (100) | 1 | 2 | |||||||||
| IMP | 3 (100) | 0.25 | 0.25 | ||||||||||
| MERO | 3 (100) | 0.25 | 0.25 | ||||||||||
| DORI | 1 (33) | 2 (100) | 0.25 | 0.25 | |||||||||
| C. freundii (5) | CAZ | 5 (100) | ≥32 | ≥32 | |||||||||
| CAZ-AVI | 2 (40) | 3 (100) | 0.5 | 0.5 | |||||||||
| ATM | 4 (80) | 1 (100) | 16 | ≥32 | |||||||||
| ATM-AVI | 4 (80) | 1 (100) | 0.25 | 0.5 | |||||||||
| ERTA | 1 (20) | 3 (80) | 1 (100) | 2 | 4 | ||||||||
| IMP | 1 (20) | 2 (60) | 2 (100) | 0.5 | 1 | ||||||||
| MERO | 3 (60) | 2 (100) | 0.5 | 1 | |||||||||
| DORI | 3 (60) | 1 (80) | 1 (100) | 0.25 | 1 | ||||||||
| S. marcescens (2) | CAZ | 2 (100) | ≥32 | ≥32 | |||||||||
| CAZ-AVI | 1 (50) | 1 (100) | 0.5 | 1 | |||||||||
| ATM | 2 (100) | ≥32 | ≥32 | ||||||||||
| ATM-AVI | 1 (50) | 1 (100) | 0.25 | 1 | |||||||||
| ERTA | 1 (50) | 1 (100)b | 1 | ≥32 | |||||||||
| IMP | 1 (50) | 1 (100)b | 0.5 | ≥32 | |||||||||
| MERO | 1 (50) | 1 (100)b | 0.25 | ≥32 | |||||||||
| DORI | 1 (50) | 1 (100)b | 0.25 | 4 | |||||||||
| P. mirabilis (1) | CAZ | 1 (100) | ≤0.06 | ≤0.06 | |||||||||
| CAZ-AVI | 1 (100) | ≤0.06 | ≤0.06 | ||||||||||
| ATM | 1 (100) | ≤0.06 | ≤0.06 | ||||||||||
| ATM-AVI | 1 (100) | ≤0.06 | ≤0.06 | ||||||||||
| ERTA | 1 (100) | ≤0.06 | ≤0.06 | ||||||||||
| IMP | 1 (100) | 8 | 8 | ||||||||||
| MERO | 1 (100) | ≤0.06 | ≤0.06 | ||||||||||
| DORI | 1 (100) | ≤0.06 | ≤0.06 | ||||||||||
| M. morganii (3) | CAZ | 1 (33) | 1 (67) | 1 (100) | 8 | ≥32 | |||||||
| CAZ-AVI | 1 (33) | 1 (67) | 1 (100) | 0.125 | 0.25 | ||||||||
| ATM | 1 (33) | 1 (67) | 1 (100) | 0.125 | 1 | ||||||||
| ATM-AVI | 3 (100) | ≤0.06 | ≤0.06 | ||||||||||
| ERTA | 3 (100) | ≤0.06 | ≤0.06 | ||||||||||
| IMP | 2 (67) | 1 (100) | 8 | 16 | |||||||||
| MERO | 3 (100) | ≤0.06 | ≤0.06 | ||||||||||
| DORI | 3 (100) | ≤0.06 | ≤0.06 | ||||||||||
CAZ, ceftazidime; CAZ-AVI, ceftazidime-avibactam; ATM, aztreonam; ATM-AVI, aztreonam-avibactam; ERTA, ertapenem; IMP, imipenem; MERO, meropenem; DORI, doripenem.
CNSE isolates with a complete loss of OmpC and OmpF porins.
True carbapenemase producers.
It is noteworthy that the level of resistance to carbapenems correlated with the number of porins expressed, except with regard to the two carbapenemase producers and the four Proteeae strains. CNSE strains resistant only to ertapenem expressed one OMP, whereas those that were resistant or had intermediate resistance to all carbapenems produced neither OmpC nor OmpF analogues. Moreover, a ≥2-fold decrease in the MIC of ertapenem in combination with the AmpC inhibitor cloxacillin was shown for all of the E. cloacae, E. aerogenes, C. freundii, and S. marcescens isolates, except the IMI-3-producing E. cloacae isolate, thus demonstrating the important contribution of class C β-lactamases to carbapenem resistance in our study.
To define the spectra of activity of ceftazidime-avibactam and aztreonam-avibactam against the CNSE isolates that were collected in our survey, we tested the susceptibilities of the 139 strains to these novel combinations. The ceftazidime-avibactam and aztreonam-avibactam combinations exhibited overall potent activities against CNSE isolates (MIC50/MIC90, 0.5/1 μg/ml and 0.5/0.5 μg/ml, respectively) compared to those with ceftazidime and aztreonam alone (MIC50/MIC90, 512/512 μg/ml and 128/512 μg/ml, respectively) or ertapenem and imipenem alone (MIC50/MIC90, 1/4 μg/ml and 0.5/8 μg/ml, respectively) (Table 2). The 123 ertapenem-nonsusceptible enterobacterial isolates exhibited ceftazidime-avibactam MICs and aztreonam-avibactam MICs of <1 μg/ml, whereas the 12 carbapenem-nonsusceptible enterobacterial strains exhibited ceftazidime-avibactam and aztreonam-avibactam MICs of ≤4 μg/ml (Table 2). The three imipenem-nonsusceptible M. morganii isolates and the P. mirabilis isolate exhibited ceftazidime-avibactam and aztreonam-avibactam MICs of ≤2 μg/ml (Table 2).
DISCUSSION
This study was intended to determine the epidemiology of CNSE clinical isolates in the northern part of France and to assess the potency of the ceftazidime-avibactam and aztreonam-avibactam combinations against these isolates. A notable feature of our interregional study is that the CNSE isolates were prospectively and exhaustively collected during a 1-year period, thus providing an accurate picture of the epidemiology in the North of France. Most CNSE isolates in this study were collected in teaching hospitals (97% of the overall strains), whereas only a few CNSE strains were recovered in general hospitals (3%). Most of the CNSE isolates included were recovered in the teaching hospitals of Amiens and Lille, which could be attributable to the local spread of ESBL- and/or AmpC-producing epidemic strains. It is possible that carbapenem-containing regimens, which were used to treat infections due to ESBL producers, might have further selected for mutants lacking outer membrane permeability. By reducing the antibiotic concentration inside the periplasm, porin changes can amplify the β-lactamase effects of ESBLs and AmpC-type enzymes on weakly hydrolyzed substrates, such as carbapenems (1, 2). An additional study is ongoing to assess the contribution of carbapenem regimens to selection of porin-deficient mutants.
AmpC enzymes appeared to contribute greatly to carbapenem resistance in our collection. The carbapenem-hydrolyzing activity of class C β-lactamases has been reported previously (1, 7). This activity could be attributable, at least in part, to the asparagine residue at position 346, which plays a role in placing the acyl enzyme intermediate of extended-spectrum cephalosporins (ESCs) in a position that is more competent for hydrolytic attack (1, 21).
In contrast, the prevalence of isolates producing true carbapenemases was low in our study. Only 2 of 139 strains produced β-lactamases (IMI-3 and OXA-48). These results are in agreement with previous studies, carried out in Chile (22), Korea (23), China (24), United Kingdom (25), Belgium (26), and France (5, 27), which showed that the emergence of carbapenem resistance in Enterobacteriaceae could be supported mainly by the combination of ESBLs or AmpC β-lactamases with porin deficiency in certain areas. Recent reports highlighted the implication of carbapenem-nonsusceptible noncarbapenemase producers in nosocomial outbreaks (28) in different countries, mostly from K. pneumoniae or Enterobacter spp. (29–31). These isolates are commonly selected in vivo during the course of carbapenem therapy, as a consequence of a lack of membrane permeability.
In 48% of the overall porin-deficient CNSE isolates, loss of membrane permeability resulted from changes in OMP structure. Genes encoding major porins were affected by mutations causing protein structure changes in the L3 loop, by premature termination of translation, or by gene disruption. The L3 domain constitutes a pore constriction region that exhibits negative charges and plays a critical role in determining the characteristics of the pore (20). The two amino acid replacements identified in our study, Y214T and A141V, which were located in OmpC-like and OmpF-like porins of E. cloacae isolates, respectively, may perturb the electrostatic field acting in the eyelet and modify the physicochemical and biological channel properties. Most of these structural alterations occurred in only one of the two major OMPs, thus affecting the susceptibility to ertapenem (1). Reduced susceptibility to other carbapenems, such as imipenem and doripenem, was achieved once both major porins were altered. It is noteworthy that no significant changes in the ompC-like and ompF-like genes were identified in 52% of the porin-deficient CNSE isolates. We assume that a loss of permeability might result from decreased porin production. Further investigations into the nature of the purported regulatory system and its mutations are ongoing.
In addition, the peculiar resistance phenotype exhibited by the M. morganii and P. mirabilis isolates, which were fully susceptible to all carbapenems but imipenem, could be related to an alteration of penicillin-binding protein 2 (PBP2), as reported previously (32, 33).
The carbapenemase threat remains rare in French hospitals compared with the ESBL endemicity. In contrast, carbapenem resistance has arisen from local spread of E. cloacae and K. pneumoniae clones associated with the dissemination of ESBL (particularly CTX-M-9 and/or SHV-12 for E. cloacae and CTX-M-15 for K. pneumoniae) and AmpC production, leaving few therapeutic options available.
Several studies, conducted on a wide range of enterobacterial clinical isolates recovered throughout the world, have already demonstrated the overall activity of the ceftazidime-avibactam combination, including activity against ESBL, AmpC, and KPC producers (ceftazidime-avibactam MIC50 of ≤0.5 μg/ml and ceftazidime-avibactam MIC90 of ≤2 μg/ml) (34–38). However, few data describe the in vitro activity of this combination against porin-deficient enterobacterial isolates. To the best of our knowledge, only two articles showed the in vitro efficiency of this combination against well-characterized porin-deficient enterobacterial clinical isolates, namely, Cedecea davisae FUR and K. pneumoniae C2/pMG247 (7, 39). Livermore et al. also reported the activity of the ceftazidime-avibactam combination against a few ertapenem-resistant enterobacterial isolates, but these isolates were not characterized on the molecular level (40).
Our study shows the in vitro activity of the ceftazidime-avibactam and aztreonam-avibactam combinations against a representative collection of well-characterized CNSE clinical isolates lacking outer membrane permeability. It appears that porin deficiency does not impair the penetration of avibactam into the periplasmic space. The highest MIC value for the ceftazidime-avibactam and aztreonam-avibactam combinations against porin-deficient CNSE isolates was 4 μg/ml. According to the U.S. FDA ceftazidime-avibactam MIC breakpoints for Enterobacteriaceae (susceptible, ≤8 μg/ml; and resistant, ≥16 μg/ml) (41), which are based on pharmacokinetic/pharmacodynamic analyses (42), these porin-deficient CNSE isolates are susceptible. Additional studies are needed to establish what the potential roles of ceftazidime-avibactam and aztreonam-avibactam might be as substitutes for carbapenems to reduce the dissemination of CNSE in the future.
Concluding remarks.
OMP deficiency in association with AmpCs and/or ESBLs was the main mechanism conferring carbapenem resistance in this survey. This study reveals the in vitro activity of the ceftazidime-avibactam and aztreonam-avibactam combinations against a representative collection of well-characterized CRE isolates with OMP deficiency.
Supplementary Material
ACKNOWLEDGMENTS
We thank AstraZeneca Pharmaceuticals (Waltham, MA) for providing avibactam, and we thank Wright Nichols for critical comments and the provision of bibliographic data before publication.
This study was supported by a Programme Hospitalier de Recherche (PHRC) of the Centre Hospitalier Universitaire d'Amiens.
We have no competing interests to declare.
Funding Statement
This work was supported by institutional funds.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01559-15.
REFERENCES
- 1.Mammeri H, Guillon H, Eb F, Nordmann P. 2010. Phenotypic and biochemical comparison of the carbapenem-hydrolyzing activities of five plasmid-borne AmpC β-lactamases. Antimicrob Agents Chemother 54:4556–4560. doi: 10.1128/AAC.01762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacoby GA, Mills DM, Chow N. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:3203–3206. doi: 10.1128/AAC.48.8.3203-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez A, Canle D, Latasa C, Poza M, Beceiro A, del Mar Tomás M, Fernández A, Mallo S, Pérez S, Molina F, Villanueva R, Lasa I, Bou G. 2007. Cloning, nucleotide sequencing, and analysis of the AcrAB-TolC efflux pump of Enterobacter cloacae and determination of its involvement in antibiotic resistance in a clinical isolate. Antimicrob Agents Chemother 51:3247–3253. doi: 10.1128/AAC.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 5.Vaux S, Carbonne A, Thiolet JM, Jarlier V, Coignard B, RAISIN and Expert Laboratories Groups. 2011. Emergence of carbapenemase-producing Enterobacteriaceae in France, 2004 to 2011. Euro Surveill 16:ppi=19880. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2012. M07-A9. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Ammenouche N, Dupont H, Mammeri H. 2014. Characterization of a novel AmpC β-lactamase produced by a carbapenem-resistant Cedecea davisae clinical isolate. Antimicrob Agents Chemother 58:6942–6945. doi: 10.1128/AAC.03237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2013. M100-S23. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Chollet R, Chevalier J, Bollet C, Pages J-M, Davin-Regli A. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob Agents Chemother 48:2518–2523. doi: 10.1128/AAC.48.7.2518-2523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okusu H, Ma D, Nikaido H. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol 178:306–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dortet L, Bréchard L, Poirel L, Nordmann P. 2014. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. 2014. J Med Microbiol 63:772–776. doi: 10.1099/jmm.0.071340-0. [DOI] [PubMed] [Google Scholar]
- 12.Dallenne C, da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 13.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolate by using multiplex PCR. J Clin Microbiol 40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aubron C, Poirel L, Ash RJ, Nordmann P. 2005. Carbapenemase-producing Enterobacteriaceae, US rivers. Emerg Infect Dis 11:260–264. doi: 10.3201/eid1102.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H, Pi B-R, Yang Q, Yu Y-S, Chen Y-G, Li L-J, Zheng S-S. 2007. Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1-blaOXA-23 genes in a Chinese hospital. J Med Microbiol 56:1076–1080. doi: 10.1099/jmm.0.47206-0. [DOI] [PubMed] [Google Scholar]
- 16.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Héritier C, Spicq C, Nordmann P. 2004. In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J Clin Microbiol 42:3831–3833. doi: 10.1128/JCM.42.8.3831-3833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol 24:330–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davin-Regli A, Monnet D, Saux P, Bosi C, Charrel R, Barthelemy A, Bollet C. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J Clin Microbiol 34:1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bornet C, Saint N, Fetnaci L, Dupont M, Davin-Régli A, Bollet C, Pagès JM. 2004. Omp35, a new Enterobacter aerogenes porin involved in selective susceptibility to cephalosporins. Antimicrob Agents Chemother 48:2153–2158. doi: 10.1128/AAC.48.6.2153-2158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahyot S, Broutin I, de Champs C, Guillon H, Mammeri H. 2013. Contribution of asparagine 346 residue to the carbapenemase activity of CMY-2 β-lactamase. FEMS Microbiol Lett 345:147–153. doi: 10.1111/1574-6968.12199. [DOI] [PubMed] [Google Scholar]
- 22.Wozniak A, Villagra NA, Undabarrena A, Gallardo N, Keller N, Moraga M, Román JC, Mora GC, García P. 2012. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol 61:1270–1279. doi: 10.1099/jmm.0.045799-0. [DOI] [PubMed] [Google Scholar]
- 23.Kim SY, Shin J, Shin SY, Ko KS. 2013. Characteristics of carbapenem-resistant Enterobacteriaceae isolates from Korea. Diagn Microbiol Infect Dis 76:486–490. doi: 10.1016/j.diagmicrobio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Yang FC, Yan JJ, Hung KH, Wu JJ. 2012. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J Clin Microbiol 50:223–226. doi: 10.1128/JCM.01263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drew RJ, Turton JF, Hill RLR, Livermore DM, Woodford N, Paulus S, Cunliffe NA. 2013. Emergence of carbapenem-resistant Enterobacteriaceae in a UK paediatric hospital. J Hosp Infect 84:300–304. doi: 10.1016/j.jhin.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Huang TD, Berhin C, Bogaerts P, Glupczynski Y, Multicentre Study Group. 2013. Prevalence and mechanisms of resistance to carbapenems in Enterobacteriaceae isolates from 24 hospitals in Belgium. J Antimicrob Chemother 68:1832–1837. doi: 10.1093/jac/dkt096. [DOI] [PubMed] [Google Scholar]
- 27.Robert J, Pantel A, Mérens A, Lavigne JP, Nicolas-Chanoine MH, ONERBA's Carbapenem Resistance Study Group. 2014. Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011–12. J Antimicrob Chemother 69:2706–2712. doi: 10.1093/jac/dku208. [DOI] [PubMed] [Google Scholar]
- 28.Novais Â, Rodrigues C, Branquinho R, Antunes P, Grosso F, Boaventura L, Ribeiro G, Peixe L. 2012. Spread of an OmpK36-modified ST15 Klebsiella pneumoniae variant during an outbreak involving multiple carbapenem-resistant Enterobacteriaceae species and clones. Eur J Clin Microbiol Infect Dis 31:3057–3063. doi: 10.1007/s10096-012-1665-z. [DOI] [PubMed] [Google Scholar]
- 29.García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. 2010. An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother 54:4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chudácková E, Bergerová T, Fajfrlík K, Cervená D, Urbásková P, Empel J, Gniadkowski M, Hrabák J. 2010. Carbapenem-nonsusceptible strains of Klebsiella pneumoniae producing SHV-5 and/or DHA-1 β-lactamases in a Czech hospital. FEMS Microbiol Lett 309:62–70. doi: 10.1111/j.1574-6968.2010.02016.x. [DOI] [PubMed] [Google Scholar]
- 31.Mena A, Plasencia V, García L, Hidalgo O, Ayestarán JI, Alberti S, Borrell N, Pérez JL, Oliver A. 2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol 44:2831–2837. doi: 10.1128/JCM.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villar HE, Danel F, Livermore DM. 1997. Permeability to carbapenems of Proteus mirabilis mutants selected for resistance to imipenem or other β-lactams. J Antimicrob Chemother 40:365–370. doi: 10.1093/jac/40.3.365. [DOI] [PubMed] [Google Scholar]
- 33.Neuwirth C, Siebor E, Duez JM, Pechinot A, Kazmierczak A. 1995. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J Antimicrob Agents 36:335–342. doi: 10.1093/jac/36.2.335. [DOI] [PubMed] [Google Scholar]
- 34.Castanheira M, Farrell SE, Krause KM, Jones RN, Sader HS. 2014. Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine US census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob Agents Chemother 58:833–838. doi: 10.1128/AAC.01896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flamm RK, Sader HS, Farrell DJ, Jones RN. 2014. Ceftazidime-avibactam and comparator agents tested against urinary tract isolates from a global surveillance program (2011). Diagn Microbiol Infect Dis 80:233–238. doi: 10.1016/j.diagmicrobio.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Flamm RK, Farrell DJ, Sader HS, Jones RN. 2014. Ceftazidime/avibactam activity tested against Gram-negative bacteria isolated from bloodstream, pneumonia, intra-abdominal and urinary tract infections in US medical centres (2012). J Antimicrob Chemother 69:1589–1598. doi: 10.1093/jac/dku025. [DOI] [PubMed] [Google Scholar]
- 37.Sader HS, Castanheira M, Flamm RK, Farrell DJ, Jones RN. 2014. Antimicrobial activity of ceftazidime-avibactam against Gram-negative organisms collected from U.S. medical centers in 2012. Antimicrob Agents Chemother 58:1684–1692. doi: 10.1128/AAC.02429-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Testa R, Cantón R, Giani T, Morosini MI, Nichols WW, Seifert H, Stefanik D, Rossolini GM, Nordmann P. 2015. In vitro activity of ceftazidime, ceftaroline and aztreonam alone and in combination with avibactam against European Gram-negative and Gram-positive clinical isolates. Int J Antimicrob Agents 45:641–646. doi: 10.1016/j.ijantimicag.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Estabrook M, Jacoby GA, Nichols WW, Testa RT, Bush K. 2015. In vitro susceptibility of characterized β-lactamase-producing strains tested with avibactam combinations. Antimicrob Agents Chemother 59:1789–1793. doi: 10.1128/AAC.04191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livermore DM, Mushtaq S, Warner M, Miossec C, Woodford N. 2008. NXL104 combinations versus Enterobacteriaceae with CTX-M extended-spectrum β-lactamases and carbapenemases. J Antimicrob Chemother 62:1053–1056. doi: 10.1093/jac/dkn320. [DOI] [PubMed] [Google Scholar]
- 41.Actavis. 2015. Ceftazidime-avibactam prescribing information 2015. http://pi.actavis.com/data_stream.asp?product_group=1957&p=pi&language=E. Accessed June 2015. [Google Scholar]
- 42.US Food and Drug Administration. 2014. Briefing package. Ceftazidime-avibactam. Division of Anti-Infective Products, Office of Antimicrobial Products, Center for Drug Evaluation and Research, US Food and Drug Administration, Silver Spring, Maryland: http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/Drugs/Anti-InfectiveDrugsAdvisoryCommittee/UCM425458.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


