Abstract
The exogenously acquired 16S rRNA methyltransferases RmtD, RmtD2, and RmtG were cloned and heterologously expressed in Escherichia coli, and the recombinant proteins were purified to near homogeneity. Each methyltransferase conferred an aminoglycoside resistance profile consistent with m7G1405 modification, and this activity was confirmed by in vitro 30S methylation assays. Analyses of protein structure and interaction with S-adenosyl-l-methionine suggest that the molecular mechanisms of substrate recognition and catalysis are conserved across the 16S rRNA (m7G1405) methyltransferase family.
TEXT
The retained potency of aminoglycoside antibiotics has renewed interest in their use in clinical practice (1). However, among several resistance mechanisms, production of acquired 16S rRNA methyltransferases has emerged as a significant threat to clinical efficacy (2, 3). These enzymes modify the aminoglycoside 16S rRNA binding pocket to confer high-level aminoglycoside resistance. The predominant 16S rRNA modification in pathogenic bacteria is methylation of the N7 position of G1405 (m7G1405), with nine distinct enzymes (ArmA and RmtA to -H) reported to date from clinical and veterinary isolates (3, 4).
Acquired 16S rRNA (m7G1405) methyltransferases are globally distributed, and the genes that encode them typically reside within mobile elements that may coharbor additional resistance determinants (3). For example, RmtD was first detected in Brazil in Pseudomonas aeruginosa coproducing SPM-1, while both RmtD and RmtG were identified in Klebsiella pneumoniae coproducing KPC-2 and CTX-M (5, 6). Both enzymes and a variant of RmtD (RmtD2) were subsequently identified in Enterobacter spp., Citrobacter freundii, and Escherichia coli isolates from South and North America (7–10).
To extend our understanding of these resistance determinants, we cloned, expressed, and purified the acquired 16S rRNA (m7G1405) methyltransferases RmtD, RmtD2, and RmtG. The genes that encode RmtD, RmtG, and RmtD2 were PCR amplified from template DNA extracted from an endemic SPM-1-producing P. aeruginosa isolate and two separate isolates from hospital-based outbreaks of K. pneumoniae in Brazil, respectively (5, 11; L.L.C. and R.C.P., unpublished data). Amplicons were cloned via the TOPO TA vector (Invitrogen) into a modified pET44a vector to generate 6×His-tagged methyltransferases with a thrombin cleavage site as described previously (12, 13). An equivalent construct was also generated by using an E. coli codon-optimized gene obtained by commercial chemical synthesis for the intrinsic 16S rRNA methyltransferase Sgm from Micromonospora zionensis for which m7G1405 activity has been directly experimentally verified (14–16).
Recombinant proteins were expressed at 37°C in E. coli BL21(DE3) using lysogeny broth (500 ml) containing ampicillin (100 μg/ml). Protein expression was induced at mid-log phase (optical density at 600 nm, 0.6 to 0.8) with 0.5 or 1.0 mM isopropyl-β-d-thiogalactopyranoside, and growth was continued for 6 h at 30°C or for 3 h at 37°C for RmtD and all other proteins, respectively. Cells were harvested by centrifugation; resuspended in lysis buffer (5 ml/g of wet cells) containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10% glycerol, and 10 mM imidazole; and lysed by sonication. Insoluble cell debris was removed by centrifugation, and target proteins were purified on an ÄKTApurifier10 system. First, Ni2+ affinity chromatography (HisTrap FF) was performed in lysis buffer with target protein elution accomplished by using a gradient of imidazole (10 to 300 mM). Target protein-containing fractions were pooled, concentrated, and further purified by gel filtration chromatography (Superdex 75 16/60) preequilibrated with 20 mM Tris buffer (pH 8.0) containing either 300 mM NaCl and 20% glycerol (RmtD and RmtD2) or 200 mM NaCl and 10% glycerol (RmtG). Sgm was purified by the same procedure but under previously established solution conditions (17). All of the proteins eluted from the gel filtration column and exhibited SDS-PAGE mobilities in good agreement with their calculated molecular weights (data not shown and Fig. 1A, respectively).
FIG 1.
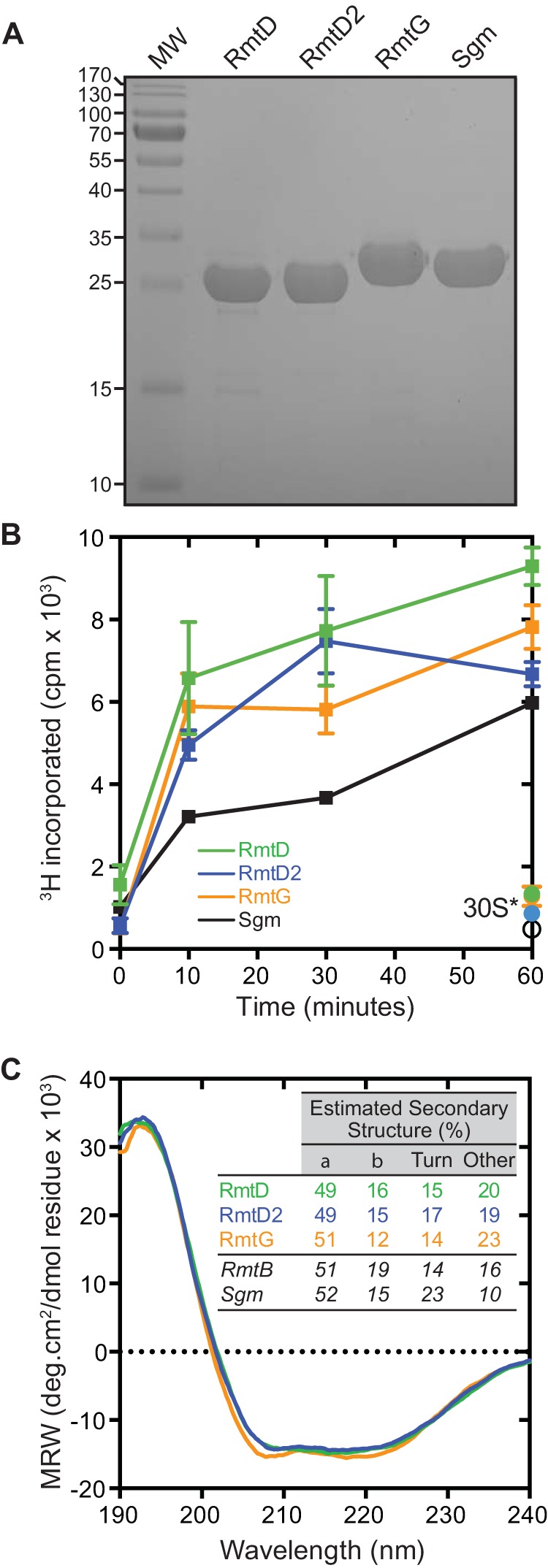
Expression and purification of active recombinant RmtD, RmtD2, and RmtG. (A) SDS-PAGE analysis of purified recombinant RmtD (29.5 kDa), RmtD2 (29.5 kDa), and RmtG (31.5 kDa). The intrinsic 16S rRNA (m7G1405) methyltransferase Sgm (32.4 kDa) is shown for comparison. MW, protein molecular weight standards (protein masses in kDa are shown to the left). (B) In vitro time course methyltransferase assays of RmtD (green), RmtD2 (blue), and RmtG (orange) using 30S subunits isolated from E. coli MRE600 in the presence of [3H]SAM. Single 60-min time points are also shown for assays using m7G1405 modified 30S (30S*) isolated from E. coli BL21(DE3) expressing Sgm. (C) Circular dichroism spectroscopy analyses of RmtD, RmtD2, and RmtG. Spectra were deconvoluted to estimate the secondary-structure content shown in the inset. Values for RmtB (Protein Data Bank code 3FRH) and Sgm (Protein Data Bank code 3LCV) were calculated from X-ray crystal structures via the STRIDE webserver (24). MRW, mean residue molar ellipticity.
Measurements of aminoglycoside MICs were made as previously described (12) in liquid cultures of E. coli BL21(DE3) transformed with the empty pET vector, pET-HTrmtD, pET-HTrmtD2, or pET-HTrmtG. All three enzymes conferred high-level resistance to gentamicin and kanamycin but not to apramycin or neomycin (Table 1), a profile consistent with the m7G1405 modification which confers resistance to 4,6-disubstituted deoxystreptamines but not other structural classes of aminoglycoside (3). All three enzymes also efficiently methylated 30S subunits in in vitro assays using S-adenosyl-l-[3H]methionine ([3H]SAM) (13), with activity comparable to that of Sgm (Fig. 1B) and other Rmt enzymes (18, 19). In contrast, in assays using 30S from Sgm-expressing cells, no additional methylation was observed (Fig. 1B, 30S*). These results demonstrate that each purified recombinant protein is active and that RmtD, RmtD2, and RmtG modify the N7 position of 16S rRNA nucleotide G1405.
TABLE 1.
Aminoglycoside MICs for E. coli harboring plasmids encoding acquired resistance methyltransferases
| Plasmid | MIC (μg/ml) |
|||
|---|---|---|---|---|
| Apramycin | Gentamicin | Kanamycin | Neomycin | |
| pET vector | 32 | 4 | 16 | 16 |
| pET-HT_rmtD | 32 | >1,024 | >1,024 | 16 |
| pET-HT_rmtD2 | 32 | >1,024 | >1,024 | 16 |
| pET-HT_rmtG | 32 | >1,024 | >1,024 | 16 |
We next used circular dichroism spectroscopy and deconvolution using the CDSSTR algorithm via DICHROWEB (20) to assess the solution structure of each methyltransferase (Fig. 1C) as previously described (12). All three methyltransferases were well folded with predicted secondary-structure contents in excellent agreement with those calculated from the high-resolution structures of Sgm and RmtB (21, 22).
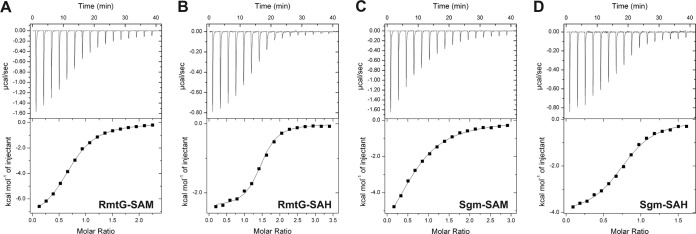
The aminoglycoside resistance methyltransferases require SAM as their obligatory cosubstrate (methyl group donor) and produce S-adenosylhomocysteine (SAH) as the methylation reaction by-product. Analyses of enzymes that catalyze the m1A1408 modification have revealed a characteristic, though not universal, higher relative affinity for SAH than for SAM (12, 23). In contrast, for Sgm, SAH binding was reported to be several hundred times weaker despite its comparable affinity for SAM (21). We therefore used isothermal titration calorimetry (ITC) to measure the affinities of RmtD, RmtD2, and RmtG for SAM and SAH (Fig. 2 and Table 2). Purified proteins were dialyzed against a mixture of 20 mM Tris (pH 8.0), 300 mM NaCl, 20% glycerol, and 10 mM β-mercaptoethanol, except Sgm, for which previously established solutions were used (17). SAM (1.0 to 1.5 mM) and SAH (0.4 to 0.8 mM) were prepared by using the final dialysis solutions, and titrations were performed on an Auto-iTC200 microcalorimeter (Malvern/Microcal) as described previously (12). All SAM binding affinities were in the low micromolar range (Table 2), consistent with previous measurements for Sgm and other RNA methyltransferases (17, 21). Surprisingly, all methyltransferases had significantly greater affinity for SAH, including Sgm, contradictory to the prior report (21). We considered whether differences in solution conditions might have affected the measured affinities. We first attempted to dialyze Sgm against the conditions used previously, where weaker SAH binding was observed (21), but found that the protein consistently precipitated from solution. As an alternative comparison, titrations were performed with Sgm under the conditions used for the acquired methyltransferases and for RmtD2 under our prior conditions for Sgm (17). Regardless of the solution conditions used, essentially identical SAM and SAH affinities for Sgm and RmtD2 were measured in each case.
FIG 2.
ITC analysis of RmtG and Sgm ligand binding. Example titrations of RmtG with SAM (A) and SAH (B) and of Sgm with SAM (C) and SAH (D). Titrations were fitted with a one binding site model to obtain binding affinities (Kd) for all protein ligand pairs (see Table 2).
TABLE 2.
Binding affinities of aminoglycoside resistance 16S rRNA (m7G1405) methyltransferases for SAM and SAH measured by ITC
| Class and enzyme |
Kd (μM)a |
Reference | |
|---|---|---|---|
| SAM | SAH | ||
| Acquired m7G1405 | |||
| RmtD | 42 ± 19 | 2.0 ± 0.2 | This study |
| RmtD2 | 29 ± 5b | 1.2 ± 0.3b | |
| RmtG | 11.4/10.0 | 3.7/1.0 | |
| Producer m7G1405 | |||
| Sgm | 25 ± 16b | 4.8 ± 2.6b | This study |
| Sgm | 17.6 | NDc | 17 |
| Sgm | 18 | 300 | 21 |
Measurements were made in triplicate with independent samples and are shown as means ± standard deviations, except for RmtG, for which only two independent measurements were made (values from both experiments are shown). Errors associated with fits to one binding site model were negligible compared to those between measurements.
Binding was assessed under two different solution conditions with essentially identical results.
ND, not determined.
Defining the molecular mechanisms of antimicrobial resistance is critical to support the development of new effective strategies to combat multidrug-resistant pathogens. Here, we have established recombinant expression of active RmtD, RmtD2, and RmtG methyltransferases, providing protein samples suitable for detailed structure-function studies. Our results show that each protein likely adopts a structure very similar to that of characterized m7G1405 enzymes and indicate that higher affinity for SAH than for SAM may be a feature common to all aminoglycoside resistance methyltransferases. For both groups of enzymes, the potential for product inhibition may contribute to the regulation of methyltransferase activity and/or control of substrate specificity. In contrast, contrary to the emerging evidence for the m1A1408 methyltransferase family, our findings suggest that among the 16S rRNA (m7G1405) methyltransferases, the molecular mechanisms of substrate recognition and catalysis are likely to be highly conserved between intrinsic and acquired enzymes. Therefore, the development of inhibitors that are broadly effective against this resistance enzyme family may be feasible.
ACKNOWLEDGMENTS
We are grateful to Christine M. Dunham and to members of the Conn and Dunham labs for many useful discussions.
Funding Statement
MCTI | Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) provided funding to Renata C. Picão and Laís L. Corrêa under grant numbers E-26/111.780/2012 and E-26/201.555/2014. L.L.C. received support from a Ciência sem Fronteiras fellowship (CNPq).
REFERENCES
- 1.Becker B, Cooper MA. 2013. Aminoglycoside antibiotics in the 21st century. ACS Chem Biol 8:105–115. doi: 10.1021/cb3005116. [DOI] [PubMed] [Google Scholar]
- 2.Doi Y, Arakawa Y. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis 45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 3.Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat 15:133–148. doi: 10.1016/j.drup.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 4.O'Hara JA, McGann P, Snesrud EC, Clifford RJ, Waterman PE, Lesho EP, Doi Y. 2013. Novel 16S rRNA methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother 57:2413–2416. doi: 10.1128/AAC.00266-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother 51:852–856. doi: 10.1128/AAC.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bueno MF, Francisco GR, O'Hara JA, de Oliveira Garcia D, Doi Y. 2013. Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum beta-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 57:2397–2400. doi: 10.1128/AAC.02108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tijet N, Andres P, Chung C, Lucero C, Group WH-A, Low DE, Galas M, Corso A, Petroni A, Melano RG. 2011. rmtD2, a new allele of a 16S rRNA methylase gene, has been present in Enterobacteriaceae isolates from Argentina for more than a decade. Antimicrob Agents Chemother 55:904–909. doi: 10.1128/AAC.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leigue L, Warth JF, Melo LC, Silva KC, Moura RA, Barbato L, Silva LC, Santos AC, Silva RM, Lincopan N. 2015. MDR ST2179-CTX-M-15 Escherichia coli co-producing RmtD and AAC(6′)-Ib-cr in a horse with extraintestinal infection, Brazil. J Antimicrob Chemother 70:1263–1265. doi: 10.1093/jac/dku520. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Labarca J, Bello H, Rioseco ML, Bernabeu S, Nordmann P. 2014. Emergence of the 16S rRNA methylase RmtG in an extended-spectrum-beta-lactamase-producing and colistin-resistant Klebsiella pneumoniae isolate in Chile. Antimicrob Agents Chemother 58:618–619. doi: 10.1128/AAC.02059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu F, Munoz-Price LS, DePascale D, Rivera JI, Doi Y. 2014. Klebsiella pneumoniae sequence type 11 isolate producing RmtG 16S rRNA methyltransferase from a patient in Miami, Florida. Antimicrob Agents Chemother 58:4980–4981. doi: 10.1128/AAC.02632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos PI, Picao RC, Almeida LG, Lima NC, Girardello R, Vivan AC, Xavier DE, Barcellos FG, Pelisson M, Vespero EC, Medigue C, Vasconcelos AT, Gales AC, Nicolas MF. 2014. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15:54. doi: 10.1186/1471-2164-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Witek MA, Conn GL. 2014. Expansion of the aminoglycoside resistance 16S rRNA (m(1)A1408) methyltransferase family: expression and functional characterization of four hypothetical enzymes of diverse bacterial origin. Biochim Biophys Acta 1844:1648–1655. doi: 10.1016/j.bbapap.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zelinskaya N, Rankin CR, Macmaster R, Savic M, Conn GL. 2011. Expression, purification and crystallization of adenosine 1408 aminoglycoside resistance rRNA methyltransferases for structural studies. Protein Expr Purif 75:89–94. doi: 10.1016/j.pep.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojic M, Topisirovic L, Vasiljevic B. 1992. Cloning and characterization of an aminoglycoside resistance determinant from Micromonospora zionensis. J Bacteriol 174:7868–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savic M, Lovric J, Tomic TI, Vasiljevic B, Conn GL. 2009. Determination of the target nucleosides for members of two families of 16S rRNA methyltransferases that confer resistance to partially overlapping groups of aminoglycoside antibiotics. Nucleic Acids Res 37:5420–5431. doi: 10.1093/nar/gkp575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cubrilo S, Babic F, Douthwaite S, Maravic Vlahovicek G. 2009. The aminoglycoside resistance methyltransferase Sgm impedes RsmF methylation at an adjacent rRNA nucleotide in the ribosomal A site. RNA 15:1492–1497. doi: 10.1261/rna.1618809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savic M, Ilic-Tomic T, Macmaster R, Vasiljevic B, Conn GL. 2008. Critical residues for cofactor binding and catalytic activity in the aminoglycoside resistance methyltransferase Sgm. J Bacteriol 190:5855–5861. doi: 10.1128/JB.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, Doi Y, Kimura K, Ike Y, Arakawa Y. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother 50:178–184. doi: 10.1128/AAC.50.1.178-184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother 48:491–496. doi: 10.1128/AAC.48.2.491-496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobley A, Whitmore L, Wallace BA. 2002. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 18:211–212. [DOI] [PubMed] [Google Scholar]
- 21.Husain N, Tkaczuk KL, Tulsidas SR, Kaminska KH, Cubrilo S, Maravic-Vlahovicek G, Bujnicki JM, Sivaraman J. 2010. Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases. Nucleic Acids Res 38:4120–4132. doi: 10.1093/nar/gkq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt E, Galimand M, Panvert M, Courvalin P, Mechulam Y. 2009. Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases. J Mol Biol 388:570–582. doi: 10.1016/j.jmb.2009.03.034. [DOI] [PubMed] [Google Scholar]
- 23.Husain N, Obranic S, Koscinski L, Seetharaman J, Babic F, Bujnicki JM, Maravic-Vlahovicek G, Sivaraman J. 2011. Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit. Nucleic Acids Res 39:1903–1918. doi: 10.1093/nar/gkq1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinig M, Frishman D. 2004. STRIDE: a Web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32:W500–W502. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]