A thaumatin-like protein regulates gene-for-gene interaction with specific rhizobial strains and controls nodulation specificity in soybeans.
Abstract
Rj4 is a dominant gene in soybeans (Glycine max) that restricts nodulation by many strains of Bradyrhizobium elkanii. The soybean-B. elkanii symbiosis has a low nitrogen-fixation efficiency, but B. elkanii strains are highly competitive for nodulation; thus, cultivars harboring an Rj4 allele are considered favorable. Cloning the Rj4 gene is the first step in understanding the molecular basis of Rj4-mediated nodulation restriction and facilitates the development of molecular tools for genetic improvement of nitrogen fixation in soybeans. We finely mapped the Rj4 locus within a small genomic region on soybean chromosome 1, and validated one of the candidate genes as Rj4 using both complementation tests and CRISPR/Cas9-based gene knockout experiments. We demonstrated that Rj4 encodes a thaumatin-like protein, for which a corresponding allele is not present in the surveyed rj4 genotypes, including the reference genome Williams 82. Our conclusion disagrees with the previous report that Rj4 is the Glyma.01G165800 gene (previously annotated as Glyma01g37060). Instead, we provide convincing evidence that Rj4 is Glyma.01g165800-D, a duplicated and unique version of Glyma.01g165800, that has evolved the ability to control symbiotic specificity.
Legumes are capable of forming a root nodule symbiosis with nitrogen-fixing soil bacteria called rhizobia. Remarkably, this symbiosis shows a high level of specificity (Broughton et al., 2000; Perret et al., 2000; Wang et al., 2012). The specificity occurs at both between- and within-species levels, such that each legume species or genotype can establish an efficient symbiosis with only a specific group of rhizobial species or strains. Genetic control of symbiosis specificity is complex, involving an exchange of multiple molecular signals between the symbiotic partners. Understanding the molecular mechanisms underlying symbiosis specificity would allow for development of tools for genetic improvement of biological nitrogen fixation in legumes.
In most but not all legumes, bacterial infection and nodule organogenesis is mediated by specific perception of bacterially derived lipo-chitooligosaccharides (called Nod factors) by the cognate plant receptors (Lerouge et al., 1990; Geurts et al., 1997; Limpens et al., 2003; Radutoiu et al., 2003, 2007). The Nod factors carry various species-specific chemical decorations, and this structural variation is widely thought to play an important role in defining the recognition specificity at the species level (Lerouge et al., 1990; Perret et al., 2000; Radutoiu et al., 2007). In addition to Nod factors, rhizobial surface polysaccharides, such as exopolysaccharides, lipopolysaccharides, capsular polysaccharides, and cyclic glucans, are also important for development of infected root nodules and for modulating host specificity (D’Haeze and Holsters, 2004; Jones et al., 2008; Deakin and Broughton, 2009). Recently, an exopolysaccharide receptor has been identified in Lotus japonicus that controls rhizobial infection and distinguishes between compatible and incompatible exopolysaccharides (Kawaharada et al., 2015).
Despite their unique attributes, the legume-rhizobial interactions share many common features with pathogenic plant-bacterial interactions (D’Haeze and Holsters, 2004; Deakin and Broughton, 2009). As such, plant immunity triggered by microbe-associated molecular patterns or bacterial effector proteins also plays a key role in regulation of strain-specific nodulation (D’Haeze and Holsters, 2004; Deakin and Broughton, 2009; Yang et al., 2010; Wang et al., 2012). It has been demonstrated in soybeans (Glycine max) that plants use classical NBS-LRR resistance genes to restrict nodulation with certain rhizobial strains (Yang et al., 2010). In this case, the host range of rhizobial symbionts is determined by the presence of type III effectors in the bacteria and the corresponding resistance genes in the plant.
In this report, we describe positional cloning of the soybean Rj4 gene. The Rj4 gene was first identified in 1972 (Vest and Caldwell, 1972) and subject to extensive study in the 1980s and 1990s (e.g. Devine and O’Neill, 1986; Devine et al., 1990; Sadowsky and Cregan, 1992). Soybean genotypes carrying an Rj4 allele restrict nodulation by many strains of Bradyrhizobium japonicum and Bradyrhizobium elkanii (Sadowsky and Cregan, 1992). B. elkanii is a poor symbiotic partner of soybeans because of its low nitrogen-fixation efficiency. In addition, many of the strains also produce rhizobitoxine, a compound that induces chlorosis in the host plant. Thus, cultivars with the Rj4 genotype are favorable in soils where the B. elkanii population is dominant because Rj4 stops those cultivars from forming invaded nodules with it. The Rj4 allele is frequently present in Gly soja, the wild progenitor of soybean, but less frequent in the modern cultivars from North America (Devine and Breithaupt, 1981). We mapped the Rj4 locus within a small genomic region on soybean chromosome 1 (Tang et al., 2014), and validated one of the candidate genes as Rj4 using both complementation tests and CRISPR/Cas9-based gene knockout experiments. We showed that Rj4 encodes a thaumatin-like protein that does not have a corresponding allele in the analyzed rj4 genetic backgrounds. This conclusion disagrees with the previous report that Rj4 is the Glyma.01G165800 gene (Hayashi et al., 2014). Instead, we provide convincing evidence that Rj4 is a duplicate copy of Glyma.01G165800.
RESULTS AND DISCUSSION
Fine Mapping of the Rj4 Locus
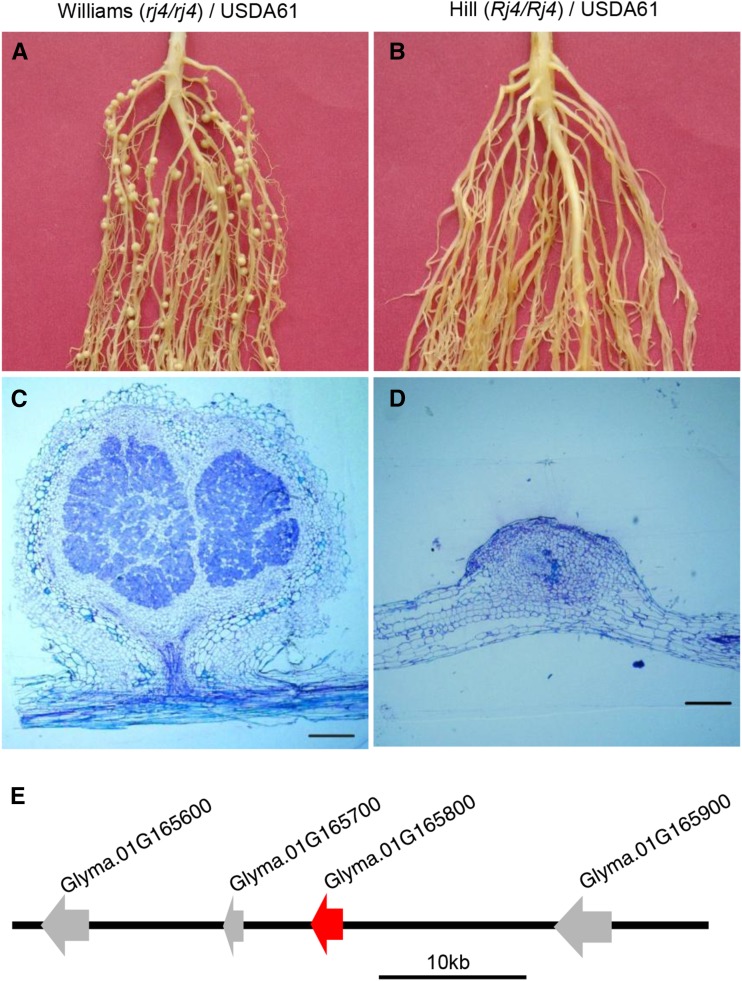
We previously mapped the dominant Rj4 locus to soybean chromosome 1 based on an F2 segregating population derived from the cross between Hill (Rj4/Rj4) and Williams (rj4/rj4) (Tang et al., 2014). Hill restricts nodulation by B. elkanii USDA61 (Nod−), whereas Williams nodulates normally with this strain (Nod+; Fig. 1, A and B). In the Rj4 background, the USDA61 strain could occasionally induce the formation of nodule primordium, but, in contrast to the compatible interaction, cortical cell division ceased at an early stage due to a lack of bacterial infection (Fig. 1, C and D). Phenotyping and genotyping of approximately 5,000 F2 plants enabled us to delimit the Rj4 locus within a 45-kb genomic region according to the reference genome sequence of Williams 82 (rj4/rj4; Schmutz et al., 2010; version Wm82.a2.v1). The 45-kb genomic sequence contains four predicted genes, including Glyma.01G165600, Glyma.01G165700, Glyma.01G165800, and Glyma.01G165900 (Fig. 1E). Sequence comparisons did not detect any nonsynonymous nucleotide substitutions between the parental alleles of Glyma.01G165600 and Glyma.01G165900, while Glyma.01G165700 represents a truncated and presumably nonfunctional gene in both parents. However, we identified five amino acid substitutions and two amino acid insertions/deletions between the two parental protein isoforms of Glyma.01G165800. These amino acid substitutions are correlated with the nodulation phenotypes based on the association analysis of 48 soybean genotypes, including 40 G. max and eight G. soja lines (Tang et al., 2014). Glyma.01G165800 (previously annotated as Glyma01g37060) encodes a thaumatin-like protein, a member of the pathogenesis-related protein family 5 (PR-5) that plays an important role in plant defense (van Loon et al., 2006). Based on these data, we suggested that the Hill allele of Glyma.01G165800 is possibly a candidate gene of Rj4 (Tang et al., 2014). Shortly after the publication of Tang et al. (2014), Hayashi et al. (2014) reported that Glyma.01G165800 was indeed the Rj4 gene. However, as described below, we were never able to validate this candidate gene.
Figure 1.
Rj4-mediated nodulation restriction in soybean and fine mapping of the Rj4 locus. A, Nod+ phenotype of Williams (rj4/rj4) by B. elkanii USDA61. B, Nod− phenotype of Hill (Rj4/Rj4) by B. elkanii USDA61. C, In the compatible Williams/USDA61 interaction, nodule developed normally and contained bacteria. D, In the incompatible interaction, occasionally nodule primordia were formed, but the nodule primordia did not contain bacteria. E, The Rj4 locus was delimited to a 45-kb genomic region on chromosome 1 containing four predicted genes, of which Glyma.01G165800 was considered as a candidate gene. Map is drawn to scale.
Complementation Tests Failed to Validate the Hill Version of Glyma.01G165800 as Rj4
To validate the candidate gene Glyma.01G165800, we developed genomic constructs that contain the Hill allele of Glyma.01G165800 under the control of its native promoter as well as driven by the Cauliflower mosaic virus 35S promoter. These gene constructs were transferred to the rj4 genetic background (Williams) via Agrobacterium rhizogenes-mediated hairy root transformation, followed by assaying the nodulation phenotypes after inoculation with B. elkanii USDA61. The transformation experiments were performed without antibiotic selection; thus, the hairy roots induced by A. rhizogenes contained both transgenic and nontransgenic, which can be readily distinguished by assaying the expression of a GUSPlus gene present in the modified binary plasmid pEarleyGate100. As shown in Figure 2, nodules were formed on the transgenic roots expressing the Hill allele of Glyma.01G165800. We repeated similar complementation tests a number of times over a 3-year period from 2012 to 2015 and obtained the same results. Thus, our results appeared not to support Rj4 as an allele of Glyma.01G165800, which contradicted the conclusion reported by Hayashi et al. (2014). As described later in this article, we further confirmed our conclusion by developing a CRISPR/Cas9 construct to knock out the Glyma.01G165800 gene in the Hill (Rj4/Rj4) background.
Figure 2.
Complementation test using the Hill allele of Glyma.01G165800. A, Introduction of the Hill allele of Glyma.01G165800 into Williams (rj4/rj4) failed to block nodulation on the transgenic hairy roots (blue) by USDA61. B, Examples of transgenic hairy roots expressing Glyma.01G165800 of Hill in Williams. The transgenic hairy roots were first identified by GUS staining of a small portion of the root segments, followed by isolation of RNA from the transgenic hairy roots.
Identification of Insertion/Deletion Polymorphisms Between the Rj4 and rj4 Genotypes
Since the genotype of the reference genome (Williams 82) is rj4/rj4, it is very likely that rj4 represents a null allele in this genetic background. To address this possibility, we screened a bacterial artificial chromosome (BAC) library derived from PI468916, a G. soja genotype harboring an Rj4 allele. BLAST search against the BAC ends database of PI468916 identified two BAC clones (GSS_Ba124A02 and GSS_Ba201P23) that cover the 45-kb reference genomic region where the Rj4 locus was mapped. Sequencing and assembly of both BACs identified numerous insertions in the PI468916 genome at this locus, including an approximately 10-kb large insertion plus several small insertions of several hundred bp (Fig. 3A). In particular, we identified a second copy of Glyma.01G165800 in the PI468916 genotype. The two duplicates are separated by approximately 7.5-kb nongenic sequences and share approximately 96% identity at the amino acid level (Fig. 3B). To distinguish between the two copies, we named the second copy of Glyma.01G165800 as Glyma.01G165800-D. Glyma.01G165800-D expressed at a very low level in the soybean root and was barely detectable under noninoculated conditions. However, its expression in the root was significantly enhanced 3 d post rhizobial inoculation (Fig. 3C). The similar expression pattern was also observed for Glyma.01G165800.
Figure 3.
Identification of a duplicate gene of Glyma.01G165800 in the Rj4 genotypes. A, Sequence analysis identified a total of approximately 13-kb insertions in the Rj4 genotype PI468916 compared to the reference genome Williams 82, including an approximately 10-kb large insertion indicated by the shaded bar plus several small insertions (not shown). The insertion contains a duplicate copy of Glyma.01G165800, which we called Glyma.01G165800-D. B, Alignment of predicted protein sequences of Glyma.01G165800 and Glyma.01G165800-D of Hill. Gray-highlighted letters indicated amino acid substitutions between the two proteins. C, RT-PCR (left) and quantitative RT-PCR (right) analyses indicated that the expression of Hill-Glyma.01G165800-D was not detectable in the noninfected roots, but the expression was induced after inoculation. Similar expression pattern was also observed for Glyma.01G165800 in Hill.
PCR amplification of genomic DNA from the same 48 Rj4 and rj4 genotypes (Tang et al., 2014) using gene-specific primers revealed that all the Rj4 genotypes possess both Glyma.01G165800 and Glyma.01G165800-D, while Glyma.01G165800-D is missing in all the tested rj4 genotypes (Supplemental Table S1). Based on these observations, we postulated that either Glyma.01G165800-D or both Glyma.01G165800 and Glyma.01G165800-D are required for Rj4-mediated nodulation restriction.
CRISPR/Cas9-Mediated Gene Knockout Revealed That Glyma.01G165800-D But Not Glyma.01G165800 Is Required for Rj4-Mediated Nodulation Restriction
We used the CRISPR/Cas9-based reverse genetics tool (Doudna and Charpentier, 2014) to knock out the Glyma.01G165800 and Glyma.01G165800-D genes in the Hill (Rj4/Rj4) genetic background. Glyma.01G165800 and Glyma.01G165800-D differ significantly at the 5′ coding region (Fig. 3B), allowing us to design gRNA vectors that specifically target individual genes. The vectors were introduced to A. rhizogenes K599 for hairy root transformation, followed by assaying the nodulation capacity of the hairy roots with B. elkanii USDA61. The transgenic roots were first identified by GUS staining, followed by DNA sequencing to validate the targeted DNA insertions/deletions. For each vector, we generated at least 100 independent transgenic roots from more than 50 plants.
For both genes, we successfully generated transgenic roots with both alleles being mutated, including homozygous roots containing two homogenous mutated alleles as well as heterozygous roots with two or more heterogeneous mutated alleles (Supplemental Text S1). Our experiments showed that the knockout of Glyma.01G165800 did not abolish Rj4 function; all the mutant roots were not able to restore nodulation with B. elkanii USDA61. In contrast, the knockout of Glyma.01G165800-D completely abolished Rj4 function, enabling all the mutant roots to form nodules after inoculation with USDA61. Examples of phenotypes are shown in Figure 4, A and B, with their mutant genotypes illustrated in Figure 4, C and D, respectively. Furthermore, we transferred the Hill version of Glyma.01G165800-D to the rj4 genetic background of Williams, and the transgenic roots successfully blocked the nodulation with USDA61 (Fig. 5). Taken together, these experiments unambiguously indicated that Glyma.01G165800-D but not Glyma.01G165800 is required for Rj4-mediated nodulation restriction.
Figure 4.
CRISPR/Cas9-mediated gene knockout of Glyma.01G165800 and Glyma.01G165800-D in the Hill (Rj4/Rj4) background. A, Knockout of Glyma.01G165800 did not affect Rj4-mediated nodulation restriction in Hill. Mutant hairy roots (blue), same as the wild-type roots, were not able to nodulate with USDA61. B, Knockout of Glyma.01G165800-D completely abolished Rj4-mediated nodulation restriction in Hill. Mutant hairy roots restored the ability to nodulate with USDA61. C, Sequence analysis indicated a single nucleotide deletion (indicated by an arrow) in one of the transgenic hairy roots shown in A. D, Sequence analysis indicated a single nucleotide insertion in one of the transgenic hairy roots (indicated by an arrow) shown in B.
Figure 5.
Complementation test using the Hill allele of Glyma.01G165800-D. A, Introduction of the Hill allele of Glyma.01G165800-D into Williams (rj4/rj4) successfully blocked the nodulation of the transgenic hairy roots (blue) by USDA61. B, Examples of transgenic hairy roots expressing Glyma.01G165800-D of Hill in Williams.
Similar to that of Glyma.01G165800, Glyma.01G165800-D contains three exons and two introns and is predicted to encode a protein of 296 amino acids. Sequence comparison identified a total of 13 amino acid substitutions between the two duplicate copies (Fig. 3B). These polymorphisms primarily occur at the N termini of the proteins, suggesting that these amino acid residues play an important role in Rj4-mediated nodulation restriction in soybeans.
CONCLUSION
We report here the isolation of the Rj4 gene that controls nodulation specificity in soybeans. We found that the Rj4 alleles are present in the Rj4 genetic backgrounds, while rj4 alleles are null alleles in the surveyed rj4 genotypes. Rj4 encodes a thaumatin-like protein but not the one previously reported by Hayashi et al. (2014). It is difficult to reconcile the different conclusions, given that we used similar experimental systems and biological materials. Resembling the soybean Rj2 gene that encodes a typical NBS-LRR protein, the Rj4-mediated nodulation restriction also relies on the bacterial type III secretion system (T3SS; Okazaki et al., 2009). Recently, a putative type III effector from B. elkanii USDA61 has been identified that is involved in the recognition process (Faruque et al., 2015). Furthermore, B. elkanii USDA61 was able to induce bacterial infection and nodule formation independent of Nod factor perception, and this ability is dependent on the T3SS of USDA61 (Okazaki et al., 2013). All these findings appeared to support that Rj4 triggers gene-for-gene resistance against the rhizobial infection through recognition of the cognate bacterial effectors. The fact that Rj4 is not an R gene but encodes a thaumatin-like protein is kind of surprising. It will be intriguing to understand how a thaumatin-like protein is involved in the effector-triggered immunity in plant-microbe interactions.
MATERIALS AND METHODS
Plant Materials and Nodulation Assay
The F2 mapping population was derived from the cross between Hill (Rj4/Rj4) and Williams (rj4/rj4). Plants were grown in sterilized perlite-turface mix in a growth chamber programmed for 16 h light at 26°C and 8 h dark at 23°C. Roots of 1-week-old seedlings were inoculated with B. elkanii USDA61. Nodulation assays were performed 2 to 3 weeks postinoculation. Genetic mapping of the Rj4 locus was described by Tang et al. (2014).
Complementation Tests and CRISPR/Cas9-Mediated Gene Knockout
The Hill versions of Glyma.01G165800 and Glyma.01G165800-D were used for complementation tests. The genomic DNA of the candidate genes, including both introns and exons, was PCR amplified and cloned into the binary vector pEarleyGate100. The expression of these genes was driven by the Cauliflower mosaic virus 35S promoter. To facilitate the identification of the transgenic hairy roots, we also pre-engineered a GUSPlus gene expression cassette (amplified from pCAMBIA1305.1) into the pEarleyGate100 vector. The primers used for amplification of the GUSPlus gene cassette were 5′GCAGGACCGGACGGGGCGAACTCGCCGTAAAGACTGG3′ and 5′TCGTCCGTCTGCGGGAGCACTGATAGTTTAATTCCCGAT3′. Purified PCR product was ligated into the AfeI and KpnI digested pEarleyGate100 DNA using the In-Fusion Advantage PCR Cloning Kits (Clontech). The primer pairs used for amplification of Glyma.01G165800 were 5′AAAAAAGCAGGCTTCCATCGTCCTTTGCCTCTTTC3′ and 5′AAGAAAGCTGGGTCTTAACAAAAGCACGGAGGGGAAATG3′, and for Glyma.01G165800-D were 5′AAAAAAGCAGGCTTCTACTTCTCCAACCCCTCACG3′ and 5′AAGAAAGCTGGGTCTCAATCTGTCCAAAGTGGGGCGTG3′. Purified PCR products were first ligated into the pENTR vector using Gateway BP clonase (Invitrogen) and then cloned into the destination vector pEarleyGate100 using Gateway LR clonase (Invitrogen).
For developing the CRISPR/Cas9 gene knockout constructs, we used the pHSE401 vector described by Xing et al. (2014). Two pairs of oligos were designed to specifically target Glyma.01G165800 and Glyma.01G165800-D, respectively. For Glyma.01G165800, we used the oligos 5′ATTGATGGGAAATTCAACTAAAA3′and 5′AAACTTTTAGTTGAATTTCCCAT3′. For Glyma.01G165800-D, we used the oligos 5′ATTGATGGCAAGTTCGACTAAAA3′ and 5′AAACTTTTAGTCGAACTTGCCAT3′. The underlined sequences represent the targeted sites. The oligo pairs were first annealed to produce a double-stranded fragment with 4-nt 5′ overhangs at both ends and then ligated into the BsaI digested pHSE401 vector. We also amplified the GUSPlus gene cassette from pCAMBIA1305.1 and ligated into the pHSE401 vector to facilitate the identification of transgenic hairy roots. For this purpose, the GUSPlus gene cassette was amplified using the primer pair 5′AATTGATTGACAACGAATTGAACTCGCCGTAAAGACTGG3′ and 5′GCTAAGATCGGCCGCAGCGCACTGATAGTTTAATTCCCGAT3′. The purified PCR product was then ligated into the EcoRI and AsiSI digested pHSE401 vector using the In-Fusion Advantage PCR Cloning Kits (Clontech).
For CRISPR/Cas9-based knockout experiments, to validate the targeted DNA insertions/deletions, the transgenic roots were first identified by GUS staining, followed by DNA isolation, PCR amplification, and DNA sequencing. If the initial sequencing indicated the p,resence of multiple heterogeneous mutant alleles, the PCR product was ligated into pGEM T-Easy Vector System (Promega), and 10 to 15 clones were selected for sequencing.
Hairy Root Transformation of Soybean
Agrobacterium rhizogenes-mediated hairy root transformation was performed according to the procedures described by Kereszt et al. (2007). Briefly, the A. rhizogenes strain K599 containing individual binary vectors was injected into the cotyledonary node of 1-week-old seedlings. The inoculated seedlings were grown in a growth chamber with at least 90% humidity. The main roots were cut when the hairy roots were long enough to support the plant growth. The composite plants were then inoculated with the rhizobial strain USDA61. Nodulation phenotypes were recorded three weeks after inoculation.
Analysis of Gene Expression
Total RNA was isolated by the Qiagen Plant RNeasy mini kit. Two micrograms of RNA was used to perform reverse transcription (RT)-PCR reactions using M-MLV reverse transcriptase (Invitrogen) in a 20-μL reaction mixture. Two microliters of the RT reaction was used as a template in a 20-μL PCR solution. The PCR primers were as follows: Glyma.01G165800 specific, 5′CATCGTGACTATGGCAACAA3′ and 5′CCGCTTCCCCTGGATATTTCTTGATA3′; Glyma.01G165800-D specific, 5′TACTTCTCCAACCCCTCACG3′ and 5′CCGCTTCCCCTGGATATTTCTTGATA3′; and GmActin, 5′GAGCTATGAATTGCCTGATGG3′ and 5′CGTTTCATGAATTCCAGTAGC3′.
Real-time quantitative PCR was carried out according to the instructions of the SsoAdvanced SYBR Green Supermix Kit (Bio-Rad) using a CFX Connect Real-Time System (Bio-Rad). The reaction mixture was heated at 95°C for 10 min and then followed by 40 cycles of 95°C for 15 s, 61°C for 15 s, and 72°C for 30 s. Three biological replicates were used in all the experiments. The ATP synthase subunit 1 (GmATS1) mRNA was amplified as a reference gene (Hayashi et al., 2014). The primers used in the analysis were as follows: for Glyma.01G165800, 5′TCGTGACTATGGCAACAACT3′ and 5′GCTGCACTTGTTGGTGATG3′; for Glyma.01G165800-D, 5′AGTGCACTTTGTGTCCCATA3′ and 5′GCTGCACTTGTTGATGATGG3′; and for GmATS1, 5′GCGATTCTTAAGCCAGCCTTT3′ and 5′ ACACACCCTGGAAACTGGTGA3′.
Anatomical Analysis
Soybean (Glycine max) roots with nodules or nodule primordia were harvested at 2 weeks after inoculation and immediately fixed in 50% ethyl alcohol, 5% glacial acetic acid, and 10% formaldehyde for 24 h at 4°C. The tissues were dehydrated in a graded ethanol series followed by a graded series of xylene. Following this, the roots were infiltrated with several changes of paraffin at 60°C and embedded in paraffin. Embedded tissues were sectioned (10 μm thick) with a microtome, stained with trypan blue, and examined with bright-field optics.
Accession Numbers
Sequence data generated from this project can be found in the GenBank/EMBL data libraries under accession numbers KU144684 to KU144689.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Presence (+) or absence (−) of Glyma.01G165800-D (Rj4) in 48 soybean genotypes.
Supplemental Text S1. Examples of the genotypes of Glyma.01G165800 and Glyma.01G165800-D knocked out hairy roots and their phenotypes
Supplementary Material
Acknowledgments
We thank Dr. Peter van Berkum (U.S. Department of Agriculture Agricultural Research Service, Beltsville, MD) for the B. elkanii strain USDA61 and Dr. Qijun Chen (China Agricultural University, Beijing, China) for providing the CRISPR/Cas9 binary vectors.
Glossary
- BAC
bacterial artificial chromosome
- RT
reverse transcription
Footnotes
This work was supported by the Kentucky Soybean Promotion Board. F.T. was supported by a graduate assistantship from Department of Plant and Soil Sciences of University of Kentucky.
References
- Broughton WJ, Jabbouri S, Perret X (2000) Keys to symbiotic harmony. J Bacteriol 182: 5641–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin WJ, Broughton WJ (2009) Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat Rev Microbiol 7: 312–320 [DOI] [PubMed] [Google Scholar]
- Devine TE, O’Neill JJ (1986) Registration of BARC-2 (Rj4) and BARC-3 (rj4) soybean germplasm. Crop Sci 26: 1263–1264 [Google Scholar]
- Devine TE, Breithaupt BH (1981) Frequencies of nodulation response alleles, Rj2 and Rj4, in soybean plant introductions and breeding lines. USDA Technical Bulletin No. 1628
- Devine TE, Kuykendall LD, O’Neill JJ (1990) The Rj4 allele in soybean represses nodulation by chlorosis-inducing bradyrhizobia classified as DNA homology group II by antibiotic resistance profiles. Theor Appl Genet 80: 33–37 [DOI] [PubMed] [Google Scholar]
- D’Haeze W, Holsters M (2004) Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol 12: 555–561 [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Faruque OM, Miwa H, Yasuda M, Fujii Y, Kaneko T, Sato S, Okazaki S (2015) Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl Environ Microbiol 81: 6710–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R, Heidstra R, Hadri AE, Downie JA, Franssen H, Van Kammen A, Bisseling T (1997) Sym2 of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis. Plant Physiol 115: 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Shiro S, Kanamori H, Mori-Hosokawa S, Sasaki-Yamagata H, Sayama T, Nishioka M, Takahashi M, Ishimoto M, Katayose Y, et al. (2014) A thaumatin-like protein, Rj4, controls nodule symbiotic specificity in soybean. Plant Cell Physiol 55: 1679–1689 [DOI] [PubMed] [Google Scholar]
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC (2008) Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci USA 105: 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, Gresshoff PM (2007) Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc 2: 948–952 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Okazaki S, Kaneko T, Sato S, Saeki K (2013) Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc Natl Acad Sci USA 110: 17131–17136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S, Zehner S, Hempel J, Lang K, Göttfert M (2009) Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol Lett 295: 88–95 [DOI] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64: 180–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky MJ, Cregan PB (1992) The soybean Rj4 allele restricts nodulation by Bradyrhizobium japonicum serogroup 123 strains. Appl Environ Microbiol 58: 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Tang F, Yang S, Liu J, Gao M, Zhu H (2014) Fine mapping of the Rj4 locus, a gene controlling nodulation specificity in soybean. Mol Breed 33: 691–700 [Google Scholar]
- van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Vest G, Caldwell BE (1972) Rj4: a gene conditioning ineffective nodulation in soybean. Crop Sci 12: 692–693 [Google Scholar]
- Wang D, Yang S, Tang F, Zhu H (2012) Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol 14: 334–342 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tang F, Gao M, Krishnan HB, Zhu H (2010) R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 107: 18735–18740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.