Abstract
Background. Currently, there are no tools to accurately predict tuberculosis relapse. This study aimed to determine whether patients who experience tuberculosis relapse have different immune responses to mycobacteria in vitro than patients who remain cured for 2 years.
Methods. Patients with an initial episode of pulmonary tuberculosis were recruited in South Africa. Diluted blood, collected at diagnosis and after 2 and 4 weeks of treatment, was cultured with live Mycobacterium tuberculosis for 6 days, and cellular RNA was frozen. Gene expression in samples from 10 patients who subsequently experienced relapse, confirmed by strain genotyping, was compared to that in samples from patients who remained cured, using microarrays.
Results. At diagnosis, expression of 668 genes was significantly different in samples from patients who experienced relapse, compared with expression in patients who remained successfully cured; these differences persisted for at least 4 weeks. Gene ontology and biological pathways analyses revealed significant upregulation of genes involved in cytotoxic cell-mediated killing. Results were confirmed by real-time quantitative reverse-transcription polymerase chain reaction analysis in a wider patient cohort.
Conclusions. These data show that patients who will subsequently experience relapse exhibit altered immune responses, including excessively robust cytolytic responses to M. tuberculosis in vitro, at the time of diagnosis, compared with patients who will achieve durable cure. Together with microbiological and clinical indices, these differences could be exploited in drug development.
Keywords: transcriptomics, microarray, drug development, blood, patient
Following antimycobacterial drug treatment, the majority of treatment-adherent patients with tuberculosis due to drug-susceptible Mycobacterium tuberculosis remain healthy and disease free. However, approximately 5% of patients experience recurrent tuberculosis within 2 years of treatment completion [1]. Tuberculosis recurrence can be due to either reinfection with a new organism or relapse with the same M. tuberculosis isolate; it is unknown why mycobacteria persist in some individuals after 6 months of treatment. In Cape Town, South Africa, a setting with a high tuberculosis burden, tuberculosis relapse predominates in the first year after tuberculosis treatment completion, whereas reinfection is more common later [2].
New tuberculosis drugs or regimens are needed, but there are no easy ways to determine the efficacy of compounds under development. The 2-year relapse rate is used to measure drug efficacy in trials of tuberculosis drugs or regimens, but demonstration of reduced relapse/enhanced efficacy will require many thousands of patients and is very time-consuming and costly. Biomarkers that predict the response to tuberculosis treatment and the risk of tuberculosis relapse early in clinical trials would greatly help to evaluate candidate drugs and select those for further development [3].
Tuberculosis recurrence is associated with the bacterial burden at the time of diagnosis and with delayed sputum conversion [4]. Early Bactericidal Assays performed during the first two weeks of treatment, and sputum conversion rates after 2 months of treatment, can indicate early drug efficacy but cannot measure the killing of persistent bacterial subpopulations later in treatment. Because bacteria with the capability of persisting and causing tuberculosis relapse cannot be detected by current techniques, identification of patients at risk of tuberculosis relapse following treatment would revolutionize clinical trials of new treatments and facilitate clinical management.
Gene expression profiling of ex vivo whole-blood specimens has revealed transcriptomic changes in patients with tuberculosis, compared with healthy people [5–7], and large-scale changes in gene expression during successful tuberculosis drug treatment [8–10]. A cross-sectional study of patients who previously experienced tuberculosis recurrence identified genes that discriminated this group from patients who remained cured after 1 tuberculosis episode [11]. Array technology has been used to investigate human phagocyte responses to pathogens, including M. tuberculosis, Toxoplasma gondii, and Leishmania species, [12, 13] and CD4+ and CD8+ T-cell responses to M. tuberculosis in vitro [14]. However, only whole-blood cultures contain all circulating leukocytes, including neutrophils, which play an important role in tuberculosis immunity and immunopathology [5, 15]. Variations in mycobacterial antigen–induced cytokine production [16, 17] or mycobacterial growth inhibition [18] among study populations are readily detectable in cultures of diluted whole-blood specimens, which traditionally include culture periods of approximately 6 days.
We hypothesized that cultures of diluted whole-blood specimens and microarray technology would enable identification of differences early during tuberculosis treatment in patients who subsequently experienced relapse, compared with patients who remained disease free for 2 years. Such differences could be developed into biomarkers to predict tuberculosis drug efficacy and could reveal why some people are more susceptible to tuberculosis relapse.
METHODS
Study Design and Sample Collection
Fourteen healthy BCG-vaccinated donors were recruited at the London School of Hygiene and Tropical Medicine. In Cape Town, 263 non–human immunodeficiency virus (HIV)-infected, smear-positive, untreated patients experiencing their first episode of pulmonary tuberculosis were recruited as part of a larger prospective cohort study [4]. Patients received conventional therapy (ie, isoniazid, rifampicin, pyrazinamide, and ethambutol for 2 months, followed by isoniazid and rifampicin for 4 months) recommended by the South African National Tuberculosis Program, and venous blood samples were collected at intervals throughout treatment. All patients were categorized as having achieved cure or completed treatment at the end of the first-episode treatment. Patients with a second tuberculosis episode within 2 years of follow-up were categorized as having tuberculosis relapse or M. tuberculosis reinfection, using restriction fragment–length polymorphism genotyping. Treatment adherence was monitored, and patients infected with drug-resistant strains were excluded. Patients gave written informed consent, with ethical approval for the study granted by the ethics committees of Stellenbosch University (Faculty of Health Sciences), the London School of Hygiene and Tropical Medicine, and the director of health of the City of Cape Town.
In Vitro Stimulation With Live Mycobacteria
One milliliter of blood, diluted 10-fold in HEPES-buffered Roswell Park Memorial Institute 1640 medium (Sigma-Aldrich, Missouri) supplemented with L-glutamine (Sigma-Aldrich), was cultured with 4 × 105 colony-forming units of M. tuberculosis H37Rv or Mycobacterium bovis bacillus Calmette-Guerin strain Glaxo-Evans, corresponding to a multiplicity of infection of 1 bacillus to 1 monocyte, assuming 4 × 105 monocytes/mL of peripheral blood [19]. After 6 days, supernatants were removed, cells were lysed in 10 mL of RNA/DNA Stabilization Reagent for Blood/Bone Marrow (Roche Applied Science, Basel, Switzerland), and specimens were frozen at −80°C.
Messenger RNA (mRNA) Isolation and Affymetrix GeneChip Hybridization
mRNA was isolated using the mRNA Isolation Kit for Blood/Bone Marrow (Roche Applied Science), quantified using the RiboGreen RNA quantitation method (Life Technologies, Paisley, United Kingdom), and processed using NuGEN Ovation reagents, to enhance sensitivity [20]. For each sample, 500 pg of mRNA was processed and hybridized to a Human Genome U133 Plus 2.0 Array GeneChip, as described previously [9].
Microarray Data Analysis
Microarray data files are deposited in the National Center for Biotechnology Information Gene Expression Omnibus Database under accession numbers GSE45386 (healthy BCG-vaccinated donors) and GSE67589 (patients with tuberculosis). Data were analyzed using GeneSpring GX 11.5 Software (Agilent Technologies). Affymetrix CEL files were processed by the GC robust multiarray average method and normalized. For the healthy BCG-vaccinated donor experiment, 47 014 of 54 675 gene entities were analyzed, and for the study of patients with tuberculosis 44 252 of 54 675 gene entities were analyzed, after filtering out genes whose expression values never exceeded the 20th percentile. Log2-transformed data are shown. Two-way analysis of variance (ANOVA), principal component analysis, and gene ontology (GO) pie chart generation were conducted using GeneSpring GX software. Further GO analyses were conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; available at: http://david.abcc.ncifcrf.gov/home.jsp [21]). The EASE score refers to a 1-tailed Fisher exact probability value in the DAVID system. Receiver-operator characteristic (ROC) curve analyses were conducted using GraphPad Prismv6.02.
Real-Time Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
mRNA was reverse transcribed in duplicate by using Superscript III (Invitrogen) according to manufacturer's instructions, with 5 ng of mRNA per 20-µL reaction. Negative controls were included without reverse transcriptase, to exclude genomic DNA contamination. Samples were diluted with 80 µL of RNAse-free water (Applied Biosystems, Cheshire, United Kingdom), and 5 µL of complementary DNA was used per 20-µL real-time qRT-PCR reaction. PCR was performed using SYBRGreen (Applied Biosystems) on an ABI Prism 7000 or 7500 Fast machine, followed by melting curve analysis: conditions were 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Absolute gene expression quantification was achieved by reference to 10-fold dilutions of PCR-generated DNA standards. Most primer pairs, purchased from Sigma-Aldrich, were designed using Primer3 software [22] as either intron-spanning sequences or as sequences binding separate exons; primer pairs for HPRT [23] and HuP0 [24] were obtained from the literature. Primer sequences are shown in Supplementary Table 1.
RESULTS
Human Transcriptomic Responses to Mycobacteria in Cultures of Diluted Whole-Blood Specimens
First, we investigated whether incubation of whole blood with live mycobacteria provides a sensitive platform to analyze global transcriptomic changes during immune responses. Preliminary flow cytometry experiments showed both M. tuberculosis and M. bovis bacillus Calmette-Guerin stimulated various lymphocyte populations in diluted whole-blood cultures after 6 days (data not shown). Diluted whole-blood specimens from 3 BCG-vaccinated donors were, for 6 days, stimulated with M. bovis bacillus Calmette-Guerin or M. tuberculosis or left unstimulated, and mRNA was analyzed by microarray. Both M. tuberculosis and M. bovis bacillus Calmette-Guerin had strong effects on gene expression, with >8000 genes differentially expressed by ≥2-fold between at least 2 conditions (Figure 1A). For approximately 2300 genes, M. tuberculosis and M. bovis bacillus Calmette-Guerin exerted a similar effect: GO analysis showed that genes involved in a broad range of biological processes, including signaling and multicellular processes, were represented among these regulated genes. Immune system genes also included a broad range of processes, including development, activation, effector process, migration, and immune response terms (Figure 1B). In contrast, M. tuberculosis and M. bovis bacillus Calmette-Guerin exerted different or opposing effects on approximately 5700 genes: these included a narrower range of gene ontologies, with overrepresentation of cell component biogenesis genes with antigen processing and presentation dominating the immune system category (Figure 1C). These may represent important immune evasion strategies.
Figure 1.
Mycobacterium tuberculosis and Mycobacterium bovis bacillus Calmette-Guerin can enhance or suppress gene expression in diluted whole-blood cultures. A, Diluted whole blood from 3 BCG-vaccinated donors was cultured for 6 days alone (control) or in the presence of live M. tuberculosis or M. bovis bacillus Calmette-Guerin. Messenger RNA was extracted and pooled for each condition, prior to hybridization to Affymetrix U133 Plus 2.0 microarrays. Data were normalized against the control sample. The heat map shows 8067 gene entities that were differentially expressed ≥2-fold between at least 2 conditions, following Euclidean complete linkage hierarchically clustering. Gene ontology analysis of the 2293 genes, which were similarly up- or down- regulated by both mycobacteria (B), or the 5774 genes, which showed opposing responses to M. tuberculosis and M. bovis bacillus Calmette-Guerin (C), are shown for gene ontology categories that were significantly overrepresented (P < 5 × 10−5). All data analysis was conducted in GeneSpring GX.
qRT-PCR assays were performed to ensure that the microarray results could be validated. Initially, reference housekeeping genes were identified. Expression of 7 potential housekeeping genes [25–28] was measured in samples generated from diluted whole-blood specimens obtained from 5 BCG-vaccinated donors and cultured for 6 days with M. tuberculosis, M. bovis bacillus Calmette-Guerin, or medium alone. The best 3 housekeeping genes, showing the highest expression stability across samples, were HuP0, cyclophilin A, and TBP (Supplementary Figure 1), determined using the GeNorm application [29].
Next, we measured expression of 2 cytokines interferon γ (IFN-γ) and interleukin 32 (IL-32) by qRT-PCR in 6 independent BCG-vaccinated donors. Upregulation of IFN-γ induced by both M. bovis bacillus Calmette-Guerin and M. tuberculosis was similar to that observed in the microarray experiment (Figure 2A), whereas potent upregulation of IL-32 specifically by M. tuberculosis was also replicated (Figure 2B). Finally, expression of 2 lysosomal enzymes, IFN γ–inducible protein 30 (IFI30) and acid phosphatase 2 (ACP2), upregulated by M. bovis bacillus Calmette-Guerin but not M. tuberculosis in the microarray experiment, was confirmed by qRT-PCR (Figure 2C and 2D). These data showed that differential responses to M. bovis bacillus Calmette-Guerin and M. tuberculosis were gene specific and not due to overall different stimulatory capacity of the mycobacteria. Biological and technical replication showed that stimulation of diluted whole-blood cultures with live mycobacteria provided a robust method to measure human transcriptomic responses. As different mycobacteria elicited strikingly different responses, subsequent work was conducted with live M. tuberculosis.
Figure 2.
Validation of diluted whole-blood assay results by real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis. Real-time qRT-PCR was conducted on samples from 6 BCG-vaccinated donors, independent from the 3 donors used in Figure 1. Results for interferon γ (IFN-γ; A), interleukin 32 (IL-32) (B), interferon gamma-inducible protein 30 IFI30 (D), and acid phosphatase 2 (ACP2; D) are shown as the number of transcript copies of the gene of interest divided by the geometric mean number of copies of the 3 housekeeping genes cyclophilin A, HuP0, and TBP, within each sample. Bars show the mean and standard error of the mean. Microarray data are shown for comparison. *P < .05 and **P < .01, by the Mann–Whitney U test. Abbreviation: CON, control.
Patients With Tuberculosis Relapse Exhibit Altered Immune Responses to M. tuberculosis In Vitro
To find biomarkers that might predict relapse, 263 HIV-negative, smear-positive patients with a first episode of tuberculosis were recruited in the Surrogate Marker study in Cape Town as previously described [4], and blood samples were collected during the first-episode treatment period. After 6 months of treatment, 191 patients from this cohort were deemed cured. However, 10 of these patients subsequently experienced relapsed during the 2-year follow-up (hereafter, “the tuberculosis-relapse group”), and their samples were analyzed in this study. From the remaining patients who were successfully cured and remained disease free for 2 years, 10 were randomly selected for comparison (hereafter, “the cured group”). Following selection, the patient groups were compared to ensure there was overall similarity in age, sex, and clinical characteristics at recruitment, to prevent confounding (Table 1).
Table 1.
Characteristics of the Patients Selected for Analysis
| Characteristic | Cured Group (n = 10) | Tuberculosis-Relapse Group (n = 10) |
|---|---|---|
| Age, y, median (range) | 31 (19–52) | 41 (22–63) |
| BMI, median (range)a | 18.3 (12.3–22.0) | 16.7 (14.2–20.4) |
| Male sex | 8/10 | 8/10 |
| Size of cavities, mm | ||
| 2–4 | 5 | 5 |
| >4 | 6 | 4 |
| Radiological extent of disease | ||
| Moderate | 3 | 3 |
| Severe | 6 | 7 |
| Sputum smear finding at 2 mo | ||
| Positive | 4 | 5 |
| Negative | 5 | 5 |
| Unknown | 1 | 0 |
a Body mass index (BMI) is calculated as the weight in kilograms divided by the height in meters squared.
To investigate whether patients with tuberculosis relapse differed in their responses to live M. tuberculosis in cultures of diluted whole-blood specimens, samples collected at diagnosis and after 2 and 4 weeks of tuberculosis treatment were analyzed by microarray. In 2-way ANOVA, 668 of 44 252 genes were significantly differentially expressed between the tuberculosis-relapse and cured groups (Supplementary Tables 2 and 3): this was substantially higher than 74, the number that would be expected by chance. When the more stringent Bonferroni multiple testing correction was applied, 18 genes were significantly differentially expressed between patients with relapse and those who were cured (Table 2), including several genes involved in cell-mediated cytotoxicity, such as perforin, granulysin, and fas ligand, which were more highly expressed in samples from patients with relapse. In accordance with previous reports using ex vivo blood [11, 12], large-scale changes in gene expression occurring during tuberculosis treatment were evident, irrespective of treatment outcome. The differences observed were not due to differential cell counts between the groups, as these were broadly similar at each time point (Supplementary Table 4).
Table 2.
Genes Most Significantly Differentially Expressed Between the Tuberculosis-Relapse and Cured Groups After Stimulation With Mycobacterium tuberculosis In Vitro
| Affymetrix Probe ID | Gene Symbol | Gene Name | Function | Patient Group With Higher Gene Expression | P Values |
|---|---|---|---|---|---|
| 239754_at | C17orf76? | C17orf76/SNORD49A/B/SNORD65 | Unknown—ambiguous probe set | Relapse | .002518 |
| 1552623_at | HSH2D | Hematopoietic SH2 domain containing | Adaptor protein, negative regulator of T-cell activation | Relapse | .011216 |
| 37145_at | GNLY | Granulysin | Cytotoxicity | Relapse | .011639 |
| 212805_at | PRUNE2 | Prune homolog 2 | Regulation of differentiation and survival of tumour cells | Relapse | .018670 |
| 209765_at | ADAM19 | Metallopeptidase domain 19 | Cell-cell and cell-matrix interaction and DC marker | Relapse | .020138 |
| 209515_s_at | RAB27A | RAB27A | Regulation of secretion including cytotoxic granules | Relapse | .030276 |
| 1553681_a_at | PRF1 | Perforin 1 | Cytotoxicity | Relapse | .042316 |
| 226863_at | FAM110C | Family with sequence similarity 110, member C | Cell cycle regulation, localized to centrosomes | Relapse | .043196 |
| 203588_s_at | TFDP2 | Transcription factor Dp-2 | Dimerization with E2F for activation of cell cycle genes | Relapse | .045783 |
| 211333_s_at | FASLG | Fas ligand | Cytotoxicity | Relapse | .049411 |
| 236858_s_at | RUNX2 | Runt-related transcription factor 2 | Osteoblastic differentiation and TGF-β signaling | Cured | .000612 |
| 1569040_s_at | ANKRD36BP2 | Ankyrin repeat domain 36B pseudogene 2 | Pseudogene (related to structural protein) | Cured | .001928 |
| 336_at | TBXA2R | Thromboxane A2 receptor | Platelet aggregation and haemostasis | Cured | .010481 |
| 227805_at | METAP1D | Methionyl amino-peptidase type 1D | Proteolysis | Cured | .015718 |
| 235085_at | SGK223 | Pragmin | Regulator of Src family kinases, signal transduction | Cured | .028989 |
| 206873_at | CA6 | Carbonic Anhydrase VI | Salivary protein | Cured | .029041 |
| 232844_at | IFT140 | Intraflagellar transport 140 homolog | Development and function of cilia | Cured | .031645 |
| 215946_x_at | IGLL3P | Immunoglobulin lambda-like polypeptide 3 | Pseudogene (related to immunoglobulin λ) | Cured | .048079 |
Gene entities that were >2-fold differentially expressed between any pair of conditions and had a P value of <.05 by 2-way analysis of variance with the Bonferroni familywise error rate correction multiple testing correction are shown.
Abbreviation: TGF-β, transforming growth factor β.
Clear differences in transcriptomic profiles were observed at the time of tuberculosis diagnosis between patients with relapse and those who were cured, with 263 genes differentially expressed by ≥2-fold (Benjamini–Hochberg corrected P < .05; Supplementary Table 5). The differential expression pattern across the first 4 weeks of treatment was strikingly consistent (Figure 3A), indicating that differences were fundamentally related to the responsiveness of patients to M. tuberculosis. In ex vivo unstimulated samples, there were no significant differences between the study group in expression of the 668 genes when a multiple testing correction was applied (data not shown).
Figure 3.
Blood gene expression patterns are consistent over 4 weeks of tuberculosis treatment and can be used to classify patient outcome. A, Expression patterns of genes differentially expressed between tuberculosis-relapse and cured groups in Mycobacterium tuberculosis–stimulated diluted whole-blood cultures over the first 4 weeks of tuberculosis treatment. The normalized hybridization intensity of expression of 381 genes found to be ≥2-fold differentially expressed (P < .05, with Benjamini–Hochberg false-discovery rate correction) at at least one time point is shown for the mean of each patient group at the indicated time points. Genes have been hierarchically clustered using a Euclidean algorithm in GeneSpring GX. B, Hierarchical clustering was performed, using a Euclidean similarity measure and centroid linkage rule, for both patients and genes, using the 309 genes that were on average ≥2-fold differentially expressed between tuberculosis-relapse and cured groups in diluted whole-blood samples collected after 4 weeks of treatment, following stimulation in vitro with live M. tuberculosis. The table shows the treatment outcome (C = cure and R = relapse), the disease severity at diagnosis (M = moderate or S = severe, based on chest radiography findings), the 2-month sputum smear result, and the ratio of lymphocytes to monocytes in blood at the time of tuberculosis diagnosis for each patient. C, Principal component analysis was performed for samples from the 10 patients with relapse and the 10 cured patients at the 3 time points, using the 18 genes that were highly significantly ≥2-fold differentially expressed, using the Bonferroni familywise error rate correction (Supplementary Table 4). D, Molecular signatures were derived for each sample by summing the normalized log2-transformed hybridization intensity data for the 8 genes that were upregulated in the cured group and the 10 genes that were upregulated in the tuberculosis-relapse group, according to analysis of variance (Table 2). Receiver operating characteristic curve analyses were conducted to determine the accuracy of the molecular signatures, to determine treatment outcome. In the treatment-relapse group, 9 had data at diagnosis, 8 had data from week 2, and 10 had data from week 4. In the cured group, 10 had data at all time points. Abbreviation: AUC, area under the curve.
Prediction of Tuberculosis Relapse, Using 668-Gene or 18-Gene Signatures in M. tuberculosis–Stimulated Samples
We investigated whether tuberculosis outcomes for individual patients could be predicted on the basis of expression of the 668-gene signature. Hierarchical clustering showed that 8 of 10 patients with relapse grouped together (Figure 3B), implying common underlying reasons for relapse, but that 2 patients may have experienced relapsed for different reasons. The patient clustering was not related to disease severity at diagnosis or to sputum smear findings at month 2. Recent studies have shown that the blood monocyte to lymphocyte (ML) ratio is associated with the risk of tuberculosis development [30]; however, the ML ratio was not related to patient clustering in this study (Figure 3B). The clustering was also unrelated to the degree of IFN-γ mRNA expression induction (data not shown). Principal component analysis using the 668-gene-signature showed that the majority of samples segregated depending on whether patients subsequently experienced relapse or remained cured (Supplementary Figure 3A) and that samples tended to group closely by patient (data not shown). Segregation was more evident when using the 18 highly significantly differentially expressed genes (Figure 3C). Classification models were also more accurate with the 18-gene signature, with an overall predictive accuracy of 0.84 with a naive Bayes model and of 0.82 with a partial least squares model, as opposed to 0.72 for each model with the 668-gene signature (Supplementary Figure 3B). Molecular signatures were derived for each sample by using the 8 genes that were upregulated in cured patients from the 18-gene signature (Supplementary Figure 3C) and the 10 genes that were upregulated in the patients who experienced relapse (Supplementary Figure 3D). ROC curve analysis showed that both the 8-gene and 10-gene molecular signatures were accurate at predicting relapse, with AUCs of 0.936 and 0.931, respectively (Figure 3D).
GO Analysis Reveals the Importance of Cytotoxicity in Relapse Discrimination
GO analysis of the 668-gene signature list showed substantial overrepresentation of genes involved in biological regulation and response to stimuli (Figure 4A). There was also significant overrepresentation of genes involved in the immune response (Figure 4A). To further understand which aspects of the immune response were differentially expressed between patients with relapse and those who were cured, the DAVID functional annotation tool was used to interrogate the biological processes GO terms more deeply. Twenty-three biological processes GO terms were identified, with ≥15 gene members and an EASE score <0.05 (Figure 4B). There was striking enrichment of genes involved in the induction of programmed cell death and apoptosis, with 9 of 23 GO terms related to this phenomenon (Figure 4B): these GO terms include the extrinsic apoptotic pathway elicited by cytotoxic immune cells, which were upregulated in the tuberculosis-relapse group. Genes involved in gene transcription were also significantly overrepresented in the 668-gene signature list (Figure 4B).
Figure 4.
Gene ontology (GO) analysis of 668-gene entities differentially expressed between the cured group and the tuberculosis-relapse group. A, GO analysis of the 668 gene entity list from the analysis of variance (Supplementary Table 2) was conducted in GeneSpring GX to grossly allocate function to the gene list. Biological processes are shown where they are overrepresented within the 668 gene entity list (P < .1). B, The same gene list was inputted into the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation tool. A total of 506 entities were mapped to unique known genes in the DAVID database, and 23 biological process GO terms, containing ≥15 entities and with an EASE score of <0.05, were identified within the gene entity list. GO terms are shown clustered by function. Fold–enrichment values, showing overrepresentation of the term within the 506 gene list, are plotted as bars on the top axis; values calculated on the basis of 1 – [EASE score] are plotted as dots on the bottom axis.
Excessive Cytolytic Gene Expression in Patients With Relapse Was Also Observed by qRT-PCR
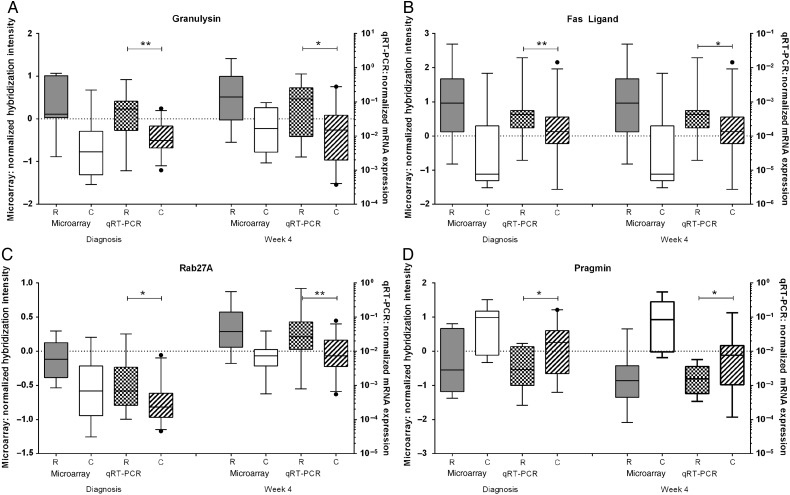
To confirm these findings, qRT-PCR experiments were conducted with mRNA samples from the original 20 patients analyzed by microarray and from 1 further patient with tuberculosis relapse who had been deemed to have completed treatment but subsequently relapsed. A further 17 successfully cured patients were also analyzed, including those with moderate and severe tuberculosis at diagnosis. The tuberculosis-relapse and cured patient groups analyzed by qRT-PCR were similar in terms of age, sex balance, month 2 sputum conversion, and proportion severely diseased at diagnosis (Supplementary Figure 2), whereas there was a higher proportion of men in these study groups than in the whole patient cohort, reflecting the association between male sex and relapse previously reported in this cohort [4]. mRNA expression of the cytotoxic effector molecules granulysin (Figure 5A), perforin (not shown), and fas ligand (Figure 5B) were significantly higher in tuberculosis-relapse patients than in cured patients, as was expression of Rab27A, which regulates secretion of cytotoxic granules (Figure 5C). In contrast, the expression of pragmin was higher in cured patients (Figure 5D), in accordance with the microarray data.
Figure 5.
Validation of microarray results by real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR). The expression of granulysin (A), fas ligand (B), rab27A (C), perforin (D), and pragmin (E) were measured by qRT-PCR in diluted whole-blood cultures stimulated for 6 days with live Mycobacterium tuberculosis H37Rv at the time of tuberculosis diagnosis and after 4 weeks of treatment. Data are for 11 patients in the tuberculosis-relapse group (2 with moderate and 9 with severe disease at the time of tuberculosis diagnosis) and for 27 patients in the cured group (6 with moderate and 21 with severe disease at the time of tuberculosis diagnosis). Data are as the number of copies of the gene of interest normalized against the geometric mean number of copies of HuP0, HPRT, and TBP. The box plots show the 25–75th percentile for the data set, with the line denoting the median, the whiskers showing the 5th–95th percentiles, and the dots showing the outliers. *P < .05 and **P < .01, by the Mann–Whitney U test. Abbreviation: mRNA, messenger RNA.
DISCUSSION
We have identified transcriptomic signatures in M. tuberculosis–stimulated whole-blood cultures that discriminated patients with tuberculosis who subsequently experienced relapse from those who remained cured for 2 years. The ability to predict which patients with tuberculosis are at risk of relapse after treatment completion would facilitate clinical management and enhance efficacy testing of new tuberculosis drugs, especially in early phase clinical trials. The majority of patients in this study (8 of 10) were correctly allocated to the tuberculosis-relapse or cured groups, based on expression of a 668-gene signature. Intriguingly, there was clear-cut discrimination of the majority of patients by principal component analysis: the distinction was present at the time of tuberculosis diagnosis, implying that these patients had inherently different immune responses to live M. tuberculosis, rather than a differential response to treatment. The misclassified patients exhibited more-complex mixed signatures. The likelihood of a relapsed, cured, or mixed phenotype in this study was not related to disease extent at diagnosis or sputum conversion at 2 months, highlighting that patients can experience relapse for different reasons. In an earlier study [11], subjects who had previously experienced recurrent tuberculosis had different blood transcriptomic profiles, compared with those who remained cured after 1 tuberculosis episode, even though blood specimens were collected from subjects while they were healthy and disease free, substantiating a role of inherent difference.
The relapse-risk gene signature included an exacerbated cytotoxicity response to M. tuberculosis in vitro, accompanied by higher expression of cell cycle genes indicative of excessive lymphocyte proliferation. Patients with tuberculosis develop peripheral CD8+ T cell cytotoxic T-lymphocyte (CTL) responses to a variety of antigens, such as Ag85A and ESAT-6 [31, 32], but the magnitude is reduced in patients with pulmonary tuberculosis, compared with latently infected healthy people [33, 34], possibly because of enhanced cell sequestration in the lungs of patients with tuberculosis or because of reduced cytolytic capacity of antigen-specific CTLs via regulatory mechanisms. Cell-mediated killing of host cells does not necessarily lead to mycobacterial killing and may drive recruitment of neutrophils. In mice, deletion of the regulatory protein PD1 leads to reduced mycobacterial control and more-rapid death [35], owing to excessive lung inflammation [36]. Here, conceivably, the inherently excessive cytotoxic inflammatory response in the patients with relapse led to exacerbated lung immunopathology, and the inflammatory response to remaining bacilli resumed once drug treatment ceased, causing further disease. It is possible that nonsterile cure can be achieved in most patients with tuberculosis, via development of an appropriately regulated effective T-cell response.
The 668-gene signature contained genes both up and down regulated in the relapse group compared to the cured group, with 356 genes more highly expressed in the cured group. Eight of the genes within the 18-gene highly significantly differentially expressed gene signature were more highly expressed in the cured group. These included Pragmin, which is involved in facilitating immune cell signaling by sequestering the negative regulator C-terminal Src kinase (Csk) in the cytoplasm [37]; downregulation in the tuberculosis-relapse group might dampen immune cell activation. The transcription factor RUNX2 was also downregulated in patients with relapse, potentially inhibiting PI3K/AKT cell signaling [38].
The inherently different immune response to M. tuberculosis in patients with relapse in this study was present at the time of tuberculosis diagnosis and during the first 4 weeks of treatment. Measuring responses later in treatment might identify patients with tuberculosis relapse for further reasons. Although the 2-month sputum conversion rate is often used as an indicator of tuberculosis drug efficacy, data regarding the association between smear/culture conversion and successful tuberculosis treatment outcome are inconsistent [39]. In 2 recent noninferiority clinical trials of gatifloxacin [40] or moxifloxacin [41] efficacy, aimed at shortening the duration of tuberculosis treatment, substantially higher recurrence and relapse rates occurred in experimental study arms despite more-rapid sputum conversion rates, compared with arms that received standard treatment. These trials demonstrate that some patients remain at risk of relapse after early effective bacterial killing, presumably owing to mycobacterial persistence in the absence of establishment of an effective immune response.
A clinically useful algorithm for predicting tuberculosis relapse or durable cure will likely include a combination of host gene expression, serum markers and microbiological readout. In a parallel study (Ronacher K, Chegou NN, Djoba Siawaya JF, et al, unpublished data), serum tumor necrosis factor β and soluble interleukin 6 receptor levels at the time of diagnosis correlated with the risk of tuberculosis relapse; these cytokines are markers of an inflammatory response. The next stage in the development of biomarkers to predict tuberculosis relapse will be testing transcriptomic signatures in much larger study cohorts. The study presented here was conducted using one of the largest cohorts currently available of patients with tuberculosis relapse, but validation in a large, independent, geographically distinct cohort would prove the ability of this transcriptomic signature for predicting tuberculosis relapse. Future relapse-prediction biomarker studies could be combined with existing tuberculosis drug and vaccine trials, to facilitate sample collection in the context of high-quality clinical and microbiological data collection from large study cohorts, including HIV-positive individuals. Although ex vivo blood samples are readily collected in field trials, it is noteworthy that the 668-gene signature that discriminated patient groups in the current study was not observed in matching ex vivo samples, and in vitro antigen-specific stimulation may add further discriminatory power to a combination biosignature. It is possible that the different patient groups responded with different kinetics following in vitro stimulation, and this could also be addressed in future studies. It may be possible to facilitate wider scale roll-out by adapting methodology using other mycobacterial preparations or antigens, such as QuantiFERON-TB Gold tube cell pellets, which might allow collection of samples after overnight culture. Measurement of protein concentrations in future studies would also provide functional rationale for inclusion in predictive biosignatures.
A composite biosignature trial end point, which accurately discriminates durable cure from risk of tuberculosis relapse, would allow more compounds and regimens to be tested more rapidly and cost-effectively. Recently, bedaquiline was conditionally approved by the Food and Drug Administration for multidrug-resistant (MDR) tuberculosis treatment; approval was based on 2-month sputum conversion data [42] with no randomized, controlled efficacy studies, as the need for new MDR tuberculosis drug need is so acute. However, there is currently no evidence of improved treatment outcome, and the use of inappropriate surrogate markers of cure could be very damaging in tuberculosis drug and regimen development [43]. The cytotoxic cell gene signature described here could be developed into one arm of a composite treatment outcome biosignature, which could be used for patient stratification in drugs trials. The discrimination of patients with relapse from those who achieved cure also indicates avenues of research for the development of host-directed therapy.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the staff of the Desmond Tutu TB Centre, under the leadership of Prof Nulda Beyers, for providing clinical samples and for database support; the clinical staff and patients at Ravensmaed, Uitsig, Elsierriver, Adriaanse, and Leonsdale, as well as the Cape Town City Health Directorate, for permitting this study; and Pauline Lukey, Ken Duncan, and Chris Clayton, for advice on study design.
Financial support. This work was supported by GlaxoSmithKline Action TB, the European and Developing Countries Clinical Trials Partnership (grants IP. 09.32040.011 and 2004.1.R.d1), the Bill and Melinda Gates Foundation (grant 48941), and EU-FP7 (TANDEM: grant 305279).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Jindani A, Nunn AJ, Enarson DA. Two 8-month regimens of chemotherapy for treatment of newly diagnosed pulmonary tuberculosis: international multicentre randomised trial. Lancet 2004; 364:1244–51. [DOI] [PubMed] [Google Scholar]
- 2.Marx FM, Dunbar R, Enarson DA et al. . The temporal dynamics of relapse and reinfection tuberculosis after successful treatment: a retrospective cohort study. Clin Infect Dis 2014; 58:1676–83. [DOI] [PubMed] [Google Scholar]
- 3.Wallis RS, Wang C, Doherty TM et al. . Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis 2010; 10:68–9. [DOI] [PubMed] [Google Scholar]
- 4.Hesseling AC, Walzl G, Enarson DA et al. . Baseline sputum time to detection predicts month two culture conversion and relapse in non-HIV-infected patients. Int J Tuberc Lung Dis 2010; 14:560–70. [PubMed] [Google Scholar]
- 5.Berry MP, Graham CM, McNab FW et al. . An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaforou M, Wright VJ, Oni T et al. . Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med 2013; 10:e1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maertzdorf J, Weiner J III, Mollenkopf HJ et al. . Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A 2012; 109:7853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom CI, Graham CM, Berry MP et al. . Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One 2012; 7:e46191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliff JM, Lee JS, Constantinou N et al. . Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis 2013; 207:18–29. [DOI] [PubMed] [Google Scholar]
- 10.Ottenhoff TH, Dass RH, Yang N et al. . Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One 2012; 7:e45839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mistry R, Cliff JM, Clayton CL et al. . Gene-expression patterns in whole blood identify subjects at risk for recurrent tuberculosis. J Infect Dis 2007; 195:357–65. [DOI] [PubMed] [Google Scholar]
- 12.Chaussabel D, Semnani RT, McDowell MA, Sacks D, Sher A, Nutman TB. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood 2003; 102:672–81. [DOI] [PubMed] [Google Scholar]
- 13.Thuong NT, Dunstan SJ, Chau TT et al. . Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog 2008; 4:e1000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cliff JM, Andrade IN, Mistry R et al. . Differential gene expression identifies novel markers of CD4+ and CD8+ T cell activation following stimulation by Mycobacterium tuberculosis. J Immunol 2004; 173:485–93. [DOI] [PubMed] [Google Scholar]
- 15.Martineau AR, Newton SM, Wilkinson KA et al. . Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest 2007; 117:1988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black GF, Weir RE, Floyd S et al. . BCG-induced increase in interferon-gamma response to mycobacterial antigens and efficacy of BCG vaccination in Malawi and the UK: two randomised controlled studies. Lancet 2002; 359:1393–401. [DOI] [PubMed] [Google Scholar]
- 17.Weir RE, Morgan AR, Britton WJ, Butlin CR, Dockrell HM. Development of a whole blood assay to measure T cell responses to leprosy: a new tool for immuno-epidemiological field studies of leprosy immunity. J Immunol Methods 1994; 176:93–101. [DOI] [PubMed] [Google Scholar]
- 18.Newton S, Martineau A, Kampmann B. A functional whole blood assay to measure viability of mycobacteria, using reporter-gene tagged BCG or M.Tb (BCGlux/M.Tb lux). J Vis Exp 2011; doi:10.3791/3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oxford Handbook of Clinical Medicine.
- 20.Vartanian K, Slottke R, Johnstone T et al. . Gene expression profiling of whole blood: comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics 2009; 10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 22.Untergrasser A, Cutcutache I, Koressaar T et al. . Primer3 - new capabilities and interfaces. Nucleic Acids Res 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burton MJ, Bailey RL, Jeffries D, Mabey DC, Holland MJ. Cytokine and fibrogenic gene expression in the conjunctivas of subjects from a Gambian community where trachoma is endemic. Infect Immun 2004; 72:7352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dheda K, Huggett JF, Chang JS et al. . The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem 2005; 344:141–3. [DOI] [PubMed] [Google Scholar]
- 25.de Kok JB, Roelofs RW, Giesendorf BA et al. . Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 2005; 85:154–9. [DOI] [PubMed] [Google Scholar]
- 26.Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004; 37:112–4, 116, 118–9. [DOI] [PubMed] [Google Scholar]
- 27.Hamalainen HK, Tubman JC, Vikman S et al. . Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal Biochem 2001; 299:63–70. [DOI] [PubMed] [Google Scholar]
- 28.Lossos IS, Czerwinski DK, Wechser MA, Levy R. Optimization of quantitative real-time RT-PCR parameters for the study of lymphoid malignancies. Leukemia 2003; 17:789–95. [DOI] [PubMed] [Google Scholar]
- 29.Vandesompele J, De Preter K, Pattyn F et al. . Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; doi:10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamath AB, Woodworth J, Xiong X, Taylor C, Weng Y, Behar SM. Cytolytic CD8+ T cells recognizing CFP10 are recruited to the lung after Mycobacterium tuberculosis infection. J Exp Med 2004; 200:1479–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lalvani A, Brookes R, Wilkinson RJ et al. . Human cytolytic and interferon gamma-secreting CD8+ T lymphocytes specific for Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 1998; 95:270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith SM, Klein MR, Malin AS et al. . Human CD8(+) T cells specific for Mycobacterium tuberculosis secreted antigens in tuberculosis patients and healthy BCG-vaccinated controls in The Gambia. Infect Immun 2000; 68:7144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caccamo N, Guggino G, Meraviglia S et al. . Analysis of Mycobacterium tuberculosis-specific CD8 T-cells in patients with active tuberculosis and in individuals with latent infection. PLoS One 2009; 4:e5528. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Madhan Kumar M, Raja A. Cytotoxicity responses to selected ESAT-6 and CFP-10 peptides in tuberculosis. Cell Immunol 2010; 265:146–55. [DOI] [PubMed] [Google Scholar]
- 35.Lazar-Molnar E, Chen B, Sweeney KA et al. . Programmed death-1 (PD-1)-deficient mice are extraordinarily sensitive to tuberculosis. Proc Natl Acad Sci U S A 2010; 107:13402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barber DL, Mayer-Barber KD, Feng CG, Sharpe AH, Sher A. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J Immunol 2011; 186:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safari F, Murata-Kamiya N, Saito Y, Hatakeyama M. Mammalian Pragmin regulates Src family kinases via the Glu-Pro-Ile-Tyr-Ala (EPIYA) motif that is exploited by bacterial effectors. Proc Natl Acad Sci U S A 2011; 108:14938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen-Solal KA, Boregowda RK, Lasfar A. RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Mol Cancer 2015; 14:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips PP, Fielding K, Nunn AJ. An evaluation of culture results during treatment for tuberculosis as surrogate endpoints for treatment failure and relapse. PLoS One 2013; 8:e63840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merle CS, Fielding K, Sow OB et al. . A four-month gatifloxacin-containing regimen for treating tuberculosis. N Engl J Med 2014; 371:1588–98. [DOI] [PubMed] [Google Scholar]
- 41.Gillespie SH, Crook AM, McHugh TD et al. . Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N Engl J Med 2014; 371:1577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diacon AH, Pym A, Grobusch M et al. . The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med 2009; 360:2397–405. [DOI] [PubMed] [Google Scholar]
- 43.Avorn J. Approval of a tuberculosis drug based on a paradoxical surrogate measure. JAMA 2013; 309:1349–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.