Abstract
Insulin-induced gene 1 (Insig-1) and Insig-2 are endoplasmic reticulum membrane–embedded sterol sensors that regulate the cellular accumulation of sterols. Despite their physiological importance, the structural information on Insigs remains limited. Here we report the high-resolution structures of MvINS, an Insig homolog from Mycobacterium vanbaalenii. MvINS exists as a homotrimer. Each protomer comprises six transmembrane segments (TMs), with TM3 and TM4 contributing to homotrimerization. The six TMs enclose a V-shaped cavity that can accommodate a diacylglycerol molecule. A homology-based structural model of human Insig-2, together with biochemical characterizations, suggest that the central cavity of Insig-2 accommodates 25-hydroxycholesterol, whereas TM3 and TM4 engage in Scap binding. These analyses provide an important framework for further functional and mechanistic understanding of Insig proteins and the sterol regulatory element–binding protein pathway.
Cholesterol homeostasis is essential for human physiology. Aberrant accumulation of sterols contributes to the initiation and progression of atherosclerosis that can lead to heart attack and stroke (1). Cellular sterol levels are monitored by several membrane-embedded proteins, including insulin-induced gene 1 (Insig-1) and Insig-2, which are essential components of the sterol regulatory element–binding protein (SREBP) pathway that controls cellular lipid homeostasis through a feedback inhibition mechanism (2–5).
SREBPs are a family of membrane-anchored transcription factors that activate genes encoding low-density lipoprotein receptor and enzymes for sterol synthesis (6–8). SREBP forms a stable complex with SREBP cleavage-activating protein (Scap) through their respective C domains (9–13). The complex is anchored on the endoplasmic reticulum (ER) through interactions between the membranous domain of Scap and Insig-1/-2 in a sterol-dependent manner (14, 15). Upon cholesterol deprivation, Scap dissociates from Insig-1/-2 and associates with COPII, which translocates the SREBP-Scap complex from the ER to the Golgi (16, 17). In the lumen of the Golgi, SREBP is cleaved by the membrane-anchored site-1 protease (S1P) and then by the intramembrane site-2 protease (S2P) (18, 19), allowing its soluble N-terminal transcription factor domain to enter the nucleus for gene activation (20–23).
Insig-1/-2 negatively regulate the cellular accumulation of sterols, mainly through two distinct mechanisms. First, upon binding to 25-hydroxycholesterol (25HC), Insig-1/-2 inhibit the exit of the SREBP-Scap complex from the ER, hence preventing transcriptional activation of genes for cholesterol synthesis and uptake (24). Second, during sterol repletion, Insig-1 recruits the protein degradation machinery to quickly destruct the rate-limiting 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase, blocking cholesterol synthesis (4, 25, 26).
The major players of the SREBP pathway have been identified for over a decade; however, the only structural information on membrane proteins of this pathway came from an archaeal ortholog of S2P (27). No bacterial homologs have been identified for SREBP, Scap, or HMG-CoA reductase. Here we report the high-resolution crystal structures of a mycobacterial homolog of Insig proteins. Guided by a homologous structural model of human Insig-2, a number of residues that may contribute to Scap association and 25HC binding were identified and biochemically confirmed.
Using human Insig-1 or -2 as a query, BLAST searches against sequenced bacterial genomes led to the identification of one protein from Mycobacterium vanbaalenii PYR-1. This protein, which we named MvINS, shares sequence identities of 23 and 26% with Insig-1 and -2, respectively (fig. S1), and sequence similarity of 40% with both proteins. Using MvINS as a query, six other proteins, all from mycobacteria, were identified (fig. S2A).
Crystals of MvINS in space group R3 appeared in several conditions under different detergents and diffracted x-rays beyond 2.0 Å resolution. The initial phases were derived from mercury-based single-wavelength anomalous dispersion (Hg-SAD) (fig. S3A). The final atomic model of MvINS was refined to 1.9 Å resolution (Fig. 1A, fig. S3B, and table S1). Consistent with the topological prediction for Insig-1/-2 (24, 28), MvINS comprises six transmembrane segments (TMs), with both the N- and C-termini located on the cytosolic side of the membrane. Given the level of sequence conservation, it is likely that all Insig proteins exhibit the same fold as MvINS.
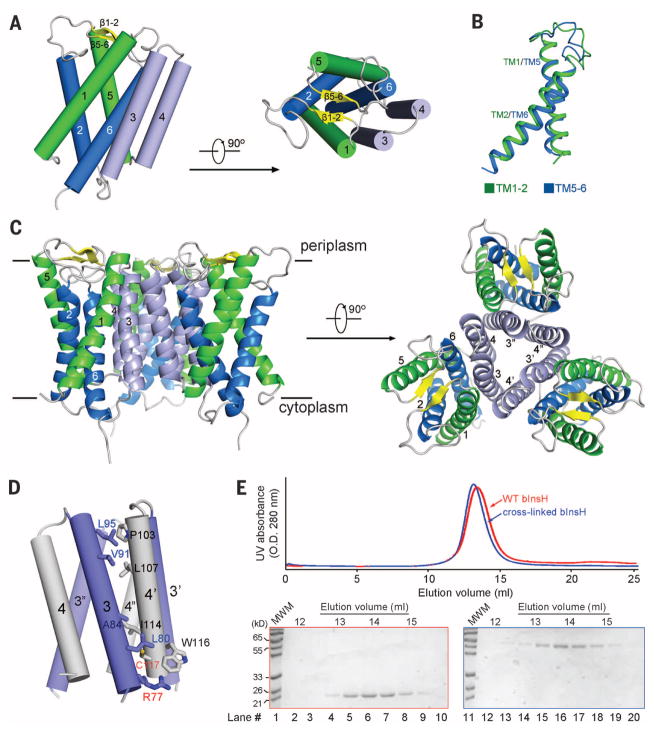
Fig. 1. The crystal structure of MvINS, a mycobacterial homolog of mammalian Insig proteins.
(A) The overall structure of an MvINS monomer. Two perpendicular views are shown. It reveals a novel fold that may be shared by all Insig proteins. (B) TM1/TM2 can be superimposed to TM5/TM6. (C) Three adjacent MvINS molecules form a homotrimer in the crystal. The side and periplasmic views of the overall trimer are shown. (D) The trimeric interface is mediated exclusively by hydrophobic residues on TM3 and TM4. Arg77 on TM3 was mutated to Cys for disulfide-bond formation with Cys117 on TM4 from the adjacent protomer. Both residues are labeled red. (E) MvINS is a trimer in solution. Wild-type and cross-linked MvINS-R77C were subjected to size exclusion chromatography. The peak fractions were applied to SDS–polyacrylamide gel electrophoresis followed by Coomassie blue staining. All structure figures were prepared with PyMol (37).
TM1 and -2, and TM5 and -6, each cross at the middle and are connected by a long segment. The four TMs together form a helical bundle that is tilted counterclockwise (Fig. 1A). The connecting sequences between TM1 and TM2 form a short β strand, which with the corresponding strand β5-6, caps the cavity enclosed by the helical bundle on the periplasmic side. TM1/TM2 and TM5/TM6 display an internal pseudo twofold symmetry around an axis that is perpendicular to the membrane plane and superimpose with root mean square deviation of 1.49 Å over 60 Cα atoms (Fig. 1B and fig. S2B). As found for the corresponding segments in human Insig-2, the N- and C-terminal fragments corresponding to TM1/TM2 and TM5/TM6 share considerable sequence similarity (fig. S2C).
There is one MvINS molecule in each asymmetric unit. Examination of the crystal lattice reveals a homotrimeric assembly involving three neighboring symmetry-related molecules (Fig. 1C and fig. S4A). The trimeric interface is mediated exclusively through van der Waals interactions between TM3 of one protomer and TM4 of the adjacent protomer (Fig. 1C, 1D). To examine whether MvINS indeed exists as a homotrimer in solution, we employed a disulfide bond–mediated cross-linking strategy.
Structural analysis showed that Arg77 on TM3 of one protomer and Cys117 on TM4 of the adjacent protomer are both on the cytoplasmic end of the helix with their Cα atoms 5.7 Å apart, ideal targets for disulfide-bond formation (Fig. 1D). We generated a MvINS variant (MvINS-R77C) with the mutation R77C and two additional cysteines mutated to alanine (C109A/C127A). Upon induction of disulfide-bond formation, MvINS formed a stable homotrimer (fig. S4B). The fully cross-linked MvINS-R77C and wild-type MvINS were eluted at almost identical volumes on size exclusion chromatography (Fig. 1E). Structural characterizations confirmed that MvINS-R77C shows the same trimer conformation as the wild type (fig. S4C and table S2).
Within each MvINS protomer, an extended cavity is formed by TM1/2/5/6 below the periplasmic β strands (Fig. 2A and fig. S5A). Modeling shows that a diacylglycerol (DAG) molecule with two 14-carbon aliphatic tails can be fitted into the V-shaped cavity. In the structure of MvINS extracted with n-dodecyl-N,N-dimethylamine-N-oxide (LDAO), two elongated strips of electron density filling two arms of the V-shaped cavity became clearly visible after most amino acids were modeled (Fig. 2A, inset). These densities probably represent aliphatic lipid or detergent molecules and occupy the DAG-binding site in the course of MvINS overexpression and/or purification.
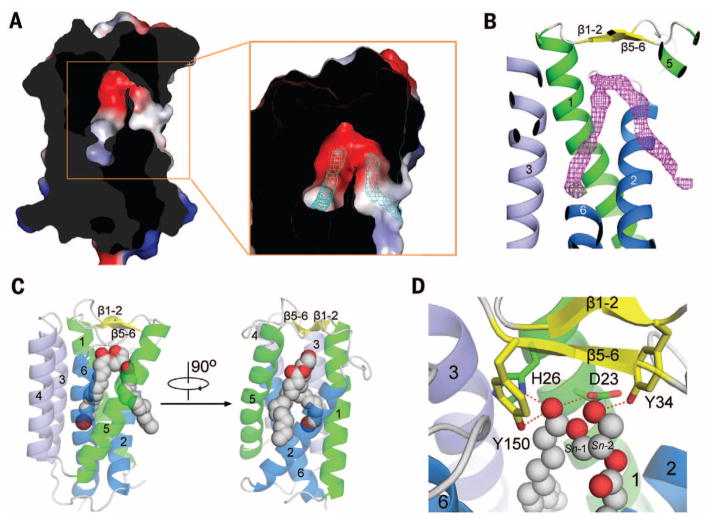
Fig. 2. Each MvINS protomer accommodates one DAG molecule.
(A) The MvINS protomer encloses a V-shaped cavity. Shown here is a cut-through view of the surface electrostatic potential (calculated in PyMol). The 2Fo-Fc electron density map, contoured at 1σ, is shown as cyan mesh in the inset. (B) Electron density for the bound DAG molecule. The 2Fo-Fc electron density for the bromine-derived DAG molecule, shown in magenta mesh, is contoured at 0.8σ. The anomalous signal for Br, shown as green mesh, is contoured at 3.5σ. (C) Overall structure of MvINS bound to DAG. The bromine (colored dark red)–derived DAG is shown in gray spheres. (D) Coordination of DAG by polar residues in MvINS. The residues that are hydrogen-bonded to DAG are shown in sticks. Hydrogen bonds are represented by red dashed lines.
To investigate whether MvINS can accommodate a DAG molecule, we synthesized a bromine-derived DAG, where the end methyl group of the Sn-1 fatty acyl chain is substituted by a bromine (Br) atom, and co-crystallized it with MvINS purified in Cymal-7. The structure was determined at 2.1 Å resolution (table S2). The V-shaped electron density, together with the Br anomalous signal, unambiguously confirmed the presence of DAG in the cavity (Fig. 2B).
The head group and the Sn-1 tail are coordinated by residues from TM1/2/3/6 and the periplasmic β strands, whereas the Sn-2 tail extends into the lipid bilayer through the cleft between TM2 and TM5 (Fig. 2C). The polar head of the DAG molecule is coordinated through hydrogen bonds by four residues close to the periplasmic side: Asp23 and His26 on TM1, Tyr34 on β1-2, and Tyr150 on β5-6 (Fig. 2D and fig. S2A). The completely buried Sn-1 aliphatic tail is surrounded by hydrophobic residues mainly from TM6 and, to a lesser extent, from TM1/2/3 (fig. S5B). The partially bound Sn-2 aliphatic chain, on the other hand, is loosely coordinated by a few amino acids, mainly from TM2/5 (fig. S5C).
On the basis of MvINS structure and the sequence similarity between MvINS and human Insig proteins (fig. S1), we generated a three-dimensional structural model for the transmembrane domain of Insig-1/-2 using the program MOE (29). As human Insig-1 and Insig-2 share 85% sequence identity at the transmembrane region, we focus here on the structural model of Insig-2 (fig. S6, A and B). The boundaries of the TMs are largely consistent with the prediction derived from biochemical analysis (24, 28), with some minor shifts. One notable difference concerns the beginning of TM2. In the homologous Insig-2 model, TM2 begins at Ser69, which adds seven extra amino acids to the extended linker sequence preceding TM2, as compared to the previous prediction (24) (fig. S6A). TM1/2/3/5/6 exhibit approximately 30% identity and 50% similarity between MvINS and Insig-2 (fig. S1). The degree of sequence similarity is probably sufficient for structural modeling of invariant and conserved residues on these five TMs (30–33).
In Insig-2, Phe115 is critical for the specific recognition of 25HC (24). Phe115 is located on the cavity-facing side of TM3 in the homologous model of Insig-2 (fig. S6C), suggesting that the central cavity might provide the accommodation for 25HC. The Insig-2 residues Gln132/Trp145/Asp149 mediate interactions with Scap but are not required for 25HC binding (24). All three residues are located on the outside surface of TM4 in the homologous model of Insig-2, suitably positioned to engage other binding partner(s) (fig. S6C). However, the corresponding residues of Gln132 and Trp145 on TM4 of MvINS are involved in homotrimerization, which may exclude its interaction with other proteins. We thus examined the oligomerization states of Insig-2. Size exclusion chromatography is consistent with Insig-2 existing as a monomer in solution; thus, TM4 of Insig-2 may be involved in hetero-oligomerization with Scap (fig. S6D).
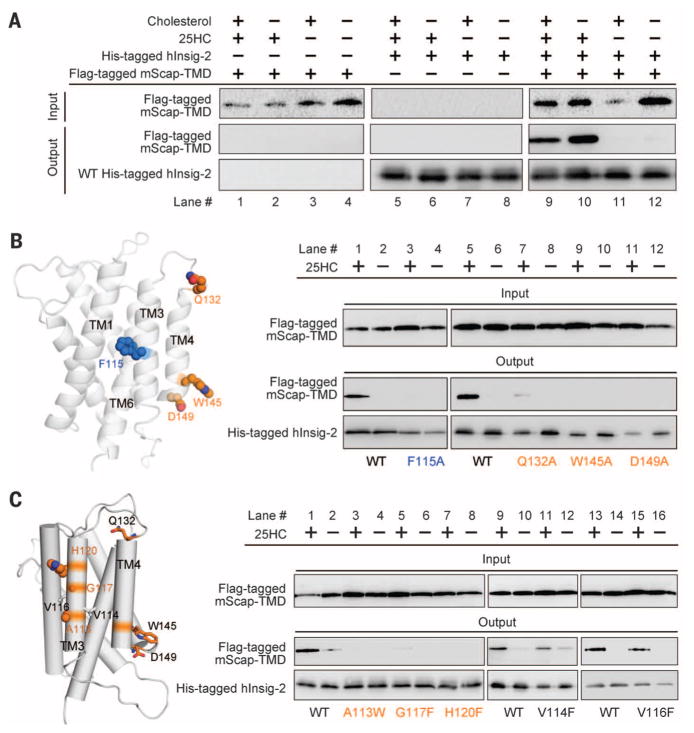
To establish the structure-function correlation, we attempted to reconstitute an assay system to examine the oxysterol-regulated interaction between Insig-2 and Scap. A pull-down assay showed 25HC-dependent complex formation between Insig-2 and Scap (Fig. 3A). Insig-2 variants that each carry a single point mutation of F115A, Q132A, W145A, or D149A failed to pull down Scap even in the presence of 25HC (Fig. 3B) (24).
Fig. 3. Structure-guided identification of functional residues in human Insig-2.
(A) The recombinantly expressed human Insig-2 interacts with the transmembrane domain of mouse Scap (mScap-TMD) in a 25HC-dependent manner. The recombinant proteins of Insig-2 and mScap-TMD were overexpressed in baculovirus-infected Sf-9 cells. (B) Examination of the previously identified functional residues using the insect cell assay system. Consistent with the previous report (24), single point mutations F115A, Q132Q, W145A, and D149A led to loss of Scap binding even in the presence of 25HC. (C) Identification of additional Insig-2 residues that are involved in the 25HC-dependent Scap binding. The three residues Ala113/Gly117/His120 are outward-facing residues on TM3. Single point mutations of these residues led to diminished Scap binding even in the presence of 25HC. In contrast, substitution of Val114 and Val116 with Phe retained complex formation with Scap. Shown here are representative results of at least three repeating experiments.
Insig-2 variants each containing a single point mutation on TM3 were generated and their interactions with Scap were examined. Replacement of the outward-protruding residues Ala113, Gly117, or His120 by aromatic residues led to complete disruption of complex formation, even in the presence of 25HC. In contrast, substitution of the residues Val114 or Val116, which appear to mediate intramolecular interactions with adjacent TMs, had no effect on Scap association (Fig. 3C). These observations suggest that TM3 and TM4 of Insig-2 contribute to Scap binding.
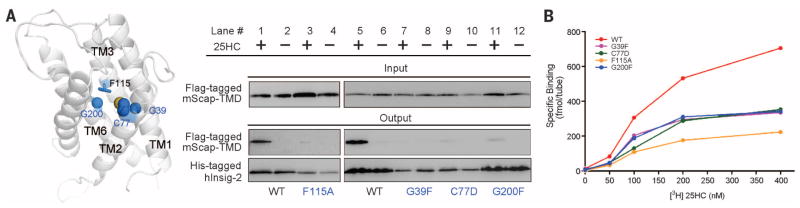
To identify additional residues for 25HC recognition, we examined the pocket residues (Fig. 4A and fig. S7). Single point mutation of residues adjacent to Phe115, including G39F, C77D, and G200F, led to abolished or markedly decreased complex formation in the presence of 25HC (Fig. 4A). These Insig-2 variants showed decreased affinity with 25HC in the direct binding assay (24) (Fig. 4B). Gly39 (TM1), Cys77 (TM2), and Gly200 (TM6) are clustered together with Phe115 (TM3) within the central cavity, enclosing a ring at approximately mid-height across lipid bilayer.
Fig. 4. Identification of residues involved in 25HC binding based on the homologous model of Insig-2.
(A) Identification of Insig-2 pocket residues that may contribute to 25HC binding. The pocket residues that are in the vicinity of Phe115 were analyzed. Single point mutations G39F, C77D, or G200F resulted in compromised mScap-TMD binding in the presence of 25HC. These three residues and Phe115 are positioned at a similar height within the central pocket of the structural model of Insig-2. (B) The measurement of direct binding between [3H]25HC and indicated Insig-2 variants. The experiments were performed following the identical protocol as reported previously (24). The data points represent the average of duplicate assays.
After the discovery of Insig proteins from mammalian cells (14, 15), their homologs have been identified in Schizosaccharomyces pombe (34), Saccharomyces cerevisiae (35), and Aspergillus fumigatus (36). Despite the moderate sequence homology between mammalian and yeast Insig proteins of typically only 20 to 30% identity, they appear to exhibit functional conservation (35). Therefore, the structural information reported here for a mycobacterial homolog is consistent with a structural and functional conservation of Insig proteins during evolution (fig. S8). However, no homologs of SREBP, Scap, or HMG-CoA reductase have been found in these mycobacteria, so the physiological function of the bacterial homolog remains a question.
Supplementary Material
Acknowledgments
We thank D. Rye, M. Brown, and J. Goldstein (University of Texas Southwestern Medical Center) for their assistance with studies on human Insig-2. We thank J. He, L. Tang, F. Yu, B. Sun, and S. Huang at the Shanghai Synchrotron Radiation Facility (SSRF) and K. Hasegawa and T. Kumasaka at the SPring-8 beamline BL41XU for onsite assistance; and B. Javid for critical discussion. This work was supported by funds from the Ministry of Science and Technology of China (grants 2015CB910101, 2011CB910501, 2014ZX09507003006) and National Natural Science Foundation of China (projects 31130002, 31125009, 91313303, and 20122209). A.R. was supported by the National Institutes of Health (HL-20948), American Heart Association (12SDG12040267), and Welch Foundation (I-1793). The research of N.Y. was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and an endowed professorship from Bayer Healthcare. The coordinates and structure factors of the MvINS proteins have been deposited in the Protein Data Bank with accession codes 4XU4, 4XU5, and 4XU6.
Footnotes
The authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Brown MS, Goldstein JL. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Shao W, Espenshade PJ. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein JL, DeBose-Boyd RA, Brown MS. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein JL, Brown MS. Cell. 2015;161:161–172. doi: 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama C, et al. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 7.Hua X, et al. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 9.Hua X, Nohturfft A, Goldstein JL, Brown MS. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 10.Sakai J, et al. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 11.Gong X, et al. Cell Res. 2015;25:401–411. doi: 10.1038/cr.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nohturfft A, Brown MS, Goldstein JL. J Biol Chem. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- 13.Yang T, Goldstein JL, Brown MS. J Biol Chem. 2000;275:29881–29886. doi: 10.1074/jbc.M005439200. [DOI] [PubMed] [Google Scholar]
- 14.Yang T, et al. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 15.Yabe D, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espenshade PJ, Li WP, Yabe D. Proc Natl Acad Sci USA. 2002;99:11694–11699. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun LP, Li L, Goldstein JL, Brown MS. J Biol Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 18.Sakai J, et al. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 19.Rawson RB, et al. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 20.Rawson RB. Nat Rev Mol Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- 21.Ikonen E. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 22.Sakai J, et al. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 23.Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo Y, Debose-Boyd RA. Crit Rev Biochem Mol Biol. 2010;45:185–198. doi: 10.3109/10409238.2010.485605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burg JS, et al. Cell Metab. 2008;8:522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng L, et al. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 28.Feramisco JD, Goldstein JL, Brown MS. J Biol Chem. 2004;279:8487–8496. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- 29.Molecular Operating Environment (MOE) Chemical Computing Group; Montreal, Canada: 2013. [Google Scholar]
- 30.Deng D, et al. Nature. 2014;510:121–125. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- 31.Sun L, et al. Nature. 2012;490:361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 32.Penmatsa A, Wang KH, Gouaux E. Nature. 2013;503:85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 34.Hughes AL, Todd BL, Espenshade PJ. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Flury I, et al. EMBO J. 2005;24:3917–3926. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborne TF, Espenshade PJ. Genes Dev. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMOL Molecular Graphics System. 2002 www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.