Abstract
The tumor suppressor p53 functions predominantly as a transcription factor by activating and downregulating gene expression, leading to cell cycle arrest or apoptosis. p53 was shown to indirectly repress transcription of the CCNB2, KIF23 and PLK4 cell cycle genes through the recently discovered p53-p21-DREAM-CDE/CHR pathway. However, it remained unclear whether this pathway is commonly used. Here, we identify genes regulated by p53 through this pathway in a genome-wide computational approach. The bioinformatic analysis is based on genome-wide DREAM complex binding data, p53-depedent mRNA expression data and a genome-wide definition of phylogenetically conserved CHR promoter elements. We find 210 target genes that are expected to be regulated by the p53-p21-DREAM-CDE/CHR pathway. The target gene list was verified by detailed analysis of p53-dependent repression of the cell cycle genes B-MYB (MYBL2), BUB1, CCNA2, CCNB1, CHEK2, MELK, POLD1, RAD18 and RAD54L. Most of the 210 target genes are essential regulators of G2 phase and mitosis. Thus, downregulation of these genes through the p53-p21-DREAM-CDE/CHR pathway appears to be a principal mechanism for G2/M cell cycle arrest by p53.
INTRODUCTION
The key functions of the tumor suppressor p53 are regulation of the cell cycle and induction of apoptosis. Its dominant role is to serve as a transcription factor by activating or downregulating expression of target genes (1,2). The gene of the cyclin-dependent kinase (CDK) inhibitor p21 (WAF1, CIP1, CDKN1A) was the first p53 transcriptional target to be identified and shown to be activated by p53 (3). Additionally, p21 was later implicated in mediating indirect transcriptional repression by p53 (4–12). A possible mechanism for indirect transcriptional repression is based on the cdk-inhibitor function of p21 which leads to hypophosphorylation of retinoblastoma tumor suppressor pRB-related pocket proteins. Low phosphorylation levels of the pRB-related p130 and p107 proteins then induce formation of a protein complex named DREAM (DP, RB-like, E2F4 and MuvB). DREAM assembles as a result of changing components of the MMB (MYB-MuvB) complex (13–15). In contrast to the activating MMB complex, DREAM is a transcriptional repressor (15–18). The p53-p21-DREAM pathway was shown to function through the cell cycle-dependent element (CDE) and cell cycle genes homology region (CHR) promoter sites (19). The switch in protein binding to the CDE and CHR elements was suggested to control transcriptional downregulation by p53 upon activation of the p53-p21-DREAM-CDE/CHR pathway (19).
Genes regulated by CDE and CHR sites are categorized as class I and class II genes. Both classes contain functional CHR elements, while class I also employs a CDE four nucleotides upstream of a CHR (20). In contrast to CDE sites, CHR elements conform to a well-defined consensus (21). Many promoters of late cell cycle genes contain functional CHR elements, but do not require CDE sites (15,21). The requirement for CHR elements stems from the binding of the MuvB complex, which forms the core of DREAM and MMB, to CHR sites (15,21). Therefore, it is reasonable to categorize genes by their CHR (15,20,21). While bound by DREAM, they mediate transcriptional repression in G0 and G1. Later during the cell cycle, DREAM-mediated repression is lost and the CHR contributes to activation of the previously repressed genes by binding MuvB complexes which recruit B-MYB (MMB) and FOXM1 proteins (15,21,22).
Previously, three target genes regulated by the p53-p21-DREAM-CDE/CHR pathway were known, namely CCNB2, KIF23 and PLK4 (19,23,24). With the small number of established targets, the general importance of the p53-p21-DREAM-CDE/CHR pathway remained to be established. Thus, we took a genome-wide approach with the aim to identify possibly all genes targeted by this pathway.
The CHR elements of CCNB2, KIF23 and PLK4 display the canonical sequence TTTGAA or its functional inverse site TTCAAA (19,23,24). Here, we included a recent compilation of CHR sites deviating from the canonical TTTGAA (21). A total of ten CHR variants are known to date with examples represented by the cell cycle genes Ccna2 (CHR sequence: CTTGAA) (25), Ccnb1 (TTTAAA) (26), Ccnb2 (TTTGAA) (15), B-Myb (Mybl2; TAGGAA) (27), Bub1 (TTCGAA), Chek2 (TTTGTA), Melk (TTTGAT), Pold1 (TTTGAG), Rad18 (TTCGAG) and Rad54l (TTCGAT) (21). Furthermore, we employed genome-wide chromatin immunoprecipitation (ChIP) data on DREAM binding (13) and genome-wide p53-dependent gene expression data from a meta-analysis (28). Our results provide a global map of 210 genes that are most likely regulated by the p53-p21-DREAM-CDE/CHR pathway. With many of these genes performing essential functions in G2 phase and mitosis, the results suggest that the p53-p21-DREAM-CDE/CHR pathway represents a key mechanism in p53-mediated cell cycle arrest.
MATERIALS AND METHODS
Computational analyses
Sources of primary data for the analyses were six studies on p53-dependent RNA expression (29–34) and ChIP results on binding of DREAM components E2F4, LIN9, LIN54 and p130 (13). A previously published list of 19 736 known protein-coding genes including their identifiers, p53 Expression Score and DREAM binding information served as a basis for the analyses (28). Furthermore, potential pathway target genes were selected for CHR sites annotated in the promoter regions of the genes. String representation for the CHR was chosen as follows: TTTGAA, TTTAAA, TTCGAA, TTTGTA, CTTGAA, TTTGAG, TTTGAT, TTCGAG, TTCGAT and TAGGAA (21). CHR elements were searched in the region of 1000 bp upstream and downstream from the transcriptional start site (TSS) on both strands that were not extended into the coding sequence or other genes located upstream of the TSS. PhastCons conservation scores (35) obtained from the multiz46 alignment of placental mammalia (36) were used to calculate average phylogenetic sequence conservation. Only those hits were listed that have an average PhastCons conservation score of at least 0.9. Results are displayed in Supplementary Table S2. Genes that are bound by DREAM, possess a conserved CHR and display a p53 Expression Score ≤ −3 are presented as strong candidate targets of the p53-p21-DREAM-CDE/CHR pathway in Table 1. Pathway enrichment analysis was carried out using the DAVID Functional Annotation tool (37).
Table 1. The p53-p21-DREAM-CDE/CHR signaling pathway regulates 210 potential target genes.
| ADSS | CDC25C | DLEU1 | IFT80 | MND1 | RAD54L |
| ANLN | CDC7 | DLGAP5 | INCENP | MTF2 | RANGAP1 |
| ANP32E | CDCA2 | ESCO2 | ING1 | MYBL2 | RBM15 |
| ARHGAP11A | CDCA3 | ESPL1 | ING3 | NASP | RBMX |
| ARHGAP11B | CDCA5 | EXOSC8 | IQGAP3 | NCAPD2 | REEP4 |
| ARL13B | CDCA8 | EXOSC9 | KIAA1731 | NCAPD3 | RIF1 |
| ARL6IP1 | CDK1 | FAM64A | KIF11 | NCAPG | RNASEH2A |
| ASF1B | CDK2 | FAM83D | KIF14 | NCAPG2 | RTKN2 |
| ASPM | CDKN3 | FANCB | KIF15 | NCAPH | SASS6 |
| ATAD2 | CENPA | FBXO5 | KIF18A | NDC80 | SCLT1 |
| AURKA | CENPE | FOXM1 | KIF20A | NEIL3 | SGOL1 |
| AURKB | CENPF | GAS2L3 | KIF20B | NET1 | SGOL2 |
| BIRC5 | CENPL | GSG2 | KIF22 | NUF2 | SHCBP1 |
| BORA | CENPM | GTSE1 | KIF23 | NUP107 | SLC25A40 |
| BUB1 | CENPN | H2AFX | KIF24 | NUP205 | SMC2 |
| BUB1B | CENPO | H2AFZ | KIF2C | NUP35 | SMC4 |
| C11orf82 | CEP152 | HAUS8 | KIF4A | NUSAP1 | SNRPA |
| C12orf32 | CEP55 | HIST1H2AE | KIFC1 | OIP5 | SP4 |
| C15orf42 | CHEK2 | HIST1H2AM | KPNB1 | ORC1 | SPAG5 |
| C2orf69 | CIT | HIST1H2BF | LIN54 | PCNT | SPC25 |
| C3orf26 | CKAP2L | HIST1H2BH | LIN9 | PLK1 | STIL |
| C9orf100 | CKAP5 | HIST1H2BI | LMNB1 | PLK4 | SUZ12 |
| CACYBP | CKS1B | HIST1H2BM | LRRC49 | POC5 | TCERG1 |
| CASC5 | CKS2 | HIST1H2BN | LSM5 | POLD1 | TMEM48 |
| CBX3 | CSE1L | HIST1H3C | MAD2L1 | POLQ | TMPO |
| CCDC150 | CSTF1 | HIST1H3D | MASTL | POP7 | TPX2 |
| CCDC18 | CTDSPL2 | HIST1H4C | MCM5 | PPIH | TRAIP |
| CCDC34 | DARS2 | HIST2H2AB | MCM7 | PRC1 | TROAP |
| CCDC99 | DBF4B | HIST2H2AC | MCM8 | PRIM2 | TTK |
| CCNA2 | DCAF16 | HJURP | MDC1 | PRPF38A | UBE2C |
| CCNB1 | DCK | HMGB2 | MELK | PRR11 | UBE2S |
| CCNB2 | DCLRE1B | HMMR | METTL13 | PTTG1 | UNG |
| CDC20 | DDX10 | HNRNPA0 | METTL4 | RACGAP1 | USP1 |
| CDC25A | DEPDC1 | HNRNPA2B1 | MIS18BP1 | RAD18 | YEATS4 |
| CDC25B | DEPDC1B | HNRNPUL1 | MKI67 | RAD21 | ZNF367 |
Cell culture and drug treatment
HCT116 wild-type and HCT116 p21−/− cells (38), HFF, and NIH3T3 cells (DSMZ, Braunschweig, Germany) were grown in Dulbecco's modified Eagle's medium (DMEM; Lonza, Basel, Switzerland) supplemented with 10% fetal calf serum (FCS) (Biochrom, Berlin, Germany) and penicillin/streptomycin and maintained at 37°C and 10% CO2. Cells were treated with DMSO (15 μl), doxorubicin (0.2 μg/ml; Medac, Wedel, Germany), nutlin-3a (10 μM; Cayman Chemicals, Ann Arbor, MI, USA) or 5-FU (25 μg/ml; Sigma, Taufkirchen, Germany) as indicated.
Flow cytometry
Cells were fixed for at least 12 h at 4°C in one volume phosphate-buffered saline/1 mM EDTA and three volumes of absolute ethanol. DNA was stained with propidium iodide at a final concentration of 10 μg/ml in presence of RNase A (10 μg/ml). DNA content per cell was measured by flow cytometry on an LSR II instrument (Becton Dickinson, Franklin Lakes, NJ, USA). Cell sorting was carried out on a FACSVantage SE (Becton Dickinson). Data analysis was carried out with WinMDI 2.9 software.
RNA extraction, reverse transcription and semi-quantitative real-time polymerase chain reaction (PCR)
Total cellular RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. One-step reverse transcription and quantitative real-time PCR were performed with an ABI 7300 Real-Time PCR System (Applied Biosystems, Forster City, CA, USA) using QuantiTect SYBRGreen PCR Kit (Qiagen, Hilden, Germany) as described previously (19,24). Primer sequences are listed in Supplementary Table S1.
Chromatin immunoprecipitation (ChIP)
HCT116 and HFF cells were cross-linked with 1% formaldehyde for 10 min at room temperature. ChIP was performed as described previously (19,24). The following antibodies were used for precipitation of transcription factors: E2F4 (C-20, Santa Cruz Biotech.), p130 (C-20, Santa Cruz Biotech.), p53 (Ab-6, DO-1, Calbiochem), LIN9 (ab62329, Abcam, Cambridge, UK). A second LIN9 antibody was a kind gift from James DeCaprio (13). A non-targeting IgG polyclonal rabbit antibody was used as a control for non-specific signals. For all precipitations 1–2 μg of antibody and 20–35 μl of Protein G Dynabead suspension (Invitrogen) were used. Immunoprecipitated DNA was used as template for quantitative real-time PCR as described previously (19,24). Primer sequences are listed in Supplementary Table S1.
RESULTS
The p53-p21-DREAM-CDE/CHR signaling pathway regulates 210 potential target genes
Three sets of information were utilized to identify possibly all genes regulated by the p53-p21-DREAM-CDE/CHR pathway: Genome-wide RNA expression dependent on p53, ChIP data on binding of DREAM components in the human genome, and genome-wide detection of phylogenetically conserved CHR elements.
Data from a meta-analysis of six genome-wide p53-dependent gene expression analyses to identify genes that are repressed by p53 formed one basis for the analysis (28–34). In each study, a gene could be identified as activated (positive score; +1) or repressed (negative score; −1) by p53. By calculating the sum over all analyses, p53-dependent Expression Scores ranging from −6 to +6 were assigned, representing direction as well as reproducibility of regulation (28). An Expression Score ≤ −3 was a criterion for a gene to be considered as repressed by p53. As a second data set, binding results of DREAM components E2F4, LIN9, LIN54 and p130 from ChIP analyses were employed to detect DREAM target genes (13). A gene was scored positive for DREAM binding if three of the four components were detected. The third selection criterion was a conserved CHR site in the promoter of a gene. CDE sites were not used because many potential target genes were expected to employ solely a CHR and not require a CDE (20). Since all CDE sites require CHR elements for their function, focusing the search on CHR sites covers all CHR- and CDE/CHR-regulated genes (20,21). CCNB2, KIF23 and PLK4, the three genes thus far shown to be regulated by the p53-p21-DREAM-CDE/CHR pathway, bind DREAM through the canonical CHR sequence TTTGAA (19,23,24). Recently, variants of CHR sites were identified in a comprehensive genome-wide screen. This yielded a list of ten functional CHR variants, namely TTTGAA, TTTAAA, TTCGAA, TTTGTA, CTTGAA, TTTGAG, TTTGAT, TTCGAG, TTCGAT and TAGGAA (21). Here, we employed all of these CHR motifs in a genome-wide search to obtain an essentially complete set of genes regulated via CHR- and CDE/CHR sites. In order to score positive as a CHR site, phylogenetic conservation of potential CHRs within 1000 bp upstream or downstream of annotated transcriptional start sites (TSS) was required.
We found 870 genes bound by DREAM in proximity to their TSS (Supplementary Table S2). Out of these 870 genes, 358 (41.1%) were repressed by p53 displaying an Expression Score of ≤ −3. When all three selection criteria were applied with binding of three out of four DREAM components, repression by p53 with an Expression Score ≤ −3, and presence of one of the ten CHR variants being phylogenetically conserved in the promoter, we identified 210 genes as targets for the p53-p21-DREAM-CDE/CHR pathway (Figure 1; Table 1, Supplementary Table S2). The three known targets of the pathway, CCNB2, KIF23 and PLK4, were detected by the analysis. This demonstrates the ability of this screening approach to identify bona fide candidates. GAS2L3 had been missed by the search because only two of the DREAM components were above the threshold (Supplementary Table S2), although it had been described as a DREAM target (39). Also, B-MYB (MYBL2) was previously shown to be a CHR-controlled gene downregulated by p53 (21,28). Therefore, GAS2L3 and MYBL2 were included in the list of genes regulated by the p53-p21-DREAM-CDE/CHR pathway (Table 1).
Figure 1.
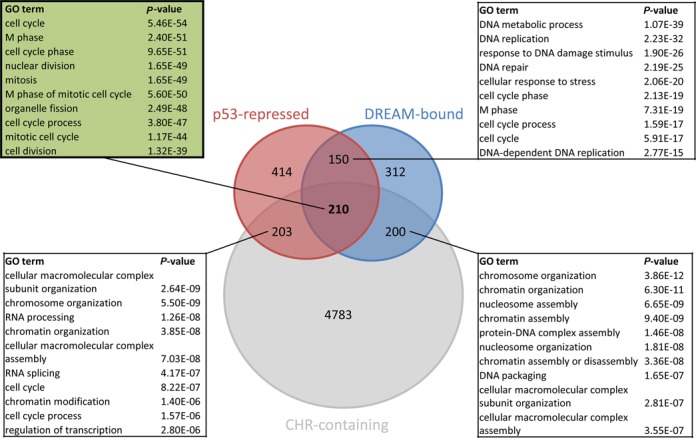
Genes associated with cell cycle and mitosis are enriched among targets of the p53-p21-DREAM-CDE/CHR pathway. Venn diagram displaying the overlap in the groups of genes found as repressed by p53, bound by DREAM, and containing a conserved CHR element. Top 10 GO terms enriched among the 210 genes displayed in Table 1 and the other group overlaps as identified using the DAVID Functional Annotation tool (Supplementary Table S3).
Repression of non-canonical CHR-containing genes by p53 requires p21 and is conserved between mouse and human
With CCNB2, KIF23 and PLK4, only genes containing canonical CHR sites had been tested in detail for their regulation through the p53-p21-DREAM-CDE/CHR pathway. In order to examine several of the new targets identified through the bioinformatic analysis for their regulation on a single-gene basis, we chose candidate genes which mostly contain CHR sites deviating from the canonical TTTGAA. We investigated p53-dependent regulation of B-MYB (MYBL2), BUB1, CCNA2, CCNB1, CHEK2, MELK, POLD1, RAD18 and RAD54L, which hold representative non-canonical CHR motifs that have been shown to bind DREAM (21). Relative mRNA expression was determined in untreated NIH3T3 cells in comparison to cells treated with the solvent DMSO, the MDM2 inhibitor nutlin-3a or the pyrimidine analogue 5-FU. We used NIH3T3 cells because cell cycle-dependent regulation of these genes was demonstrated in this cell line in a previous study (21) and results from this mouse cell line would complement the data from the human cell systems (Supplementary Table S2). Indeed, we found all genes with non-canonical CHR elements to be downregulated upon p53 activation (Figure 2A, Supplementary Figure S1). Next, we investigated p53-dependent regulation of the human gene orthologs in HCT116 wild-type, HCT116 p21-negative and non-cancerous HFF cells (Figure 2B–D, Supplementary Figure S1). Again, we found all genes containing a non-canonical CHR to be repressed following p53 activation in HCT116 and HFF cells, similar to the observations in NIH3T3 cells. These results support the data from our genome-wide screening and provide evidence that p53-dependent repression of these genes is conserved between mouse and human. Importantly, p53-dependent repression is essentially lost in HCT116 p21−/− cells (Figure 2C). In agreement with our data, p21-dependent downregulation has been reported for four of the nine genes tested here, namely CCNA2 (4,6,11,40), CCNB1 (6–8,11), POLD1 (11) and RAD54L (7). These results are consistent with a mechanism involving indirect, p21-mediated repression upon p53 stabilization. Based on these findings, we conclude that p53-dependent repression of these CHR-containing genes requires p21.
Figure 2.
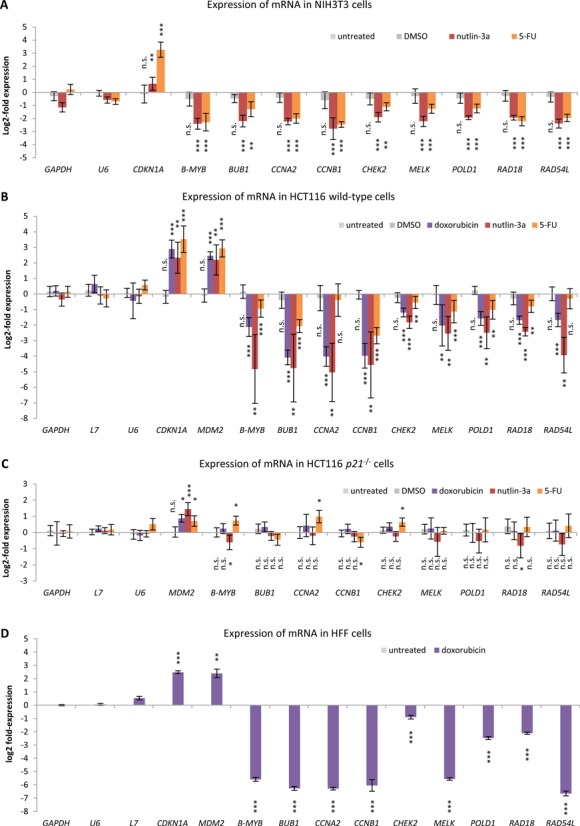
Repression of non-canonical CHR-containing genes by p53 requires p21 and is conserved between mouse and human. The log2-fold change of mRNA expression from treated compared to untreated (A) NIH3T3, (B) HCT116 wild-type, (C) HCT116 p21−/− and (D) HFF cells is displayed. Cells were treated with doxorubicin, nutlin-3a or 5-FU for 24 h. Untreated cells and cells treated with DMSO served as controls. Normalization was carried out against measurements from untreated cells. GAPDH, L7 and U6 served as negative controls for p53 response, while CDKN1A and MDM2 were employed as positive controls. (A–C) Experiments were performed with two biological replicates and three technical replicates each (n = 6). (D) Experiments were performed with three technical replicates (n = 3). Significance of changes in expression levels was tested against GAPDH expression levels using the unpaired Student's t-test; n.s. not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
p53 does not bind to promoters of genes harboring CHR sites
The finding that all genes tested require p21 for p53-dependent repression contrasts the previously proposed direct repression by p53 for two of the genes, CCNB1 and POLD1 (41–44). Thus, we tested for p53 binding to the promoters of CCNB1 and POLD1 as well as B-MYB (MYBL2), BUB1, CCNA2, CHEK2, MELK, RAD18 and RAD54L in HCT116 wild-type, HCT116 p21−/−, or HFF cells, untreated or treated with doxorubicin (Figure 3). Binding of p53 to promoters with CHR sites was not observed. These findings do not support a mechanism of direct repression by p53 (41–44), but are consistent with the indirect p53-p21-DREAM-CDE/CHR pathway which requires p21 (Figure 2).
Figure 3.
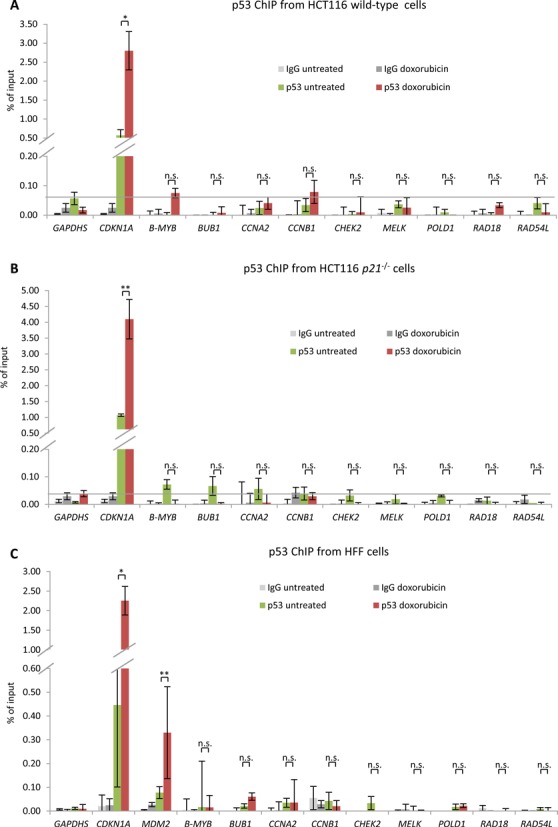
p53 does not bind to promoters of genes harboring CHR sites. Protein binding to promoters of the indicated genes was tested by ChIP in (A) HCT116 wild-type cells, (B) HCT116 p21−/− and (C) HFF cells either left untreated or treated with doxorubicin for 24 h (HFF) or 48 h (HCT116) followed by real-time PCR. The CDKN1A promoter served as a positive control for p53 binding; the GAPDHS promoter which does not bind p53 was used as a negative control. One representative experiment with three technical replicates (n = 3) is displayed.
p53 induces p21-dependent binding of DREAM to genes with CHR sites
The CHR sites in the B-MYB (MYBL2), BUB1, CCNA2, CCNB1, CHEK2, MELK, POLD1, RAD18 and RAD54L genes have previously been shown by DNA affinity purification assays to bind DREAM in vitro (21). Here, we tested for differential DREAM binding by ChIP in cells before and after p53 activation. Binding of the DREAM components LIN9, E2F4 and p130 to the promoters of these genes were examined upon p53 induction by doxorubicin. Binding of the MuvB core component LIN9 was found to be enriched at some but not all promoters (Figure 4A). In contrast, the DREAM components E2F4 and p130 showed increased binding at all CHR-containing promoters in HCT116 wild-type cells treated with doxorubicin compared to untreated cells (Figure 4A). These findings are consistent with the model developed from the regulation of CCNB2, KIF23 and PLK4, which contain canonical CHR sites (19,23,24). Moreover, DREAM binding to the promoters was generally reduced in HCT116 p21−/− cells and did not increase after doxorubicin treatment of HCT116 p21−/− cells (Figure 4B). Interestingly, binding of E2F4 was observed for most genes to be reduced in doxorubicin-treated compared to untreated HCT116 p21−/− cells (Figure 4B). A likely explanation is the cell cycle shift toward G2/M upon doxorubicin treatment that causes the expected decrease in DREAM stability when p21 cannot inhibit the CDKs (Figure 4B and C). Thus, the p53-p21 pathway appears to stabilize formation of DREAM and enhances its binding to promoters in wild-type cells despite the doxorubicin-induced G2/M cell cycle shift. Taken together, the observations on these nine individual genes suggest that p21 is required for DREAM binding to CHR promoters upon p53 activation.
Figure 4.
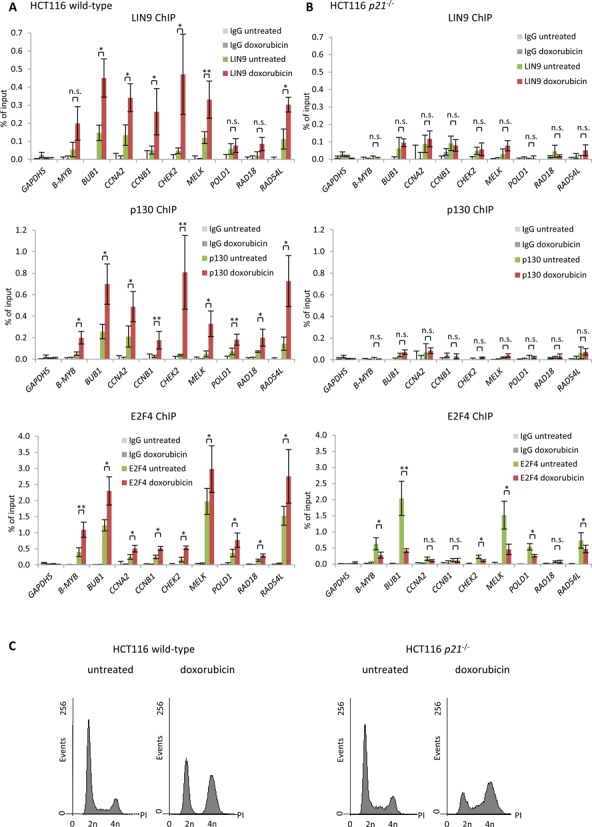
p53 induces p21-dependent binding of DREAM to genes with CHR sites. Protein binding to CHR-containing promoters in untreated or 48 h doxorubicin-treated HCT116 (A) wild-type or (B) p21−/− cells was tested by ChIP followed by real-time PCR. Protein binding to the GAPDHS promoter served as negative control. One representative experiment with three technical replicates (n = 3) is displayed. Significance was tested using the paired Student's t-test; n.s. not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (C) Flow cytometry of cells used in Figure 4; cells were stained with propidium iodide (PI).
G2/M cell cycle genes are among the targets of the p53-p21-DREAM-CDE/CHR signaling pathway
We performed a Gene Ontology analysis of the 210 target genes using the DAVID Functional Annotation tool (37). Cell cycle-associated terms were prominently represented in this set of genes (Figure 1, Supplementary Table S3). Particularly, genes encoding proteins required in late phases of the cell cycle from G2 through mitosis comprised the largest fraction in the analysis. When comparing the 210 genes to an analysis of four genome-wide cell cycle expression studies (21), it was observed that 158 (75.2%) of the CHR genes show cell cycle-dependent regulation. More precisely, the vast majority of genes regulated by the p53-p21-DREAM-CDE/CHR pathway displayed their peak expression in G2 phase or mitosis.
DISCUSSION
In this study, a list of target genes for the p53-p21-DREAM-CDE/CHR signaling pathway is presented. Selection of the 210 candidate targets was performed using stringent criteria to minimize the number of false-positives. Identification of the known p53-p21-DREAM-CDE/CHR targets CCNB2, KIF23 and PLK4 demonstrates the ability of the screening approach to identify bona fide candidates. These three genes harbor the canonical CHR sequence TTTGAA in their promoters (19,23,24). Notably, the non-canonical CHR elements TTTAAA, TTCGAA, TTTGTA, CTTGAA, TTTGAG, TTTGAT, TTCGAG, TTCGAT and TAGGAA had not yet been tested for their function in the p53-p21-DREAM-CDE/CHR pathway (21). Thus, B-MYB (MYBL2), BUB1, CCNA2, CCNB1, CHEK2, MELK, POLD1, RAD18 and RAD54L as representative genes harboring the nine non-canonical CHR variants were tested for their response to p53 activation. Detailed experiments with all of these genes confirmed results from the genome-wide analyses and supported the notion that the genes are regulated by this pathway (Figures 2–4). DREAM, represented by its components p130 and E2F4, was shown to be recruited to their promoters (Figure 4) (19). DREAM binding to the specific CHR sites has also been established (21). In agreement with our observations, it has been reported that p53-dependent downregulation of the mouse homologs Bub1, Ccna2 and Ccnb1 requires p107 and p130 (45). These pRB-related pocket proteins were later identified as components of the DREAM complex (13,14,18). This leads to the conclusion that the p53-p21-DREAM-CDE/CHR pathway functions through all known CHR variants and is largely conserved between mouse and human.
Furthermore, there are two indicators that threshold settings for the genome-wide analysis were adjusted to yield high-confidence candidates. One indicator is that all candidates tested in detail for their regulation by the pathway were confirmed (Figures 2–4). This suggests that the rate of false-positives may be low. The other observation is that GAS2L3 was not selected as a pathway gene. Since the DREAM binding threshold required three of the four DREAM components tested to be detected in the ChIP screen. However, in the case of GAS2L3 only two components were found positive for binding in the screen (Supplementary Table S2) (13), although GAS2L3 had been reported to be a DREAM target (39). Taken together, these observations indicate that threshold settings were so stringent that the computational analysis rather missed candidates than to include false-positive genes. This suggests that the 210 genes in Table 1 are indeed strong candidate targets of the p53-p21-DREAM-CDE/CHR pathway but that some additional target genes may have been missed.
GO term enrichment analysis of the 203 genes that contain a conserved CHR and are repressed by p53, but do not bind DREAM, shows a small enrichment also for cell cycle genes (Figure 1). Considering that GAS2L3 belonged to that group before we manually assigned it as DREAM target, it is likely that more currently unknown DREAM target genes can be found among this group of genes. The group of 200 genes binding DREAM, possessing a conserved CHR, and that are not found to be repressed by p53 likely contains genes that either are repressed by p53 but were missed due to the high stringency criteria in our analysis or genes that might be regulated solely by DREAM in the cell cycle. Interestingly, GO term analysis of the 150 genes that bind DREAM and are repressed by p53, but do not possess phylogenetically conserved CHR elements, shows high enrichment for biological processes of early (G1/S) cell cycle genes, such as DNA replication and DNA metabolism (Figure 1, Supplementary Table S3).
Cell cycle genes are general targets for p53-mediated downregulation independent of cell type or method of p53 activation. In regard to the underlying mechanism, either no or contradictory details have been reported. Several cell cycle genes among the 210 targets have been suggested to be directly repressed by p53. Examples include ANLN (46), AURKA (29), CDC20 (47), CDC25B (48), CDK1 (CDC2) (49), PRC1 (50) or PTTG1 (51). However, these genes were not found to be directly repressed by p53 in several genome-wide studies (52–54). Moreover, recent evidence suggests that p53 does not directly repress target genes (28). These observations are consistent with our data, which provide no evidence for direct repression of CCNB1 and POLD1, although these genes had been reported as direct p53 targets (Figures 2–4) (41–44). Additionally, for a large number of cell cycle genes the mechanism underlying p53-dependent repression remained unresolved, e. g. CDC25A (55), CKS1B (56), CKS2 (57) or HMMR (RHAMM) (58). However, for several other genes indirect repression via p21 was reported, including BUB1B (59,60), CENPA (61), CENPE (61), CENPF (59), FOXM1 (62) and MAD2L1 (59,60). All of these genes can be found among the 210 target genes (Table 1). Two previous studies had suggested that many cell cycle genes are repressed by p53 via p21 and E2F4, with the earlier of the two reports showing DREAM components and CHR sites as being part of this regulation and the later comparing E2F4 binding with p53-dependent expression data (19,63). Here, we took a genome-wide approach combining p53-dependent expression, binding of several DREAM components and selection of CHR-containing genes. We provide evidence that the E2F4-containing DREAM complex represses many, if not all, of these genes upon p53 activation and that this complex is targeted to many promoters via CDE/CHR motifs. Thus, the present analysis offers a mechanism for p53-dependent repression of these genes, resolving contradictions between earlier reports.
Interestingly, expression of most of the 210 candidate genes varies during the cell cycle. Expression levels peak in the late cell cycle phases, which correspond to the function of their encoded proteins in G2 and mitosis. Thus, the results suggest that repression of these cell cycle genes serves as a mechanism for p53 to stop cell division, particularly in G2/M. This mechanism of p53-dependent cell cycle control is exemplified by downregulation of key cell cycle regulators such as the cyclins CCNA2, CCNB1 and CCNB2, the cyclin-dependent kinases CDK1 and CDK2, as well as the kinases and phosphatases AURKA, AURKB, PLK1, PLK4, CHEK2, CDC25A and CDC25C. In addition to fast responses which stop cell cycle progression such as transcriptional suppression by cyclin F (64), the p53-p21-DREAM-CDE/CHR pathway may contribute to a permanent cell cycle arrest. In agreement with this model, the DREAM complex was shown to be important for permanent proliferation arrest during senescence (65).
Taken together, we establish a target list of the p53-p21-DREAM-CDE/CHR signaling pathway. The results also suggest a mechanism for the regulation of cell cycle genes that are targeted by this pathway. Most of the genes are dominantly expressed in G2 phase and mitosis and essential for the progression through the late cell cycle. Thus, downregulation of these genes through the p53-p21-DREAM-CDE/CHR pathway appears to be a principal mechanism for G2/M cell cycle arrest by p53.
Supplementary Material
Acknowledgments
We are indebted to Carola Koschke and Andrea Rothe for expert technical assistance as well as Kathrin Jäger, Andreas Lösche and Knut Krohn for performing DNA sequencing and flow cytometry. We thank Bert Vogelstein for the kind gift of HCT116 cell lines, Larissa Litovchick and James DeCaprio for LIN9 antibody and Christine Engeland for critical reading of the manuscript. This work was supported by a postdoctoral fellowship provided by the Fritz Thyssen Foundation (to M.F.), a graduate fellowship provided by the Freistaat Sachsen (to M.Q.) and the grant ‘Origins and Evolution of Regulation in Biological Systems’ (Grant ID: 24332) by the John Templeton Foundation (to L.S.). The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the funders.
Author Contributions: M.F. conceived and K.E. supervised the study. M.F. and M.Q. performed the experiments. L.S. and M.F. performed the computational analyses. M.F. and K.E. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Present address: Martin Fischer, Department of Medical Oncology, Dana–Farber Cancer Institute and Department of Medicine, Brigham and Women's Hospital, and Harvard Medical School, Boston, MA, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fritz Thyssen Foundation [to M.F.]; Freistaat Sachsen [to M.Q.]; John Templeton Foundation [Grant ID 24332 to L.S.]. Funding for open access charge: German Research Foundation (DFG) and University of Leipzig within the program of Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 4.Spitkovsky D., Schulze A., Boye B., Jansen-Durr P. Down-regulation of cyclin A gene expression upon genotoxic stress correlates with reduced binding of free E2F to the promoter. Cell Growth Differ. 1997;8:699–710. [PubMed] [Google Scholar]
- 5.Azzam E.I., deToledo S.M., Pykett M.J., Nagasawa H., Little J.B. CDC2 is down-regulated by ionizing radiation in a p53-dependent manner. Cell Growth Differ. 1997;8:1161–1169. [PubMed] [Google Scholar]
- 6.de Toledo S.M., Azzam E.I., Keng P., Laffrenier S., Little J.B. Regulation by ionizing radiation of CDC2, cyclin A, cyclin B, thymidine kinase, topoisomerase IIalpha, and RAD51 expression in normal human diploid fibroblasts is dependent on p53/p21Waf1. Cell Growth Differ. 1998;9:887–896. [PubMed] [Google Scholar]
- 7.Chang B.D., Watanabe K., Broude E.V., Fang J., Poole J.C., Kalinichenko T.V., Roninson I.B. Effects of p21(Waf1/Cip1/Sdi1) on cellular gene expression: Implications for carcinogenesis, senescence, and age-related diseases. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4291–4296. doi: 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flatt P.M., Tang L.J., Scatena C.D., Szak S.T., Pietenpol J.A. p53 regulation of G(2) checkpoint is retinoblastoma protein dependent. Mol. Cell Biol. 2000;20:4210–4223. doi: 10.1128/mcb.20.12.4210-4223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottifredi V., Karni-Schmidt O., Shieh S.S., Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol. Cell Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H., Chang B.D., Uchiumi T., Roninson I.B. Identification of promoter elements responsible for transcriptional inhibition of polo-like kinase 1 and topoisomerase IIalpha genes by p21(WAF1/CIP1/SDI1) Cell Cycle. 2002;1:59–66. [PubMed] [Google Scholar]
- 11.Löhr K., Möritz C., Contente A., Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J. Biol. Chem. 2003;278:32507–32516. doi: 10.1074/jbc.M212517200. [DOI] [PubMed] [Google Scholar]
- 12.Shats I., Milyavsky M., Tang X., Stambolsky P., Erez N., Brosh R., Kogan I., Braunstein I., Tzukerman M., Ginsberg D., et al. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 2004;279:50976–50985. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- 13.Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Schmit F., Korenjak M., Mannefeld M., Schmitt K., Franke C., von Eyss B., Gagrica S., Hanel F., Brehm A., Gaubatz S. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 15.Müller G.A., Quaas M., Schümann M., Krause E., Padi M., Fischer M., Litovchick L., DeCaprio J.A., Engeland K. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40:1561–1578. doi: 10.1093/nar/gkr793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mannefeld M., Klassen E., Gaubatz S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res. 2009;69:4073–4080. doi: 10.1158/0008-5472.CAN-08-4156. [DOI] [PubMed] [Google Scholar]
- 17.Calvisi D.F., Simile M.M., Ladu S., Frau M., Evert M., Tomasi M.L., Demartis M.I., Daino L., Seddaiu M.A., Brozzetti S., et al. Activation of v-Myb avian myeloblastosis viral oncogene homolog-like2 (MYBL2)-LIN9 complex contributes to human hepatocarcinogenesis and identifies a subset of hepatocellular carcinoma with mutant p53. Hepatology. 2011;53:1226–1236. doi: 10.1002/hep.24174. [DOI] [PubMed] [Google Scholar]
- 18.Sadasivam S., DeCaprio J.A. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quaas M., Müller G.A., Engeland K. p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle. 2012;11:4661–4672. doi: 10.4161/cc.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller G.A., Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277:877–893. doi: 10.1111/j.1742-4658.2009.07508.x. [DOI] [PubMed] [Google Scholar]
- 21.Müller G.A., Wintsche A., Stangner K., Prohaska S.J., Stadler P.F., Engeland K. The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42:10331–10350. doi: 10.1093/nar/gku696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X., Müller G.A., Quaas M., Fischer M., Han N., Stutchbury B., Sharrocks A.D., Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell Biol. 2013;33:227–236. doi: 10.1128/MCB.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer M., Grundke I., Sohr S., Quaas M., Hoffmann S., Knörck A., Gumhold C., Rother K. p53 and cell cycle dependent transcription of kinesin family member 23 (KIF23) is controlled via a CHR promoter element bound by DREAM and MMB complexes. PLoS One. 2013;8:e63187. doi: 10.1371/journal.pone.0063187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer M., Quaas M., Wintsche A., Müller G.A., Engeland K. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements, repressed by DREAM through CDE/CHR sites and deregulated by HPV E7 protein. Nucleic Acids Res. 2014;42:163–180. doi: 10.1093/nar/gkt849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwicker J., Lucibello F.C., Wolfraim L.A., Gross C., Truss M., Engeland K., Müller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasner M., Tschöp K., Spiesbach K., Haugwitz U., Johne C., Mössner J., Mantovani R., Engeland K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003;536:66–70. doi: 10.1016/s0014-5793(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 27.Liu N., Lucibello F.C., Zwicker J., Engeland K., Müller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer M., Steiner L., Engeland K. The transcription factor p53: Not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikulenkov F., Spinnler C., Li H., Tonelli C., Shi Y., Turunen M., Kivioja T., Ignatiev I., Kel A., Taipale J., et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death. Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Böhlig L., Friedrich M., Engeland K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res. 2011;39:440–453. doi: 10.1093/nar/gkq796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashi-Elkeles S., Elkon R., Shavit S., Lerenthal Y., Linhart C., Kupershtein A., Amariglio N., Rechavi G., Shamir R., Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol. Oncol. 2011;5:336–348. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozenblatt-Rosen O., Deo R.C., Padi M., Adelmant G., Calderwood M.A., Rolland T., Grace M., Dricot A., Askenazi M., Tavares M., et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kracikova M., Akiri G., George A., Sachidanandam R., Aaronson S.A. A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 2013;20:576–588. doi: 10.1038/cdd.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldstein I., Ezra O., Rivlin N., Molchadsky A., Madar S., Goldfinger N., Rotter V. p53, a novel regulator of lipid metabolism pathways. J. Hepatol. 2012;56:656–662. doi: 10.1016/j.jhep.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang d.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Waldman T., Kinzler K.W., Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 39.Wolter P., Schmitt K., Fackler M., Kremling H., Probst L., Hauser S., Gruss O.J., Gaubatz S. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J. Cell Sci. 2012;125:2393–2406. doi: 10.1242/jcs.097253. [DOI] [PubMed] [Google Scholar]
- 40.Jackson J.G., Pereira-Smith O.M. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol. Cell. Biol. 2006;26:2501–2510. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B.Q., Lee M.Y.W. Transcriptional regulation of the human DNA polymerase delta catalytic subunit gene POLD1 by p53 tumor suppressor and Sp1. J. Biol. Chem. 2001;276:29729–29739. doi: 10.1074/jbc.M101167200. [DOI] [PubMed] [Google Scholar]
- 42.Innocente S.A., Lee J.M. p53 is a NF-Y- and p21-independent, Sp1-dependent repressor of cyclin B1 transcription. FEBS Lett. 2005;579:1001–1007. doi: 10.1016/j.febslet.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 43.Ceribelli M., Alcalay M., Vigano M.A., Mantovani R. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle. 2006;5:1102–1110. doi: 10.4161/cc.5.10.2777. [DOI] [PubMed] [Google Scholar]
- 44.Lipski R., Lippincott D.J., Durden B.C., Kaplan A.R., Keiser H.E., Park J.H., Levesque A.A. p53 Dimers associate with a head-to-tail response element to repress cyclin B transcription. PLoS One. 2012;7:e42615. doi: 10.1371/journal.pone.0042615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson M.W., Agarwal M.K., Yang J., Bruss P., Uchiumi T., Agarwal M.L., Stark G.R., Taylor W.R. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J. Cell Sci. 2005;118:1821–1832. doi: 10.1242/jcs.02307. [DOI] [PubMed] [Google Scholar]
- 46.Mirza A., Wu Q., Wang L., McClanahan T., Bishop W.R., Gheyas F., Ding W., Hutchins B., Hockenberry T., Kirschmeier P., et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22:3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- 47.Banerjee T., Nath S., Roychoudhury S. DNA damage induced p53 downregulates Cdc20 by direct binding to its promoter causing chromatin remodeling. Nucleic Acids Res. 2009;37:2688–2698. doi: 10.1093/nar/gkp110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalvai M., Mondesert O., Bourdon J.C., Ducommun B., Dozier C. Cdc25B is negatively regulated by p53 through Sp1 and NF-Y transcription factors. Oncogene. 2011;30:2282–2288. doi: 10.1038/onc.2010.588. [DOI] [PubMed] [Google Scholar]
- 49.Le Gac G., Esteve P.O., Ferec C., Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J. Biol. Chem. 2006;281:24161–24170. doi: 10.1074/jbc.M603724200. [DOI] [PubMed] [Google Scholar]
- 50.Li C., Lin M., Liu J. Identification of PRC1 as the p53 target gene uncovers a novel function of p53 in the regulation of cytokinesis. Oncogene. 2004;23:9336–9347. doi: 10.1038/sj.onc.1208114. [DOI] [PubMed] [Google Scholar]
- 51.Kho P.S., Wang Z., Zhuang L., Li Y., Chew J.L., Ng H.H., Liu E.T., Yu Q. p53-regulated transcriptional program associated with genotoxic stress-induced apoptosis. J. Biol. Chem. 2004;279:21183–21192. doi: 10.1074/jbc.M311912200. [DOI] [PubMed] [Google Scholar]
- 52.Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 53.Menendez D., Nguyen T.A., Freudenberg J.M., Mathew V.J., Anderson C.W., Jothi R., Resnick M.A. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41:7286–7301. doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlereth K., Heyl C., Krampitz A.M., Mernberger M., Finkernagel F., Scharfe M., Jarek M., Leich E., Rosenwald A., Stiewe T. Characterization of the p53 cistrome—DNA binding cooperativity dissects p53's tumor suppressor functions. PLoS Genet. 2013;9:e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rother K., Kirschner R., Sänger K., Böhlig L., Mössner J., Engeland K. p53 downregulates expression of the G(1)/S cell cycle phosphatase Cdc25A. Oncogene. 2007;26:1949–1953. doi: 10.1038/sj.onc.1209989. [DOI] [PubMed] [Google Scholar]
- 56.Rother K., Li Y.Y., Tschöp K., Kirschner R., Müller G.A., Mössner J., Engeland K. Expression of cyclin-dependent kinase subunit 1 (Cks1) is regulated during the cell cycle by a CDE/CHR tandem element and is downregulated by p53 but not by p63 or p73. Cell Cycle. 2007;6:853–862. doi: 10.4161/cc.6.7.4017. [DOI] [PubMed] [Google Scholar]
- 57.Rother K., Dengl M., Lorenz J., Tschöp K., Kirschner R., Mössner J., Engeland K. Gene expression of cyclin-dependent kinase subunit Cks2 is repressed by the tumor suppressor p53 but not by the related proteins p63 or p73. FEBS Lett. 2007;581:1166–1172. doi: 10.1016/j.febslet.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 58.Sohr S., Engeland K. RHAMM is differentially expressed in the cell cycle and downregulated by the tumor suppressor p53. Cell Cycle. 2008;7:3448–3460. doi: 10.4161/cc.7.21.7014. [DOI] [PubMed] [Google Scholar]
- 59.Tabach Y., Milyavsky M., Shats I., Brosh R., Zuk O., Yitzhaky A., Mantovani R., Domany E., Rotter V., Pilpel Y. The promoters of human cell cycle genes integrate signals from two tumor suppressive pathways during cellular transformation. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100030. 2005.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schvartzman J.M., Duijf P.H., Sotillo R., Coker C., Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scian M.J., Carchman E.H., Mohanraj L., Stagliano K.E., Anderson M.A., Deb D., Crane B.M., Kiyono T., Windle B., Deb S.P., et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27:2583–2593. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- 62.Barsotti A.M., Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benson E.K., Mungamuri S.K., Attie O., Kracikova M., Sachidanandam R., Manfredi J.J., Aaronson S.A. p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene. 2014;33:3959–3969. doi: 10.1038/onc.2013.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klein D.K., Hoffmann S., Ahlskog J.K., O'Hanlon K., Quaas M., Larsen B.D., Rolland B., Rosner H.I., Walter D., Kousholt A.N., et al. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat. Commun. 2015;6:5800. doi: 10.1038/ncomms6800. [DOI] [PubMed] [Google Scholar]
- 65.Litovchick L., Florens L.A., Swanson S.K., Washburn M.P., DeCaprio J.A. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–813. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


