Abstract
Chlamydia trachomatis is an intracellular gram-negative bacteria which causes several clinically important diseases. T-cell-mediated immunity and the production of gamma interferon (IFN-γ) are known to be essential for the clearance of the bacteria in vivo. Here we have investigated CD8+-T-cell responses to C. trachomatis in patients with previous episodes of chlamydia infection. To isolate C. trachomatis-specific CD8+-T-cell lines, dendritic cells (DC) were infected with C. trachomatis and cocultured with purified CD8+ T cells to generate C. trachomatis-specific CD8+-T-cell lines which were then cloned. Two patterns of recognition of C. trachomatis-infected cells by CD8+-T-cell clones were identified. In the first, C. trachomatis antigens were recognized in association with classical class I HLA antigens, and responses were inhibited by class I HLA-specific monoclonal antibodies. The second set of clones was unrestricted by classical HLA class I, and further studies showed that CD1 molecules were also not the restriction element for those clones. Both types of clones produced IFN-γ in response to C. trachomatis and were able to lyse C. trachomatis-infected target cells. However, unrestricted clones recognized C. trachomatis-infected cells at much earlier time points postinfection than HLA-restricted clones. Coculture of C. trachomatis-infected DC with the C. trachomatis-specific clones induced DC activation and a rapid enhancement of interleukin-12 (IL-12) production. Early production of IL-12 during C. trachomatis infection, facilitated by unrestricted CD8+-T-cell clones, may be important in ensuring a subsequent Th1 T-cell-mediated response by classical major histocompatibility complex-restricted CD4+ and CD8+ T cells.
Chlamydia trachomatis is an intracellular gram-negative bacteria which is a major cause of sexually transmitted disease (12, 16, 21, 50), and in less developed countries, ocular infection by C. trachomatis (trachoma) is the leading cause of preventable blindness (52). In addition, 2 to 5% of patients infected by C. trachomatis develop an inflammatory arthritis, reactive arthritis.
Both humoral and cellular responses can be readily detected in patients suffering from C. trachomatis infection. The antibody responses are largely directed against the major outer membrane protein (MOMP) but are serovar specific and, therefore, afford little protection against repeated infection (2, 4). In addition, since C. trachomatis causes intracellular infection, neutralizing antibodies have little relevance to resolving established infection even if they confer some immunity to subsequent infection. As with other intracellular infections, clearance of the organism requires T-cell-mediated immunity, and there is good evidence that T-cell responses are critical in host resistance to C. trachomatis. In animal models, transfer of T lymphocytes from infected or immunized mice can facilitate clearance of infection in T-cell-deficient mice, and this effect has been demonstrated for both CD4+ and CD8+ T lymphocytes (30, 40, 45, 46, 49). However, the protection conferred by a CD8+-T-cell clone in murine experiments was dependent on its ability to produce gamma interferon (IFN-γ) rather than its cytolytic activity, and in this respect the protection resembled that conferred by CD4+ T cells (27, 30).
In humans both T-cell subsets can be detected at the site of C. trachomatis infection, but most work on defining the immune response to C. trachomatis in humans has concerned CD4+ T cells. A number of C. trachomatis antigens which can be recognized by human CD4+ T cells have been identified, including MOMP (36, 37), the 60-kD cysteine-rich outer membrane protein Omp2 (14), polymorphic membrane protein D (PmpD) (15), heat shock protein 60 (hsp60) (8), the histone-like protein Hc1 (15), and enolase (15); several epitopes have been mapped within these antigens. In contrast, much less is known about the antigenic specificity and roles of CD8+ T cells during human C. trachomatis infection. In recent years human CD8+ T cells which are able to recognize MOMP (18, 24, 25) or hsp60 (18, 26) have been isolated from infected humans, but the approach in each case was to identify peptides in C. trachomatis proteins which would be predicted to bind to common class I HLA alleles and to determine whether infected subjects had CD8+ T cells able to recognize these peptides. T-cell lines and clones were isolated by stimulation with synthetic peptides.
In the present study we have generated chlamydia-specific CD8+-T-cell lines from which we isolated and functionally characterized 31 C. trachomatis-specific CD8+-T-cell clones. To obtain C. trachomatis-specific lines, CD8+ T cells from infected subjects were stimulated with C. trachomatis-infected dendritic cells (DC), mimicking the likely situation in vivo and thus avoiding the need to select candidate antigens. Clones generated from C. trachomatis-specific lines showed two distinct patterns of recognition of C. trachomatis-infected cells. One group of clones recognized C. trachomatis antigens in the context of classical class I HLA, whereas the recognition of C. trachomatis-infected cells by the other group of clones was not restricted by classical HLA-class I antigens. These findings are in general agreement with the recently published data by Gervassi et al. (13). Both kinds of clones produced large amounts of IFN-γ in response to C. trachomatis-infected cells, and coculture of C. trachomatis-infected DC with C. trachomatis-specific CD8+ clones resulted in a significant increase in interleukin-12 (IL-12) production by the DC. These activities suggest that the CD8+-T-cell response to C. trachomatis may play an important part in control of this important human pathogen.
MATERIALS AND METHODS
Human subjects.
Peripheral blood was obtained from four patients, three male and one female, known to have been previously infected by C. trachomatis and from control subjects. Three out of the four patients were asymptomatic at the time when blood samples were obtained for T-cell cloning. Tissue typing showed that all of these patients were HLA-A2+ and three of them were HLA-B27+. All human studies have been approved by Addenbrooke's Hospital Local Research Ethical Committee.
Cell lines.
A number of antigen-presenting cells (APC) were used to study their ability to present chlamydial antigens to C. trachomatis-specific CD8+-T-cell clones. These were either autologous cells (DC, peripheral blood mononuclear cells [PBMC], and Epstein-Barr virus-transformed B-cell lines) HLA-matched PBMC or allogeneic cell lines, including the myeloid cell line U937 and U937 transfected with HLA-A2, HLA-B27, or HLA-B8, and the lymphoblastoid cell line C1R transfected with CD1a, CD1b, or CD1c.
To infect U937 and C1R- and Epstein-Barr virus-transformed B cell lines, cells were plated in 24-well plates (Gibco BRL) in 2-ml volumes at 0.5 × 106 to 1 × 106 cells per ml and C. trachomatis were added at a 10:1 ratio (bacteria to cell). Control wells lacking C. trachomatis cells (mock infected) were also included. The plates were centrifuged at 1,400 × g for 30 min at 37°C.
Preparation of DC and infection with C. trachomatis.
DC were differentiated from CD14+ cells as described previously (44). In brief, PBMC were purified on Ficoll gradients (Amersham Pharmacia Biotech AB) and labeled with anti-CD14 monoclonal antibody conjugated to microbeads (Miltenyi Biotec); the labeled cells were then purified on separation columns (Miltenyi Biotec). CD14+ monocytes were cultured for 7 to 8 days in complete RPMI medium (Gibco) containing 10% human serum, 10 mM HEPES buffer, 1% nonessential amino acids, 1% sodium pyruvate, and 2 mM l-glutamine (Sigma). No antibiotic was added to the medium. Cultures were supplemented with 50 ng of human granulocyte-macrophage colony-stimulating factor (Novartis) per ml and 1,000 U of human recombinant IL-4 (Pharmingen) per ml. Granulocyte-macrophage colony-stimulating factor and IL-4 were added to the cultures every 4 days.
C. trachomatis serovar L2 was used throughout this study and was purified from infected HeLa cells as previously described (5). DC were infected with C. trachomatis (50 bacteria per cell) at day 7 of culture.
Antibodies and flow cytometry and in vitro blocking.
Anti-chlamydia lipopolysaccharide (LPS) antibody (fluorescein isothiocyanate [FITC]-conjugated) was obtained from DAKO. To detect intracellular bacteria, cells were fixed and permeabilized by using Perm/Fix solution (Pharmingen) prior to staining. Any surface-bound C. trachomatis organisms were visualized on infected cells fixed in 4% paraformaldehyde prior to staining.
Unconjugated or conjugated (conjugate indicated in parentheses)monoclonal antibodies specific for T-cell receptor (TCR)-α/β (FITC), TCR-γ/δ, perforin (phycoerythrin [PE]), granzyme A (FITC), CD69 (FITC), CD40L (FITC), CD1a (FITC), CD1b (FITC), CD1c, CD1d (PE), and the appropriate immunoglobulin G (IgG) isotype control antibody were obtained from Pharmingen-BD (Oxford, United Kingdom). Anti-CD4 (FITC) and -CD8 (PE) were purchased from DAKO. Anti-CD14 (FITC) was purchased from BioSource. The w6/32 (anti-HLA-A/B/C) hybridoma was a kind gift from P. Lehner (Cambridge Institute of Medical Research, University of Cambridge), the MA2.1 (anti-HLA-A2) hybridoma was provided by F. Hall (Addenbrooke's Hospital, Cambridge, United Kingdom), and the ME1 (anti-HLA-B27) hybridoma was a kind gift from R. Colbert (Children's Hospital Medical Center, Cincinnati, Ohio). All antibodies were purified from hybridoma supernatants on protein G columns (Amersham Biosciences UK, Ltd., Buckinghamshire, United Kingdom). The purified anti-CD1a, -CD1b, -CD1c, and -CD1d antibodies used for blocking experiments (51) were generously provided by M. Vincent (Brigham and Women's Hospital, Boston, Mass.). Goat anti-mouse immunoglobulin (FITC) F(ab)2 fragments were used to detect unconjugated antibodies (DAKO). Cell staining was analyzed on a FACScan (Becton Dickinson).
For in vitro blocking experiments antibodies were used at the following concentrations: w6/32 at 30 μg/ml; MA2.1 at 10 μg/ml; ME1 at 10 μg/ml; and CD1a, CD1b, CD1c, and CD1d at 20 μg/ml.
Generation of chlamydia-specific CD8+-T-cell clones.
DC from a chlamydia-infected patient were generated in vitro as described above. Cells were infected with live C. trachomatis at day 7 of culture and cultured for an additional 24 h. DC were harvested, washed, irradiated, and plated at 104 cells/well in a 96-well plate. CD8+ T cells were purified from PBMC by negative selection by using a cocktail of antibodies: anti-CD4, -CD19, -CD14, -CD56, and anti-TCR-γ/δ. Cells were then incubated with anti-mouse IgG conjugated to microbeads (Miltenyi Biotec). CD8+ T cells were eluted on a separation column (Miltenyi Biotec). Autologous purified CD8+ T cells were added to DC cultures at 104 cells/well and cultured for 2 weeks in the presence of 50 U of recombinant human IL-2 (rIL-2) (Chiron, Uxbridge, United Kingdom) per ml. Cultures were supplemented with rIL-2 every 7 days. After 2 weeks of culture, the CD8+-T-cell line was further purified by negative selection as described above to obtain a pure CD8+ population for cloning. Following two rounds of purification, the T-cell lines contained 98% CD8+ T cells. The cells were then seeded at 0.3 cells/well in 96-well plates with 105 allogeneic-PBMC (allo-PBMC)/well. Cells were cultured in 10% complete medium supplemented with 1 μg of phytohemagglutinin (Sigma) per ml, 50 U of rIL-2 per ml, and 0.5 ng of rIL-7 (Pharmingen) per ml. Cultures were supplemented with fresh rIL-2 and rIL-7 every 5 days. After 2 to 3 weeks the clones were established and were tested by flow cytometry for the surface expression of CD8 and TCR-α/β and then examined for their ability to recognize C. trachomatis-infected cells by using an enzyme-linked immunosorbent assay (ELISA) to detect IFN-γ and assays of cytotoxicity.
Cytokine detection by ELISA.
APC were irradiated, pulsed with C. trachomatis, and cultured at 105 cells/well in 96-well plates. CD8+-T-cell clones were added at 104 cells/well. Culture medium was supplemented with 25 U of rIL-2 per ml. After 72 h of culture, supernatants were collected, and the production of IFN-γ was measured by ELISA.
DC were pulsed with C. trachomatis or LPS or were used as immature cells. They were seeded at 104 cells/well. CD8+-T-cell clones were added to DC cultures at 2 × 104 cells/well, and culture supernatants were collected at days 1, 2, and 3. The production of IFN-γ and IL-12 was measured by ELISA.
The human IFN-γ and IL-12p70 ELISA kits (Human CytoSets Antibody Pairs) were obtained from Biosource International (Nivelles, Belgium). ELISA staining was carried out as recommended by the manufacturers.
Cytotoxicity assay.
Lysis by CD8+-T-cell clones was measured by using a lactate dehydrogenase (LDH) release assay (Promega). Work in our laboratory has previously shown that the results obtained from LDH assays are comparable to those from 51Cr release assays. CD8+-T-cell clones were cocultured with infected target cells for 6 h. Six hours of incubation was found to be optimal for the CD8+ clones. The percent lysis was measured by quantifying the amount of LDH in the supernatant as per the manufacturer's instructions. The percent lysis was calculated as follows: % Lysis = [(Total lysis − Effector spontaneous lysis − Target spontaneous lysis)/(Target maximum lysis − Target spontaneous lysis)] × 100. To determine the level of infection of the target cells used in the cytotoxicity assays, U937 cells were stained for intracellular C. trachomatis by using mouse anti-C. trachomatis antibody. The proportion of cells infected ranged from 35 to 60% in different assays. It was therefore critical to correlate the level of infection with the percentage of lysis obtained with particular T-cell clones. The data shown in our figures represent the lysis obtained in the assays without adjustment for the proportion of target cells infected. Taking this into account, 80 to 100% C. trachomatis-infected cells were killed by the CD8+ clones at an effector-to-target cell (E:T) ratio of 10:1.
RESULTS
CD8+-T-cell responses to C. trachomatis.
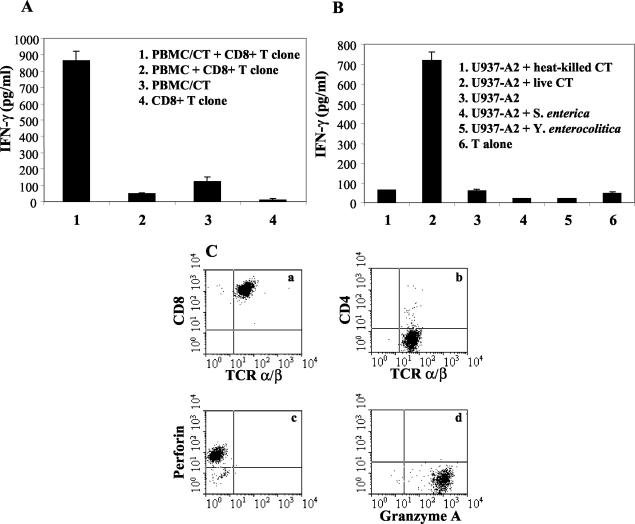
DC infected with live C. trachomatis were used to expand C. trachomatis-specific CD8+ T cells purified from patients with a history of chlamydia infection. Expanded CD8+-T-cell lines were then used to clone C. trachomatis-specific T cells. In the first instance, the clones were selected for further analysis on the basis of their ability to produce IFN-γ in response to C. trachomatis-infected, but not uninfected, autologous PBMC (Fig. 1A).
FIG. 1.
Characterization of a C. trachomatis-specific CD8+-T-cell clone. (A) IFN-γ production by the clone in response to C. trachomatis-infected autologous PBMC. (B) IFN-γ production in response to U937-A2 infected with live or heat-killed C. trachomatis, live S. enterica and live Y. enterocolitica. (C) Surface expression of CD8, CD4, and TCR-α/β and intracellular expression of perforin and granzyme A by a C. trachomatis-specific CD8+-T-cell clone, analyzed by using two-color flow cytometric analysis (a and b) and a single-color flow cytometric analysis (c and d). These data are representative of 31 C. trachomatis-specific T-cell clones studied. Each experiment was carried out at least three times. CT, C. trachomatis.
In addition to responses to C. trachomatis-infected PBMC, clones from HLA-A2-positive individuals were found to be able to recognize the cell line U937-A2 (i.e., transfected with HLA-A2) infected by C. trachomatis. These experiments also showed that these clones produced no IFN-γ when U937-A2 cells were infected with heat-inactivated C. trachomatis, showing that infection with live organisms is essential for the generation of CD8+-T-cell responses to chlamydia (Fig. 1B). Specificity for chlamydia antigens was also demonstrated, since the clones did not recognize U937-A2 infected with Yersinia enterocolitica or Salmonella enterica (Fig. 1B).
C. trachomatis-responsive clones were tested for surface expression of TCR-α/β and CD8 by flow cytometry. All clones were TCR-α/β+ CD8+ CD4− and positive for perforin and granzyme A, thus showing that they contained classical perforin granules (Fig. 1C). A total of 31 C. trachomatis-specific CD8+-T-cell clones from four C. trachomatis-infected subjects were selected and analyzed.
Two patterns of chlamydia recognition by CD8+ T cells.
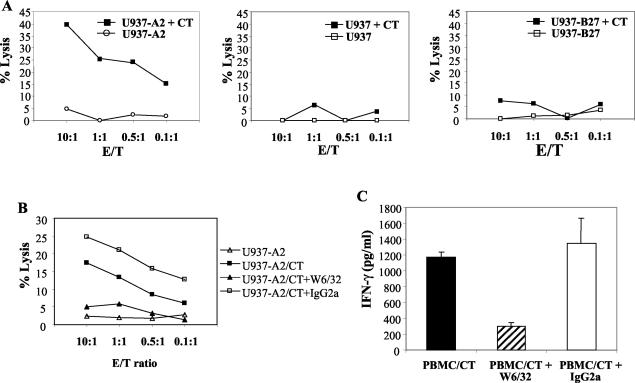
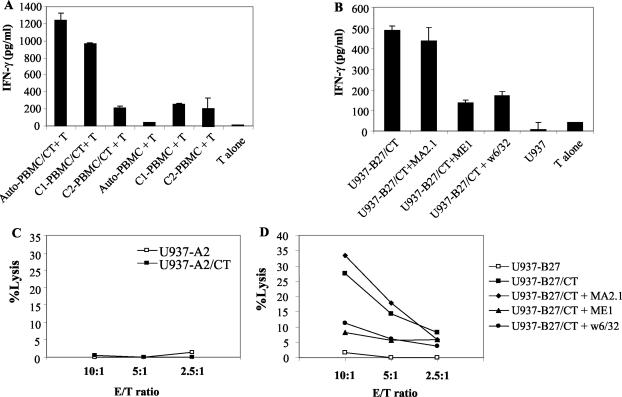
Figure 2 shows a clone which lysed C. trachomatis-infected U937 cells transfected with HLA-A2 (U937-A2), but not uninfected cells, infected parental U937 cells, or C. trachomatis-infected U937 cells transfected with HLA-B27 (U937-B27) (Fig. 2A). The lysis of infected U937-A2 cells was inhibited by w6/32 (Fig. 2B). Likewise, IFN-γ production by this clone, following stimulation with autologous PBMC infected with C. trachomatis, was inhibited with w6/32. (Fig. 2C). These properties are consistent with restriction by HLA-A2. Similarly, Fig. 3 shows a clone generated from an HLA-B27+ ankylosing spondylitis patient with a history of chlamydia infection; this clone produced IFN-γ when cocultured with C. trachomatis-infected autologous PBMC or allo-PBMC which also expressed HLA-B27 (C1) but not with allo-PBMC which were HLA-B27 negative (C2) (Fig. 3A). IFN-γ production was inhibited by approximately 70% by w6/32 or the HLA-B27-specific antibody (ME1) but not by the HLA-A2-specific antibody MA2.1 (Fig. 3B). The HLA-B27-restricted nature of the response was also confirmed in cytotoxicity assays where only C. trachomatis-infected U937 was lysed, and the lysis was inhibited by ME1 and w6/32 but not by MA2.1 (Fig. 3C and D).
FIG. 2.
A C. trachomatis-specific CD8+-T-cell clone which recognizes C. trachomatis in association with HLA-A2. (A) Specific lysis of C. trachomatis-infected U937-A2, U937, and U972-B27 cells and of uninfected cells. (B) Lysis of C. trachomatis-infected U937-A2 cells in the presence of w6/32 (anti-HLA class I MAb) or an IgG2a isotype control. Each experiment was carried out three times. (C) IFN-γ production in response to autologous PBMC infected with C. trachomatis and with the addition of w6/32 (anti-HLA class I MAb) or IgG2a isotope control antibody. No IFN-γ was obtained in cocultures with uninfected U937-A2 or from C. trachomatis-infected U937 or T cells cultured alone (data not shown). CT, C. trachomatis.
FIG. 3.
A C. trachomatis-specific CD8+-T-cell clone which recognizes C. trachomatis in association with HLA-B27. (A) IFN-γ production in response to C. trachomatis-infected autologous PBMC and two HLA-matched allo-PBMC. C1 PBMC were matched for HLA-A2 and HLA-B27, while C2 PBMC were matched for HLA-A2 but were HLA-B27 negative. (B) IFN-γ production by a C. trachomatis-specific CD8+-T-cell clone in the presence of w6/32, ME1 (anti-HLA-B27), and MA2.1 (anti-HLA-A2) antibodies. (C and D) Specific lysis of C. trachomatis-infected and uninfected U937-A2 target cells and of C. trachomatis-infected U937-B27 alone or in the presence of w6/32, ME1, or MA2.1. Each experiment was repeated three times. CT, C. trachomatis.
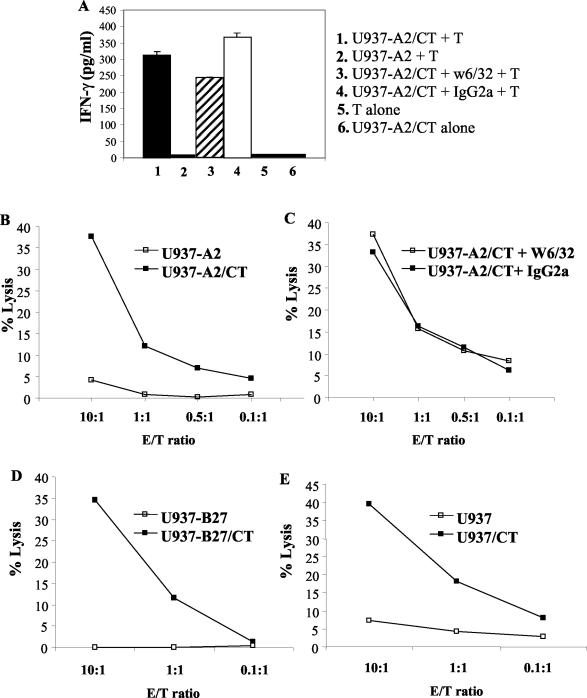
In striking contrast to these results, for a large number of additional CD8+-T-cell clones, neither IFN-γ production nor specific lysis of infected target cells was inhibited by w6/32 (Fig. 4A and C). Thus, these cells showed no evidence of HLA restriction and were able to lyse all C. trachomatis-infected cell lines tested, i.e., U937-A2 (Fig. 4B), U937-B27 (Fig. 4D), untransfected U937 (Fig. 4E), and U937-B8 (data not shown). However, there was no lysis of uninfected cells, showing that the lysis required the C. trachomatis infection of target cells. Likewise, these clones produced IFN-γ when cocultured with C. trachomatis-infected autologous or allo-PBMC (data not shown) but not with APC infected with live Y. enterocolitica and S. enterica.
FIG. 4.
A C. trachomatis-specific CD8+-T-cell clone which recognizes C. trachomatis in an HLA class I-unrestricted manner. IFN-γ production in response to C. trachomatis-infected U937-A2 cells and with the addition of w6/32 or IgG2a isotope control antibody (A). No IFN-γ was obtained in cocultures with uninfected U937-A2 cells or from C. trachomatis-infected U937 or T cells cultured alone. Specific lysis of uninfected or C. trachomatis-infected U937-A2 cells alone (B) or in the presence of w6/32 or IgG2a isoptype control antibody (C). Specific lysis of uninfected or C. trachomatis-infected U937-B27 cells (D) and of uninfected or C. trachomatis-infected U937 cells (E). CT, C. trachomatis.
Table 1 shows the summary of the HLA class I-restricted and -unrestricted clones for each patient studied. As can be seen, the number of HLA class I-unrestricted clones was higher than the number of HLA class I-restricted clones. Table 2 shows the surface phenotype of the unrestricted clones. Interestingly, they did not express CD40L, even when activated. Likewise, there was no expression of cytotoxic-T-lymphocyte-associated antigen 4 (CTLA-4) on resting T cells and only a low level of intracellular CTLA-4 following cell activation. On the other hand, these clones expressed some NK cell markers, e.g., NKG2D and CD56 but not CD94. HLA class I-unrestricted clones were negative for CD161 (a marker of NK T cells).
TABLE 1.
Properties of C. trachomatis-specific CD8+-T-cell clones isolated from four patients
| Patient | No. of HLA class I-restricted clones | No. of HLA class I-unrestricted clones |
|---|---|---|
| 1 | 1 | 3 |
| 2 | 12 | 16 |
| 3 | 2 | 6 |
| 4 | 4 | 23 |
TABLE 2.
Phenotype of CD8+ HLA-unrestricted clones
| Surface expression of: | Relative level of expression ona:
|
|
|---|---|---|
| Resting T cells | Activated T cells | |
| Differentiation antigens | ||
| CD8 | ++ | ++ |
| CD4 | − | − |
| TCR-α/β | ++ | ++ |
| CD3 | ++ | ++ |
| Activatory or costimulatory receptors | ||
| CD25 | − | + |
| CD28 | − | − |
| CD30 | − | − |
| CD30L | − | ++ |
| CD69 | − | ++ |
| CD40L | − | − |
| CTLA-4 | − | + |
| Perforin granules | ||
| Perforin | ++ | ++ |
| Granzyme A | ++ | ++ |
| NK and NKT receptors | ||
| CD94 | − | − |
| CD56 | + | + |
| NKG2D | ++ | ++ |
| CD161 | − | − |
+, weak staining (low level of expression); ++, strong staining (high level of expression); −, no surface expression.
HLA class I-restricted and -unrestricted clones recognize C. trachomatis-infected cells at different times following infection.
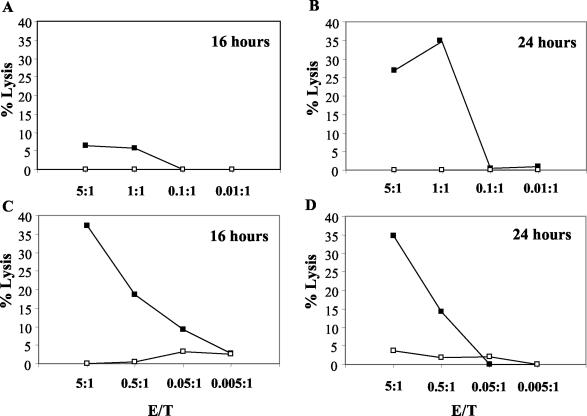
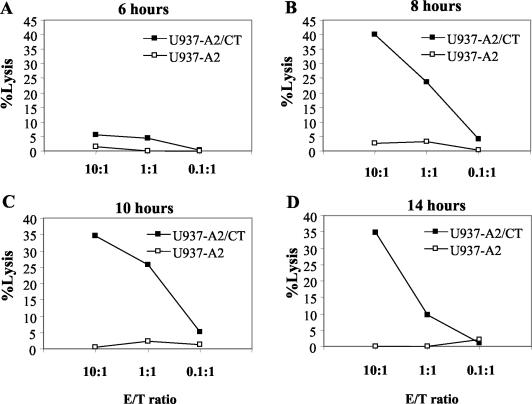
APC were infected with C. trachomatis and used at different times postinfection as target cells in a 6-h cytotoxicity assay. The time course studied was from 30 min to 24 h postinfection (Fig. 5 and Fig. 6). Figure 5 shows that whereas both unrestricted and restricted clones lysed C. trachomatis-infected cells 24 h after infection, only the unrestricted clone produced lysis at the 16-h time point. Figure 6 extends the analysis of the time course for an unrestricted clone and shows that significant lysis was seen as early as 8 h following infection but not at the 6-h time point and remained at the same level at later time points.
FIG. 5.
Time course of specific lysis of C. trachomatis-infected U937-A2 cells. Specific lysis of uninfected or C. trachomatis-infected U937-A2 cells by an HLA-A2-restricted clone (A and B) and by an HLA class I-unrestricted clone (C and D) at 16 h and 24 h postinfection. This experiment was carried out three times with equivalent results in each case.
FIG. 6.
Time course of specific lysis of C. trachomatis-infected U937-A2 cells. Specific lysis of uninfected or C. trachomatis-infected U937-A2 cells by an HLA class I-unrestricted clone is shown at various time points following infection. This experiment was carried out three times with equivalent results in each case. CT, C. trachomatis.
CD1 is not the restriction element for the HLA class I-unrestricted CD8+-T-cell clones.
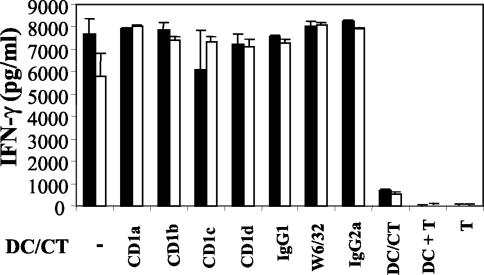
The U937 monocytic cell line does not express measurable levels of CD1a, CD1b, CD1c, and CD1d when tested by flow cytometry (data not shown). Since all the class I major histocompatibility complex-unrestricted clones which we studied were routinely tested for IFN-γ production and lysis when cocultured with C. trachomatis-infected U937 cells (Fig. 3), it was unlikely that CD1 molecules were presenting C. trachomatis antigens to these clones. However, to evaluate this possibility further, lymphoblastoid C1R cell lines transfected with CD1a, CD1b, or CD1c molecules were infected with C. trachomatis and cocultured with HLA class I-unrestricted CD8+-T-cell clones. There was no IFN-γ production in any of these cultures. Next, we used antibodies directed against CD1a, CD1b, CD1c, and CD1d to try to block responses to C. trachomatis-infected DC; these antibodies have been previously shown to block CD1-restricted responses (51). Figure 7 shows that there was no inhibition by any of these antibodies, which is again consistent with the idea that CD1 molecules are not the restricting elements used by these clones.
FIG. 7.
IFN-γ production by HLA-unrestricted CD8+-T-cell clones was not inhibited by antibodies to CD1. IFN-γ production by two HLA-unrestricted clones (clone 1, white bars; clone 2, black bars) in responses to autologous C. trachomatis-infected DC, alone or in the presence of antibodies specific for CD1a, CD1b, CD1c, CD1d, or w6/32. IgG1 and IgG2a isotype control antibodies were also included. Minimal IFN-γ was seen in cultures containing T cells and uninfected DC or in C. trachomatis-infected DC and T cells cultured separately. The experiments were carried out three times for each clone tested. CT, C. trachomatis.
Enhancement of IL-12 production by C. trachomatis-specific CD8+-T-cell clones.
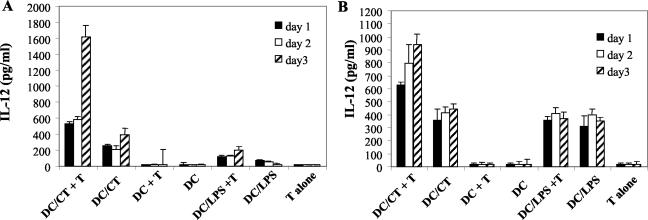
DC were infected with live C. trachomatis and cocultured with C. trachomatis-specific CD8+-T-cell clones. The cultures were set up immediately after infection, supernatants were collected daily for 4 days, and IFN-γ and IL-12 were measured by ELISA. Figure 8A shows results from an HLA class I-unrestricted clone, and Fig. 8B shows the results for an HLA class I-restricted clone. For uninfected DC, IL-12 production was minimal in the presence of either type of clone and comparable when stimulated with LPS whether or not either type of clone was present. However, IL-12 production in cultures with C. trachomatis-infected DC was substantially increased by the presence of either HLA-restricted or -unrestricted C. trachomatis-specific CD8+-T-cell clones. A twofold increase in IL-12 was already evident at day 1 of culture, and further increases were seen at later time points. To determine whether the increase of IL-12 production was the result of early IFN-γ release by these T cells, various amounts of recombinant human IFN-γ were added to cultures containing C. trachomatis-infected DC, and LPS-activated DC were used as controls. A significant increase in IL-12 production was detected with as little as 300 pg of IFN-γ per ml, with a further increase when a higher dose of IFN-γ was added (data not shown). The amount of IL-12 produced by the IFN-γ-treated DC was comparable to that produced by DC cocultured with CD8+ T cells, suggesting that the IFN-γ produced by CD8+ T cells was sufficient to account for the increase in IL-12 production seen in the cocultures of T-cell clones and C. trachomatis-infected DC.
FIG. 8.
Increase in IL-12 production by C. trachomatis-infected DC cocultured with a CD8+-T-cell clone. IL-12 levels in supernatants from uninfected or C. trachomatis-infected DC cocultured with either a C. trachomatis-specific HLA-unrestricted clone (A) or a C. trachomatis-specific HLA-A2-restricted clone (B). Also shown are IL-12 levels from cultures of uninfected or C. trachomatis-infected DC in the absence of T cells, T cells alone, LPS-stimulated DC, and LPS-stimulated DC cocultured with T-cell clones. IL-12 production was measured after days 1, 2, and 3. Note the difference in scale (vertical axis) between panels A and B. The figure shows the results from one of three similar experiments. CT, C. trachomatis.
DISCUSSION
We have shown that CD8+ T cells specific for C. trachomatis are present in the PBMC of patients with previous episodes of C. trachomatis infection. The clones could be subdivided into two groups; the first recognized C. trachomatis antigens in association with classical HLA class I, whereas the second did not show evidence of HLA restriction. Unlike HLA class I-restricted clones which only lysed C. trachomatis-infected target cells 24 h postinfection, HLA-unrestricted clones lysed infected cells within 8 h postinfection. Although the antigens recognized by these HLA-unrestricted clones require infection by C. trachomatis and are not expressed by cells infected by other gram-negative bacteria (Salmonella and Yersinia), their nature and the identity of the molecule which presents these antigens remain unclear. An obvious possibility to consider was the group of CD1 molecules which have the ability to present nonpeptide antigens such as glycolipids from intracellular organisms, particularly mycobacteria to TCR-α/β and γδ T cells (17, 31, 39). However, our experimental data showed that CD1 molecules are not the restriction elements for the HLA class I-unrestricted clones. Importantly, U937, which does not express CD1 molecules, was consistently recognized, and the requirement for infection with live C. trachomatis would be unusual for CD1-restricted T cells where the lipid antigens can be derived from organism cell walls. In addition, the experiments with C1R transfected with different CD1 molecules were negative, although it must be conceded that these experiments are not conclusive, since there is no proof that lymphoblastoid cells such as C1R can process C. trachomatis antigens. Other candidate-presenting molecules such as HLA-E are now under investigation. Note, however, that the present study does not exclude the existence of CD1-restricted C. trachomatis-specific CD8+ T cells as an additional component of the response to C. trachomatis in infected patients. Preliminary results show that adding anti-CD1 antibodies to C. trachomatis-infected PBMC reduces IFN-γ production (data not shown). These responses are now being further investigated.
The importance of CD8+-T-cell responses to intracellular pathogens has previously been reported for several bacteria including Listeria monocytogenes (6, 9), S. enterica serovar Typhimurium (29, 43), and Mycobacterium tuberculosis (28), but CD8+-T-cell responses to C. trachomatis have not been extensively studied. Starnbach et al. (45, 46) reported murine C. trachomatis-specific CD8+ T cells, but these were derived following intravenous challenge of mice with C. trachomatis rather than the usual mucosal route of infection. Nevertheless, these CD8+ T cells were able to confer resistance to C. trachomatis-induced disease. Two C. trachomatis antigens, Cap1 and CrpA, recognized by murine CD8+-T-cell clones have recently been identified (11, 46). Both of these antigens have been shown to be associated with the chlamydia inclusion membrane. As noted previously, human CD8+ T cells able to recognize C. trachomatis MOMP or hsp60 have been generated from infected individuals by using synthetic peptides (18, 24-26). The identity of the antigen(s) recognized by HLA-restricted T cells isolated in our laboratory is as yet unknown but can be revealed by approaches such as expression library screening, as recently described for the identification of Cap1 and CrpA and C. trachomatis antigens recognized by CD4+ T cells (15).
The C. trachomatis-specific T cells that we isolated were able to lyse C. trachomatis-infected cells, but it is unclear how important lysis is in comparison to cytokine production for host resistance to infection. The effectiveness of transfer of a murine C. trachomatis-specific CD8+ clone was entirely dependent on its ability to produce IFN-γ and on the ability of the recipient to respond to IFN-γ. Likewise, infection of gene-targeted mice that lack perforin resulted in unimpaired clearance of the bacteria, suggesting that perforin-dependent killing was not critical.
Very recently Gervassi et al. have reported CD8+ T cells specific for C. trachomatis and also noted both classical HLA class I-restricted and -unrestricted populations (13). They estimated the frequency of these cells in six patients, and found that the frequency of HLA class I-unrestricted CD8+ T cells was higher than that of classical HLA class I restriction. These results are in agreement with our studies. However, we found certain functional differences between the C. trachomatis-specific CD8+-T-cell clones raised in our laboratory and those which they reported. HLA class I-restricted clones were reported not to kill C. trachomatis-infected target cells, but we found that our HLA class I-restricted clones readily lysed C. trachomatis-infected U937 cells as long as they expressed the appropriate HLA class I restriction element. It is possible that the difference between the two sets of results is explained by the use of different C. trachomatis-infected target cells, since different infected cells may be more or less susceptible to lysis. Thus, Passmore et al. (38) have shown differences in the lysis of M. tuberculosis-infected U937 cells and macrophages. Gervassi et al. used E:T ratios of more than 100:1 in cytolysis assays, whereas we observed significant levels of lysis at E:T ratios of 1:1 or even 0.1:1, ratios which are commonly used in assessing cytolytic T-cell clones. This suggests that the clones which we generated are efficient in bringing about cytolysis of C. trachomatis-infected U937 cells. However, since the clones described by Gervassi et al. were capable of efficient antigen-independent killing, it may be that the difference in results is mainly related to the target cells tested. Fan et al. (10) have shown that intracellular C. trachomatis can inhibit the induction of host cell apoptosis, but this effect varied according to time after infection and number of chlamydia infecting the cells. It is possible, therefore, that U937 cells (used in our experiments) and fibroblasts (used by Gervassi et al.) differ in the time taken for chlamydiae to render cells resistant to apoptosis triggered by cytotoxic T lymphocytes or the numbers of chlamydiae typically present in each cell.
In the present study we have evaluated differences not only in the antigen restriction element of the two sets of C. trachomatis-specific CD8+ T cells but also in the time required for the antigens recognized by each subset to be presented. We have shown that HLA class I-unrestricted clones can kill infected target cells as early as 8 h postinfection, whereas the expression of C. trachomatis antigens in association with HLA class I required more than 20 h of processing. The early response by unrestricted clones may be important in producing conditions which skew subsequent HLA-restricted C. trachomatis-specific T-cell responses towards the Th1 phenotype (see below).
The strategy employed by Gervassi et al. and by us permits identification of the predominant antigens which elicit CD8+-T-cell responses when the immune system is challenged by live organisms. Approaches using synthetic peptides can only identify the HLA-restricted subset of C. trachomatis-specific CD8+ T cells, since peptides are tested on the basis of their ability to bind to common class I HLA alleles. The relative importance of HLA-restricted and HLA-independent CD8+-T-cell responses to C. trachomatis is unknown, but concentrating solely on HLA-restricted, peptide-specific responses may misrepresent the physiologic response to C. trachomatis by CD8+ T cells.
A common feature of both types of C. trachomatis-specific clones was the production of IFN-γ. IFN-γ is critical for the clearance of C. trachomatis in vivo (23). In animal models, treatment with neutralizing antibodies to IFN-γ or disruption of either the IFN-γ gene or the IFN-γ receptor gene substantially increased host susceptibility to C. trachomatis infection (7, 22). Furthermore, IFN-γ inhibits the growth of C. trachomatis in cell culture (33, 34, 41). C. trachomatis infection can be affected by IFN-γ in several ways: it activates APCs which may harbor C. trachomatis organisms (1, 42, 53); it induces the enzyme indoleamine 2,3 dioxygenase which degrades intracellular tryptophan, an amino acid which many biovars of chlamydiae are unable to synthesize de novo (3, 33, 35, 48); and lastly, in murine cells it induces nitric oxide production which has been shown to be bactericidal to chlamydiae (19, 20). In passive transfer experiments, CD4+ T cells which produce IFN-γ have been shown to provide protection against C. trachomatis in rodents (47), but clearance of the pathogen was slower in comparison to animals which received transfer of both CD4+ and CD8+ T cells (30, 49), suggesting that additional IFN-γ might be produced by the CD8+ subset. Interestingly, we found a much higher quantity of IFN-γ produced by clones which exhibit HLA class I-unrestricted recognition of C. trachomatis. Given the important role of IFN-γ in the clearance of C. trachomatis infection, the rapid production of large quantities of IFN-γ by the HLA-independent CD8+ subset could represent an important immune response to C. trachomatis.
C. trachomatis-specific CD8+ T cells also play a role in the activation of DC. These clones, when cocultured with infected DC, induced a rapid increase in IL-12 production by DC. Since IL-12 is critical for the stimulation of Th1 responses and, therefore, the expansion of IFN-γ-producing CD4+ T cells, the effects of CD8+ T cells may be to obtain an appropriate Th1-polarized CD4+-T-cell response to C. trachomatis in addition to their own ability to produce IFN-γ. The increase in IL-12 production following coculture of infected DC with C. trachomatis-specific CD8+-T-cell clones was not dependent on CD40-CD40L interactions since HLA class I-unrestricted clones did not express CD40L, either in their resting state or following activation. On the other hand, we have shown that the addition of IFN-γ to C. trachomatis-infected DC cultures results in an increase in IL-12 production. It has previously been shown that DC infected with C. trachomatis produce high levels of tumor necrosis factor alpha (32), and these levels were also increased and sustained by the addition of IFN-γ. Taking these findings together strongly suggests that the increase in IL-12 production was mainly mediated by the synergistic effect of tumor necrosis factor alpha produced by infected DC and of IFN-γ produced by T cells, allowing the development of strong Th1-polarized immune responses.
In summary, we have generated and characterized C. trachomatis-specific CD8+ T cells from C. trachomatis-infected individuals. The approach taken in our study should allow us to study the full repertoire of CD8+-T-cell responses to C. trachomatis, including those which are not restricted by classical HLA class I. The identification of the antigens recognized by both CD8+ and CD4+ T cells in patients with C. trachomatis infection, both those with uncomplicated local infection and those who develop inflammatory sequelae, is a vital step in the development of a safe and effective vaccine.
Acknowledgments
This work was supported by the Medical Research Council, London, United Kingdom.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Airenne, S., H. M. Surcel, A. Bloigu, K. Laitinen, P. Saikku, and A. Laurila. 2000. The resistance of human monocyte-derived macrophages to Chlamydia pneumoniae infection is enhanced by interferon-gamma. APMIS 108:139-144. [DOI] [PubMed] [Google Scholar]
- 2.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatty, W. L., T. A. Belanger, A. A. Desai, R. P. Morrison, and G. I. Byrne. 1994. Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62:3705-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunham, R. C., J. Kimani, J. Bwayo, G. Maitha, I. Maclean, C. Yang, C. Shen, S. Roman, N. J. Nagelkerke, M. Cheang, and F. A. Plummer. 1996. The epidemiology of Chlamydia trachomatis within a sexually transmitted diseases core group. J. Infect. Dis. 173:950-956. [DOI] [PubMed] [Google Scholar]
- 5.Brunst, M., M. Shahmanesh, A. Sukthankar, J. H. Pearce, and J. S. Gaston. 1998. Isolation and characterisation of T lymphocytes from the urethra of patients with acute urethritis. Sex. Transm. Infect. 74:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch, D. H., and E. G. Pamer. 1999. T lymphocyte dynamics during Listeria monocytogenes infection. Immunol. Lett. 65:93-98. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, T. W., K. H. Ramsey, G. S. Miranpuri, C. E. Poulsen, and G. I. Byrne. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect. Immun. 65:2145-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deane, K. H., R. M. Jecock, J. H. Pearce, and J. S. Gaston. 1997. Identification and characterization of a DR4-restricted T cell epitope within chlamydia heat shock protein 60. Clin. Exp. Immunol. 109:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finelli, A., K. M. Kerksiek, S. E. Allen, N. Marshall, R. Mercado, I. Pilip, D. H. Busch, and E. G. Pamer. 1999. MHC class I restricted T cell responses to Listeria monocytogenes, an intracellular bacterial pathogen. Immunol. Res. 19:211-223. [DOI] [PubMed] [Google Scholar]
- 10.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome C release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fling, S. P., R. A. Sutherland, L. N. Steele, B. Hess, S. E. D'Orazio, J. Maisonneuve, M. F. Lampe, P. Probst, and M. N. Starnbach. 2001. CD8+ T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 98:1160-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaston, J. S. 2000. Immunological basis of Chlamydia induced reactive arthritis. Sex. Transm. Infect. 76:156-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gervassi, A. L., P. Probst, W. E. Stamm, J. Marrazzo, K. H. Grabstein, and M. R. Alderson. 2003. Functional characterization of class Ia- and non-class Ia-restricted Chlamydia-reactive CD8+ T cell responses in humans. J. Immunol. 171:4278-4286. [DOI] [PubMed] [Google Scholar]
- 14.Goodall, J. C., H. Beacock-Sharp, K. H. Deane, and J. S. Gaston. 2001. Recognition of the 60 kilodalton cysteine-rich outer membrane protein OMP2 by CD4(+) T cells from humans infected with Chlamydia trachomatis. Clin. Exp. Immunol. 126:488-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodall, J. C., G. Yeo, M. Huang, R. Raggiaschi, and J. S. H. Gaston. 2001. Identification of Chlamydia trachomatis antigens recognized by human CD4+ T lymphocytes by screening an expression library. Eur. J. Immunol. 31:1513-1522. [DOI] [PubMed]
- 16.Guaschino, S., and F. De Seta. 2000. Update on Chlamydia trachomatis. Ann. N. Y. Acad. Sci. 900:293-300. [DOI] [PubMed] [Google Scholar]
- 17.Guermonprez, P., J. Valladeau, L. Zitvogel, C. Thery, and S. Amigorena. 2002. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 20:621-667. [DOI] [PubMed] [Google Scholar]
- 18.Holland, M. J., D. J. Conway, T. J. Blanchard, O. M. Mahdi, R. L. Bailey, H. C. Whittle, and D. C. Mabey. 1997. Synthetic peptides based on Chlamydia trachomatis antigens identify cytotoxic T lymphocyte responses in subjects from a trachoma-endemic population. Clin. Exp. Immunol. 107:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, J., F. J. DeGraves, S. D. Lenz, D. Gao, P. Feng, D. Li, T. Schlapp, and B. Kaltenboeck. 2002. The quantity of nitric oxide released by macrophages regulates Chlamydia-induced disease. Proc. Natl. Acad. Sci. USA 99:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igietseme, J. U. 1996. Molecular mechanism of T-cell control of Chlamydia in mice: role of nitric oxide in vivo. Immunology 88:1-5. [PMC free article] [PubMed] [Google Scholar]
- 21.Inman, R. D., J. A. Whittum-Hudson, H. R. Schumacher, and A. P. Hudson. 2000. Chlamydia and associated arthritis. Curr. Opin. Rheumatol. 12:254-262. [DOI] [PubMed] [Google Scholar]
- 22.Johansson, M., K. Schon, M. Ward, and N. Lycke. 1997. Genital tract infection with Chlamydia trachomatis fails to induce protective immunity in gamma interferon receptor-deficient mice despite a strong local immunoglobulin A response. Infect. Immun. 65:1032-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson, M., K. Schon, M. Ward, and N. Lycke. 1997. Studies in knockout mice reveal that anti-chlamydial protection requires TH1 cells producing IFN-gamma: is this true for humans? Scand. J. Immunol. 46:546-552. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S. K., M. Angevine, K. Demick, L. Ortiz, R. Rudersdorf, D. Watkins, and R. DeMars. 1999. Induction of HLA class I-restricted CD8+ CTLs specific for the major outer membrane protein of Chlamydia trachomatis in human genital tract infections. J. Immunol. 162:6855-6866. [PubMed] [Google Scholar]
- 25.Kim, S. K., L. Devine, M. Angevine, R. DeMars, and P. B. Kavathas. 2000. Direct detection and magnetic isolation of Chlamydia trachomatis major outer membrane protein-specific CD8+ CTLs with HLA class I tetramers. J. Immunol. 165:7285-7292. [DOI] [PubMed] [Google Scholar]
- 26.Kuon, W., R. Lauster, U. Bottcher, A. Koroknay, M. Ulbrecht, M. Hartmann, M. Grolms, S. Ugrinovic, J. Braun, E. H. Weiss, and J. Sieper. 1997. Recognition of chlamydial antigen by HLA-B27-restricted cytotoxic T cells in HLA-B*2705 transgenic CBA (H-2k) mice. Arthritis Rheum. 40:945-954. [DOI] [PubMed] [Google Scholar]
- 27.Lampe, M. F., C. B. Wilson, M. J. Bevan, and M. N. Starnbach. 1998. Gamma interferon production by cytotoxic T lymphocytes is required for resolution of Chlamydia trachomatis infection. Infect. Immun. 66:5457-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewinsohn, D. M., L. Zhu, V. J. Madison, D. C. Dillon, S. P. Fling, S. G. Reed, K. H. Grabstein, and M. R. Alderson. 2001. Classically restricted human CD8+ T lymphocytes derived from Mycobacterium tuberculosis-infected cells: definition of antigenic specificity. J. Immunol. 166:439-446. [DOI] [PubMed] [Google Scholar]
- 29.Lo, W. F., H. Ong, E. S. Metcalf, and M. J. Soloski. 1999. T cell responses to Gram-negative intracellular bacterial pathogens: a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. J. Immunol. 162:5398-5406. [PubMed] [Google Scholar]
- 30.Loomis, W. P., and M. N. Starnbach. 2002. T cell responses to Chlamydia trachomatis. Curr. Opin. Microbiol. 5:87-91. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda, J. L., and M. Kronenberg. 2001. Presentation of self and microbial lipids by CD1 molecules. Curr. Opin. Immunol. 13:19-25. [DOI] [PubMed] [Google Scholar]
- 32.Matyszak, M. K., J. L. Young, and J. S. H. Gaston. 2002. Uptake and processing of Chlamydia trachomatis by human dendritic cells. Eur. J. Immunol. 32:742-751. [DOI] [PubMed] [Google Scholar]
- 33.Mehta, S. J., R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-gamma: role of tryptophan catabolism. J. Infect. Dis. 177:1326-1331. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, R. P. 2000. Differential sensitivities of Chlamydia trachomatis strains to inhibitory effects of gamma interferon. Infect. Immun. 68:6038-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, H. W., A. Szuro-Sudol, D. Wellner, M. J. Oca, A. M. Granger, D. M. Libby, C. D. Rothermel, and B. Y. Rubin. 1989. Role of tryptophan degradation in respiratory burst-independent antimicrobial activity of gamma interferon-stimulated human macrophages. Infect. Immun. 57:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortiz, L., M. Angevine, S. K. Kim, D. Watkins, and R. DeMars. 2000. T-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humans. Infect. Immun. 68:1719-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortiz, L., K. P. Demick, J. W. Petersen, M. Polka, R. A. Rudersdorf, B. Van der Pol, R. Jones, M. Angevine, and R. DeMars. 1996. Chlamydia trachomatis major outer membrane protein (MOMP) epitopes that activate HLA class II-restricted T cells from infected humans. J. Immunol. 157:4554-4567. [PubMed] [Google Scholar]
- 38.Passmore, J. S., P. T. Lukey, and S. R. Ress. 2001. The human macrophage cell line U937 as an in vitro model for selective evaluation of mycobacterial antigen-specific cytotoxic T-cell function. Immunology 102:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porcelli, S. A., and R. L. Modlin. 1999. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 17:297-329. [DOI] [PubMed] [Google Scholar]
- 40.Ramsey, K. H., and R. G. Rank. 1991. Resolution of chlamydial genital infection with antigen-specific T-lymphocyte lines. Infect. Immun. 59:925-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapoza, P. A., S. G. Tahija, J. P. Carlin, S. L. Miller, M. L. Padilla, and G. I. Byrne. 1991. Effect of interferon on a primary conjunctival epithelial cell model of trachoma. Investig. Ophthalmol. Vis. Sci. 32:2919-2923. [PubMed] [Google Scholar]
- 42.Rothermel, C. D., B. Y. Rubin, and H. W. Murray. 1983. Gamma-interferon is the factor in lymphokine that activates human macrophages to inhibit intracellular Chlamydia psittaci replication. J. Immunol. 131:2542-2544. [PubMed] [Google Scholar]
- 43.Salerno-Goncalves, R., M. F. Pasetti, and M. B. Sztein. 2002. Characterization of CD8(+) effector T cell responses in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 169:2196-2203. [DOI] [PubMed] [Google Scholar]
- 44.Sallusto, F., and A. Lanzavecchia. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starnbach, M. N., M. J. Bevan, and M. F. Lampe. 1994. Protective cytotoxic T lymphocytes are induced during murine infection with Chlamydia trachomatis. J. Immunol. 153:5183-5189. [PubMed] [Google Scholar]
- 46.Starnbach, M. N., W. P. Loomis, P. Ovendale, D. Regan, B. Hess, M. R. Alderson, and S. P. Fling. 2003. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8(+) T cell response. J. Immunol. 171:4742-4749. [DOI] [PubMed] [Google Scholar]
- 47.Su, H., and H. D. Caldwell. 1995. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect. Immun. 63:3302-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, M. W., and G. S. Feng. 1991. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5:2516-2522. [PubMed] [Google Scholar]
- 49.Thoma-Uszynski, S., U. Simnacher, R. Marre, and A. Essig. 1998. Clearance of Chlamydia trachomatis-induced polyserositis in SCID mice requires both CD4+ and CD8+ cells. Med. Microbiol. Immunol. 187:71-78. [DOI] [PubMed] [Google Scholar]
- 50.Toivanen, A., and P. Toivanen. 2000. Reactive arthritis. Curr. Opin. Rheumatol. 12:300-305. [DOI] [PubMed] [Google Scholar]
- 51.Vincent, M., D. Leslie, D. Gumperz, X. Xiong, E. Grant, and M. Brenner. 2002. CD1-dependent dendritic cell instruction. Nat. Immunol. 3:1163-1168. [DOI] [PubMed] [Google Scholar]
- 52.West, S. K., P. Rapoza, B. Munoz, S. Katala, and H. R. Taylor. 1991. Epidemiology of ocular chlamydial infection in a trachoma-hyperendemic area. J. Infect. Dis. 163:752-756. [DOI] [PubMed] [Google Scholar]
- 53.Zhong, G. M., and L. M. de la Maza. 1988. Activation of mouse peritoneal macrophages in vitro or in vivo by recombinant murine gamma interferon inhibits the growth of Chlamydia trachomatis serovar L1. Infect. Immun. 56:3322-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]