Abstract
Aging is a multifactorial process during which physiological alterations occur in all tissues. A decline in mitochondrial function plays an important role in the process of aging and in aging-associated diseases. The mitochondrial genome encodes 13 essential subunits of protein complexes belonging to the oxidative phosphorylation system, while most of the mitochondria-related genes are encoded by the nuclear genome. Coordination between the nucleus and mitochondria is crucial for the regulation of mitochondrial biogenesis and function. In this review, we will discuss aging-related mitochondrial dysfunction in various tissues and its implication in aging-related diseases and the aging process.
Keywords: Review, Mitochondria, Aging, Aging-associated diseases, Tissue-specific, mtDNA mutation, Mitochondrial regulation, Review
2. AGING AND TISSUE-SPECIFIC AGING
As a consequence of rising life expectancy and declining birth rates, population aging is taking place globally. Humans have never before reached such old ages as they do now. In fact, aging is the major risk factor for many degenerative diseases, metabolic diseases and death. Worldwide, about 100,000 people die of aging-related causes each day.
As organisms age, all tissues start to decline in their functions and thus exhibit various associated anomalous features at different aging time points. In humans, subtle irreversible changes occur by the third and fourth decades of life in most tissues and progressively deteriorate with further aging. The rate of aging-associated decline varies among different tissues (1), and the aging rate differs from person to person. Nevertheless, some universal molecular events occur, in particular within the same tissues.
Age is the major risk factor for neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis. Autopsy and MRI studies have revealed that brain volume and weight decrease, accompanied by increases in ventricular volume and cerebrospinal fluid spaces, in individuals over 60 (2). There are other changes like loss of neural circuits, brain plasticity and thinning of the cortex in old people. Meanwhile, there are notable cellular changes occurring in aging brains. Lipofuscin, which contains peroxidised protein and lipids, accumulates in some neurons (2). Granulovacuolar degeneration and Hirano bodies are usually found in pyramidal cells in the hippocampus and increase with age. Neurofibrillary tangles, which are aggregates of hyperphosphorylated tau protein, and senile plaques, extracellular deposits mainly composed of small peptides containing 39–43 amino acids known as amyloid β-peptide (Aβ), are neuropathological hallmarks of Alzheimer’s disease, which has been considered as accelerated brain aging (2, 3).
It is well established that the age-dependent decline in body mass is largely due to loss and atrophy of muscle cells (4). Age-related reductions in muscle mass (sarcopenia) are a direct cause of the declines in muscle strength and disability observed in older adults. Muscle mass is maintained mostly through the balance between protein synthesis and degradation, and the rates of skeletal muscle protein synthesis decline with age. Muscle changes usually start in the fourth decade of life which include reductions in muscle fiber number, decreases in the size of fast-twitch type II fibers and motor unit number and size, and increases in noncontractile structures such as fat and connective tissue (5, 6).
Fibroblasts are the most common cells of connective tissues, providing structure and regulatory signals. By continuously secreting precursors of the extracellular matrix, fibroblasts are important in maintaining the structural integrity of connective tissues. In response to a wound or a need for tissue remodeling, fibroblasts also secrete enzymes to degrade the matrix. Fibroblasts were among the earliest cell types to be cultured in vitro and were the first cell-culture model for aging research (7). Such studies led to the important discovery of limited replicative life span of cells because fibroblasts from older donors have a shorter replicative life span than those from younger donors (8). Underin vitro culture conditions, ‘aged’ or senescent fibroblasts are characterized by growth arrest, resistance to apoptosis and altered morphology. Fibroblasts also undergo progressive telomere shortening. The dermis is mainly composed of fibroblasts. It plays important roles in wound-healing by secreting dermal collagens, elastin and other extracellular matrix components. With aging, the dermis becomes thinner with significant loss of collagen and other fibers, and wound healing slows down as well. Senescent fibroblasts appear to increase with age in human dermis and secrete factors that degrade dermal collagens and elastin (9).
The liver is a vital organ with a wide range of functions, including detoxification and production of enzymes necessary for digestion. Liver weight decreases about 20% after the age of 50 and the hepatocytes accumulate lysosomes, residual bodies and lipofuscin. Although the majority of liver functions seem well maintained, there is a dramatic age-associated loss of its regenerative capacity after partial hepatectomy or chemical injury. It was reported that 99% of hepatocytes of young mice proliferate after partial hepatectomy, whereas only 30% of hepatocytes in old livers undergo such proliferation (10). Another age-related liver functional change concerns drug metabolism. It was observed that many drugs, such as diazepam and antipyrine, were metabolized much more slowly by the liver in elderly people (10, 11).
Blood, which makes up of about 8% of an adult’s body weight, is composed of plasma and cells including red and white blood cells and platelets. Blood plays many critical roles such as transporting nutrients and oxygen to various tissues and removing metabolic wastes from them, defending against infections and other pathogenic agents, and regulating temperature and pH of the body. With aging, both the blood volume and the number of red blood cells decrease (1). While the number of white blood cells remains relatively unchanged, the lymphocyte number and infection resistance also decrease (11). At the same time, increases in blood viscosity, fibrinogen level and red blood cell rigidity have also been recorded during aging. All these changes contribute to an increased incidence of hypertension, clot formation and atherosclerosis in older people (11).
3. TISSUE-SPECIFIC METABOLIC PATTERNS
The metabolic patterns of brain, muscle, liver and other tissues are strikingly different. Glucose is virtually the sole fuel for the human brain except after prolonged starvation. Since the brain stores only a very small amount of glycogen, it requires a continuous daily supply of about 120g glucose, accounting for 60% of the glucose utilization of the whole body in its resting state. In contrast to many other tissues, fatty acids do not serve as fuel for the brain as they cannot traverse the blood-brain barrier (12). The brain also consumes as much as 20–25% of the body’s oxygen for oxidative phosphorylation (OXPHOS).
In contrast to brain tissue, muscles can use glucose, fatty acids and ketone bodies as energy sources. Skeletal muscle is composed of different fiber types. Mitochondria are abundant in type I, IIA and IIX fibers which produce ATP by OXPHOS. Type IIB fibers have few mitochondria and rely on glycolysis for ATP production (13). In resting skeletal muscle, fatty acids are the major fuel and the ATP produced by OXPHOS meets 85% of the energy needs. The energy demand is met by both OXPHOS and glycolysis in moderate exercise. In actively contracting skeletal muscle, the rate of glycolysis far exceeds that of the citric acid cycle, and much of the pyruvate is reduced to lactate (13). Unlike skeletal muscle, cardiac muscle functions almost exclusively aerobically, as evidenced by the high density of mitochondria. Fatty acids are the heart’s main source of fuel (14).
Liver is the metabolic hub providing fuel to the brain, muscle and other peripheral organs. The maintenance of a fairly steady blood glucose level is one of the liver’s main functions. When the blood glucose is low, the liver meets its own energy needs through OXPHOS and releases glucose into the blood. In the case of high blood glucose, the liver converts absorbed glucose to glucose 6-phosphate, most of which is converted into glycogen while the rest is utilized to meet its own energy demands through glycolysis (15).
Red blood cells, which have no mitochondria, can only use glycolysis to meet their energy demands (16).
4. REGULATION AND TISSUE-SPECIFIC REGULATION OF MITOCHONDRIAL FUNCTION
Mitochondria play critical roles in regulating energy production, metabolism, signal transduction and apoptosis (17). Therefore, tight and delicate regulation of mitochondrial functions is vital. Mitochondrial dysfunction has been associated with various age-related human diseases and the aging process itself (18). Most of the mitochondrial proteins are encoded by the nuclear genome (nDNA), while the mitochondrial DNA (mtDNA) encodes 13 essential peptides that all are subunits of the OXPHOS system, 22 tRNAs and 2 rRNAs (18). Regulation of mitochondrial functions requires coordination of the two genomes.
Replication of mtDNA is independent of the cell cycle. A mtDNA copy may be replicated many times or not at all as a cell divides, and there is continuous mtDNA replication in postmitotic cells (18). This indicates that a single mtDNA mutation can be expanded clonally or may be lost when a cell divides.
The mitochondrial transcription machinery is composed of a single RNA polymerase (POLRMT), initiation (TFB2M) and stimulatory (Tfam, for example) transcription factors and termination factors (mTERFs family) (19). The long-term adaptive regulation of mitochondria is achieved by mitochondrial biogenesis, whereas transient changes can be met by altered expression of a subset of mitochondrial genes or of a subset of critical regulators (20). Such regulation requires a highly interconnected network of nDNA-binding transcription factors and co-regulators. Strikingly, about half of the mitochondrial genes are expressed in a tissue-specific pattern to meet the demand of specialized functions in different tissues (21).
Nuclear respiratory factors NRF-1 and NRF-2 are the first identified transcription factors that recognize conserved motifs of the promoters of mitochondrial OXPHOS genes (22). NRF-1 binds to a palindromic sequence in the promoter of cytochrome c and is associated with many genes required for expression and function of the respiratory chain (22). NRF-2 is a multi-subunit activator of respiratory complex IV (22). These two factors often work in conjunction (23). NRF-1 and -2 are also parts of the defensive mechanisms against oxidative stress. Inactive NRF-2 is retained in the cytosol by association with a cytoskeletal protein Keap1. Cytosolic NRF-2 is phosphorylated and then translocates into the nucleus in response to PKC activation through MAPK pathways. In the nucleus, NRF-2 regulates antioxidant response element (ARE) mediated expression of detoxifying and antioxidant enzymes (24). Constitutive expression of NRF-2 increases tolerance to oxidative stress and promotes longevity in worms and fruitflies (25). In mice, p62, an autophagy regulatory protein, plays an important role in ensuring longevity by activating NRF-2 to maintain mitochondrial integrity (25). Interestingly, NRF-1 has also been demonstrated to bind to ARE and regulate the expression of glutathione biosynthesis and other oxidative defense enzymes (26). NRF-1 null mice show early embryonic lethality and their blastocysts have reduced mtDNA content and mitochondrial membrane potential (20). Similarly, NRF-2 disruption causes preimplantation lethality (20). Additional nuclear factors, such as CREB, YY1, MEF-2/EBOX, c-myc, ERRα and MT-1 to -4, have also been linked to the control of mitochondrial biogenesis and respiratory function (23).
Despite all the complexity of the signaling pathways regulating mitochondrial biogenesis, it is believed that the PPARγ coactivator-1 family (PGC-1α, -1β, and PRC) plays a paramount role by integrating and coordinating the activities of multiple transcription factors (20). PGC-1α was identified as a transcriptional regulator of PPARγ during adaptive thermogenesis in brown fat (27). PGC-1α binds not only a large complement of nuclear receptors, including PPARγ, HNF4α, GR and ERRα, but also a number of transcription factors, such as NRF-1, MEF-2C, FOXO1 and YY1 (23). All these factors either act directly on the expression of components of the respiratory chain or support the respiratory machinery through other pathways. PGC-1α is therefore considered as the master regulator for mitochondrial biogenesis. PGC-1α is induced by cold exposure, short-term exercise and other conditions that increase tissue demands on mitochondrial function (28). PGC-1β and PRC are homologues of PGC-1α (29). PGC -1β and PGC-1α are all highly expressed in tissues with abundant mitochondria, such as heart, skeletal muscle and brown fat, whereas the expression of PRC is not associated with mitochondrial abundance (30, 31). Like PGC-1α, PGC-1β can bind to NRF-1, NRF-2, PPARs, ERRs and YY1 (20). Specifically, PGC-1 docks on transcription factors that bind to their downstream targets, and enables the recruitment of histone acetyltransferases and the mediator complex, thereby enhancing transcription initiation and/or elongation of critical mitochondrial genes (20).
In particular, muscle specific expression of PGC-1α or PGC-1β can increase mitochondrial content and the expression of mitochondrial genes, and enhances exercise performance in mice (20). However, the consequence of PGC-1α or PGC-1β knockout alone is subtle. PGC-1α disruption only causes mild to modest mitochondrial dysfunction, a modest decrease in mitochondrial gene expression, and decreased mitochondrial enzymatic activities (32). Similarly, PGC-1β null mice are generally viable and metabolically healthy with decreased mitochondrial gene expression and defects in thermogenesis and cardiac performance (33). These results suggest that PGC-1α and PGC-1β may compensate for each other’s loss in vivo. This was validated by the finding that most of the PGC-1α and PGC-1β double knockout mice died of cardiac failure within 24h and none survived beyond 14 days (34). The expression of many mitochondrial respiratory genes was significantly reduced and mitochondrial volume and cristae density were markedly decreased in the double knockout mice (34).
The expression of PGC-1α is regulated by many other molecules, including members of the TORC family, calmodulin kinase, calcineurin, CREB, AMPK, nitric oxide, SIRT1 and insulin (23). All these factors form a fine-tuned network that is vital for the regulation of mitochondrial biogenesis and function. For example, PGC-1α is phosphorylated and inhibited by AKT in response to insulin increase in liver and heart (35). Mitochondrial dysfunction has been associated with insulin resistance which is also a prevalent phenotype of aging. The binding activity of mtTFA to the D-loop region of mtDNA decreased in diabetic rat heart, and the transcription activity significantly reduced as a consequence (36, 37). The role of PGC-1α in insulin resistance is tissue-specific. PGC-1α level increased in pancreas of obese mouse, and overexpression of PGC-1α induced the defect in glucose regulated insulin secretion (38). On the other hand, PGC-1α level decreased in skeletal muscle in human diabetics, and muscle specific knockout of PGC-1α causes reduced insulin tolerance (39). Moreover, the up-regulation of PGC-1α was reported to regulate the expression of TRB3, an inhibitor of insulin-dependent AKT activation, in liver of diabetes mouse model (40).
Numerous environmental toxins, such as dioxins, polychlorinated biphenyls (PCBs) and others, have been suggested to cause mitochondrial dysfunction and contribute to the aging process and aging-associated diseases (41). It was reported that many persistent organic pollutants (POPs) were detectable in over 60% of population (42, 43). Among five major POPs subclasses, organochlorine (OC) pesticides were associated with metabolic syndrome. Interestingly, different POPs were associated with different types of metabolic syndrome (43). It was reported that hypertension had gender-specific associations with POPs. Polychlorinated dibenzofurans and polychlorinated dibenzo-p-dioxins were associated with prevalent but newly diagnosed hypertension among women, but not among men, whereas PCBs showed positive association with hypertension only among men (42). OC pesticides were not associated with hypertension in either gender (42). In another report, Mokdad et al found that the heavy usage of atrazine, an extensively used herbicide, may increase the risk of obesity (44). The direct toxic effects of these environmental toxins on mitochondria have been noticed. For example, Atrazine can inhibit mitochondrial function and induce insulin resistance in cultured cells and in rats (45). 2,3,7,8-tetrachlorodibenzodioxin (TCDD), one of the most important herbicides and industrial waste products, can inhibit mitochondrial function when injected into mice (46).
5. MITOCHONDRIAL ALTERATIONS DURING AGING
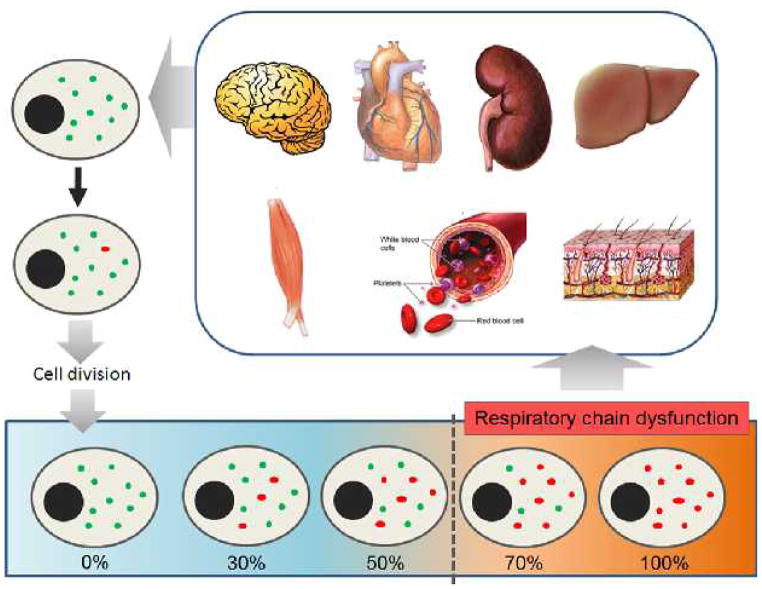
Mitochondrial functional capacity, including transcription, translation, enzyme activity and ATP synthesis, declines with aging. In particular, mtDNA is particularly susceptible to reactive oxygen species (ROS), which are the byproduct of OXPHOS, due to its proximity and less robust protective system compared with that of the nuclear DNA (47). An age-related increase in oxidative damage to mtDNA has been reported in various systems (17). Such damage can generate point mutations, deletions and strand breaks in the mitochondrial genome. The mtDNA is present in multiple copies within each cell, which means mtDNA mutations can be co-existed with wild type molecules in a situation known as heteroplasmy (48). Replication of mtDNA is independent to the cell cycle. A particular mtDNA copy may be replicated many times or not at all as a cell divides (18). This indicates that a single mtDNA mutation can be expanded clonally or be lost when cell divides, and there is continuous mtDNA replication in postmitotic cells (Figure 1). Although the overall level of mtDNA mutations seems relatively low to result in tissue dysfunction, based on the detection of specific mutations in tissue homogenates, investigation of mtDNA mutations in individual cells indicated that particular mtDNA mutations could accumulate during aging to a significant level (49). In fact, respiratory chain dysfunction will occur when heteroplasmic mtDNA mutations reach a certain threshold level, ranging from 50–60% for deleted mtDNA and as high as 90% for some tRNA mutations, varying with the mutation type and the type of affected tissue (48, 50).
Figure 1.
Tissue-specific clonal expansion of mutated mtDNA. As there is no synchronization between cell division and mtDNA replication, a mutated mtDNA (red dots) of different tissue cells may be replicated many times. This will lead to accumulation of mtDNA mutations and impaired respiratory chain function, which in turn cause decline in tissue functions.
Numerous reports recorded the accumulation of mtDNA mutations in different tissues in human aging. Most of these studies only showed the correlation between mtDNA mutations and human aging, but the causality is unclear. The mtDNA-mutator mice, which have a proofreading-deficient version of Polγ and thus show accelerated mtDNA mutation level, displayed a premature aging reminiscent of human aging (51). The accumulation of mtDNA mutations in mutator mice has been associated with alteration of metabolic pattern, such as enhanced glycolysis and lactate production. Interestingly, such changes have also been observed when cells became tumorigenic once a mtDNA mutation was introduced (52–54). Aging-related metabolic changes caused by mtDNA mutations have also been observed in c. elegens (55).
The free radical or mitochondrial theory of aging is one of the best accepted among all proposed explanations for the aging process (56, 57). Key to this theory is the idea that the aging process is driven by a vicious cycle of mitochondrial damage resulting in dysfunction. Briefly, ROS generated during OXPHOS may cause somatic mtDNA mutations, and these mutations may lead to the production of dysfunctional proteins. Dysfunctional proteins may cause defective respiration which in turn generates more ROS. In support of this theory, various genetic alterations in mitochondria have been found to cause phenotypes that resemble premature aging, and there seems a direct link between somatic mtDNA mutations and mammalian aging (58). However, this theory has also been challenged over whether ROS is the proximate cause of aging (57, 59, 60). It is possible that other consequences of mitochondrial DNA alterations, such as deficient ATP production, increased NADH/NAD+ ratios and alterations in Ca2+ homeostasis contribute to the aging process (61).
Another fundamental question about the role of mitochondrial dysfunction in aging is when mtDNA mutations are generated and how they accumulate. However, the accumulation of mtDNA mutations during aging and its effects show a tissue-specific pattern, which may be related to the physiology, bioenergetics and metabolic characteristics of the tissue (62, 63).
5.1. Mitochondrial alterations during aging in the brain
Aging of the mammalian brain is associated with a continuous decrease in ATP production by OXPHOS. Mitochondria from old brains show decreased rates of electron transfer in complexes I and IV, decreased membrane potential, increased content of oxidation products and increased size and fragility (64, 65). There is growing evidence that mitochondrial dysfunction and impairment of the respiratory complexes are associated with neuronal loss, which is a hallmark of human neurodegenerative diseases such as Parkinson’s, Huntingdon’s and Alzheimer’s diseases. Parkinson’s disease is associated with mitochondrial dysfunction and oxidative damage. Reduced complex I dysfunction has been reported in the substantia nigra and frontal cortex in Parkinson’s disease patients (66). Inhibition of complex I in mice and other animals can reproduce the clinical symptoms of Parkinson’s disease (67). Cytochrome c oxidase (COX) deficiency has also been reported in both platelets (68) and post-mortem brain samples from Parkinson’s disease patients (69).
The expression of a subset of mitochondrial genes decreases with age in human brains (70). Accordingly, it has been reported that expression of the PGC-1 family is down-regulated in mammalian aging brains (70, 71). In addition, the activities of SIRT1, calcineurin and AMPK, which are all important regulators of PGC-1 family, are also reduced in aging brains (71). SIRT1 and its homolog have been implicated in longevity in yeast, worms and flies (72). SIRT1 can deacetylate and activate PGC-1α. SIRT1 null mice show similar transcriptional changes to those seen during aging in the brain, and overexpression of SIRT1 can suppress these age-dependent transcriptional changes (73). AMPK serves as a sensor of cellular energy status. AMPK activation can induce transcription of PPARα and PGC-1α, and regulate mitochondrial biogenesis and function (74). Thus, down-regulation of the PGC-1 family and its activators may contribute to mitochondrial transcriptional dysregulation in the process of brain aging.
Dysregulation of PGC-1α is involved in neurodegeneration. In Huntington’s disease patients, expression of PGC-1α is suppressed by a mutant huntingtin. Expression of PGC-1α can provide neuroprotection and thus partially reverse the deleterious effects in huntingtin mutant mice (75). Furthermore, PGC-1α induces the expression of many anti-oxidant genes, including GPx1 and SOD2, and thus protects neural cells from oxidative damages (76).
mtDNA mutations, including point mutations and deletions, have been well-reported to accumulate during aging in brains in different species. The ‘common deletion’ refers to the most frequently observed age-related deletion, spanning 4977 nucleotides in humans, which removes all or parts of the genes for NADH dehydrogenase or complex I subunits 3, 4, 4L, and 5, cytochrome c oxidase or complex IV subunit III and ATP synthase or complex V subunits 6 and 8 (77). In an effort to determine levels of the common deletion in 12 different brain regions, the highest level of accumulation was found in the caudate, putamen and substantia nigra (78), which may be due to increased oxidative stress during the metabolism of dopamine. The lowest level was detected in the white matter and cerebellum. Using a single-molecule PCR approach, up to 60% of mtDNA deletions were found in aged human substantia nigra (79). Further investigation showed a good correlation between high levels of mtDNA deletion and cytochrome c oxidase (COX) deficiency. These results suggest that mtDNA deletions may be directly responsible for impaired cellular respiration and thus play important role in the process of brain aging and its related diseases. Interestingly, Ochoa et al. found that the age-related increase in mtDNA deletion can be prevented by a monounsaturated fatty acid diet (80).
In spite of many studies implicating mtDNA mutations in aging and aging-associated neurodegenerative diseases, few if any causative mutations have been established. The levels of point mutations increased with age in the frontal cortex but remained similar in the substantia nigra (81), while Parkinson’s patients showed no significant differences compared with their age-matched controls in somatic mtDNA mutation levels. Horvath et al. observed an A8344G mutation in the tRNALys gene of the mtDNA in a 66 year old man with dopa-responsive Parkinsonism (82), but this mutation does not exist in any of 159 Italian patients with Parkinson’s disease (83). Several novel mtDNA mutations have been reported in Parkinson’s disease (84), but their pathological significance has not been determined. Nevertheless, it was reported that a heteroplasmic mutation in a specific region of ND5 can largely segregate Parkinson’s disease patients from controls and thus may be of pathogenic importance (50).
While the etiology of Alzheimer’s disease (AD) remains unclear, it is well documented that mitochondrial dysfunction and oxidative stress occur in brains of AD patients (85). The expression of several metabolic enzymes decreased and energy production shifted to ketone body metabolism, and this decline in metabolism occurred years before diagnosis of clinical changes in AD patients (12). Regional reduction of the glucose metabolism rate in cerebral cortex has been related to the clinical disabilities of AD patients and can also precede the clinical symptoms by decades (86). Accordingly, the expression of nDNA encoded mitochondrial electron transport chain subunits is significantly decreased in the posterior cingulate cortex, the middle temporal gyrus, hippocampal CA1 cells, the entorhinal cortex and other brain areas (87). Morphological alterations of neuronal mitochondria such as enlargement, swelling, and disruption of the cristae or osmiophilic inclusions have all been reported (88). Accumulation of the somatic mtDNA common deletion has been reported (89). In the mtDNA control region, AD brains show significant increases in heteroplasmic mtDNA mutations compared with controls. In particular, T414C and T477C mutations are frequent in AD brains (89, 90). These mutations locate in mtDNA control region and may account for the reduction of brain OXPHOS caused by the decrease of mtDNA L-strand ND6 transcript and mtDNA/nDNA ratio in AD patients (90), and thus may be responsible for the reduction of OXPHOS in AD patients. However, other reports failed to observe differences in mtDNA mutation frequency between autopsied AD brain samples and normal controls (91).
5.2. Mitochondrial alterations during aging in muscle
As people age, the strength of skeletal muscles decreases at a rate of 1–2% per year after the age of 50 and will reduced by 30–40% at the age of 70 (4). The loss of strength is accounted by reduced muscle mass, which is known as senile sarcopenia. As the largest oxygen consumer in the body, the aging of skeletal muscle has been well associated with the decline in mitochondrial functions (6). The mtDNA copy number and expression of mitochondrial mRNA decrease with age, and the oxidative capacity of elderly subjects is nearly 50% lower than that of younger subjects (92, 93).
The expression of PGC-1 family members is down-regulated in muscles of type 2 diabetes mellitus patients and in the muscles of denervated mice (13). PGC-1α is preferentially expressed in type I muscle fibers, and its overexpression enhances mitochondrial biogenesis and induces muscle fibers to shift to oxidative type I and IIA fibers through upregulation of MEF2 (30). In contrast, muscle-specific disruption of PGC-1α induces fibers shift to glycolytic fibers with reduced expression of mitochondrial genes (94).
As a master regulator of skeletal muscle metabolism, increased expression of PGC-1α in skeletal muscle protected aging mice from loss of muscle integrity and function, and improved their whole-body health (95). Increased PGC-1α can also postpone the onset of myopathy and rescue COX deficiency in mouse models (96, 97). Furthermore, increased expression of PGC-1 coactivators can also protect skeletal muscle from protein degradation and atrophy (13).
Accumulation of mitochondrial DNA deletions has long been observed during aging in skeletal muscle of several species including mice, rhesus monkeys and humans (98, 99). In human vastus lateralis muscle, the COX-deficient muscle fibers increased from 6% at age 49 to 31% at age 93, and each COX-deficient fiber harbored a clonal mtDNA deletion (100). Similarly, an investigation on 46 human extraocular muscle samples aged 3–96 years indicated that the COX-negative fibers first occurred at the age of 30, after which there was a significant age-related increase. Most COX-negative fibers harbored high levels (>70%) of mtDNA deletions (101).
The tRNALys A8344G and tRNALeu A3243G mutations are among the earliest identified pathogenic mitochondrial mutations (102). A comprehensive study on isolated COX-deficient muscle fibers from old individuals has identified relatively high levels of mutations including A3243G and A8344G in several mt-tRNA genes. Their focal accumulation is associated with cytochrome c oxidase deficient and may cause significant mitochondrial dysfunction in individual muscle cells (103). Meanwhile, Cybrid cells harboring either A3243G or A3243G mutation are more susceptible to apoptosis when treated with various apoptotic inducers (104).
To study the consequences of accumulated muscle-specific mtDNA deletions and mutations, we transferred muscle mtDNA from young and old mice to mtDNA-less (ρ0) cells by cytoplast-cell fusion, and generated mouse cybrids carrying skeletal muscle mtDNA. We found a significant decrease in oxidative phosphorylation coupling and regulation capacity in cybrids carrying mtDNA from skeletal muscle of old mice compared to muscle mtDNA from young mice (105). These findings indicate that mitochondrial dysfunction caused by mtDNA mutations leads to decreases in both the capacity and regulation of OXPHOS.
5.3. Mitochondrial alterations during aging in fibroblast
Mitochondrial dysfunction has been well defined in aging fibroblasts, including increases in oxidative damage and lipid peroxidation, and decreases in respiratory rate and mtDNA content (106). A comprehensive investigation of mitochondrial biochemical and bioenergetics properties of fibroblasts derived from 55 individuals ranging in age from 20 wk fetal stage to 103 years old revealed significant decreases in mitochondrial protein synthesis rate, respiration rate and OXPHOS capacity during aging. Interestingly, skin fibroblasts from individuals older than 90 years exhibited an exceptionally high respiration rate, which may be a consequence of a compensatory effect (106).
Overexpression of PGC-1α in human 3T3 fibroblast cells can induce the expression of mtTFA and cytochrome c and increase mtDNA copy number, which are all hallmarks of mitochondrial biogenesis (107). Meanwhile, increased PGC-1α levels also confer on fibroblasts higher ATP production levels and resistance to oxidative damage (107).
An age-related increase in mtDNA deletion levels was found in human skin fibroblasts from fetal, young and old donors (108). In another study, it was found that photoaged human skin contains increased amounts of large-scale mtDNA deletions, and repetitive sublethal exposure to UVA radiation induces mtDNA mutations in dermal fibroblasts (109).
mtDNA point mutations in the control region increase during aging in skin fibroblasts, with several specific mutations occurring at high levels (110). Among them, a frequency of up to 50% of a T414G transversion, which lies in the middle of the promoter for primer synthesis for H-strand replication and L-strand transcription, was found in the skin fibroblasts of individuals over 65 (110). Moreover, this mutation was present at high levels in sun-exposed skin, suggesting it might be related to increased oxidative stress (111). However, it has also been reported that T414G mutation does not directly correlate with alteration in mtDNA copy number, lipofuscin and ROS production in five separate fibroblast cultures from elderly individuals. (112).
5.4. Mitochondrial alterations during aging in the liver
The liver plays important roles in integrating energy metabolism, detoxification and immunity. Although the majority of liver functions seem to be maintained relatively well during aging, there is a dramatic age-associated loss of its regenerative capacity and drug metabolism (10, 11). Mitochondrial dysfunction associated with aging in liver includes a decline in OXPHOS capacity, decreased expression of electron transport chain complexes, and accumulation of oxidative damage products (47, 113).
The expression of PGC-1α is relatively low in liver (114). Rosiglitazone, a ligand of PPARγ that is capable of activating PGC-1α, can attenuate age-associated liver pathology in a nonalcoholic steatohepatitis mouse model (115). Progressive age-related damage specific to mtDNA was reported in a reported in a quantitative investigation on the levels of 8-oxo-dG, an indicator of oxidative damage to DNA, in mtDNA and nDNA derived from rat livers at different ages (116). Moreover, an increase with age in the incidence of large mtDNA deletions has been observed in human liver mtDNA (117). Interestingly, it was also observed that the mtDNA deletion levels in liver mitochondria from rats fed with sunflower oil (rich in polyunsaturated linoleic acid) were higher than those from mice fed with virgin olive oil (rich in the monounsaturated oleic acid) (118).
Furthermore, the mtDNA point mutation load was also reported to increase with age in liver (119). Particularly, the pathogenic A3243G mutation, which is associated with cytochrome c oxidase deficient was found in human liver and its content increased with age (120).
5.5. Mitochondrial alterations during aging in the blood cells
An early electron microscopic study showed that mitochondrial damage in human peripheral lymphocytes increased from the age of 50 years to 80 years, but declined after 80 years (121). A significant decrease in complex II + III activity was reported with aging in blood (122).
The mtDNA common deletion has been detected in blood mtDNA. Li et al. reported an accumulation of the common deletion as a function of age in blood samples from 110 healthy individuals (123). However, others found no such accumulation (124, 125). Another report claimed that the individual common deletion load was much lower in nonagenarians and centenarians compared with other age groups, although the proportions of the common deletion carrier were very similar (126).
6. PERSPECTIVE
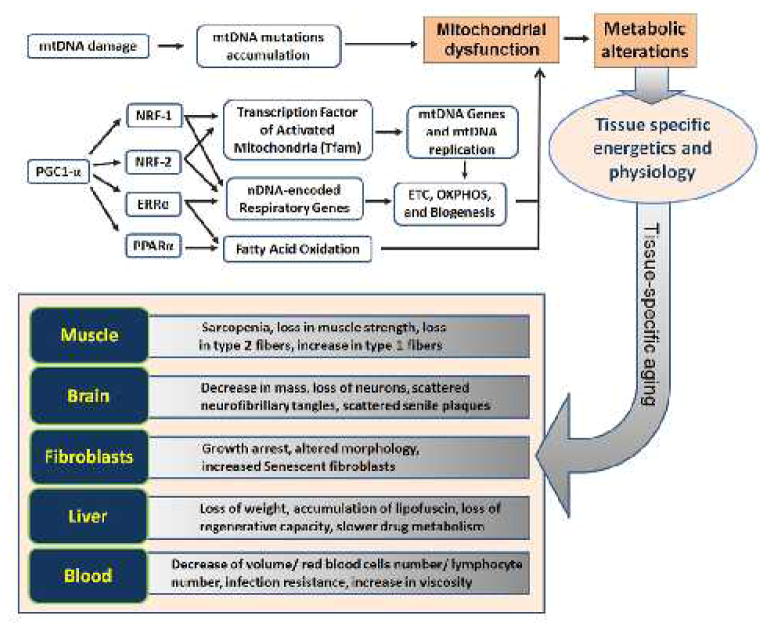
As people age, physiological changes occur in all tissues at varying rates. Most noticeably, alteration in body composition, energy production/utilization imbalance, neurodegeneration and homeostatic dysregulation are among the hallmarks of aging. To greater or less extents, mitochondrial dysfunction is involved in all of these processes. As discussed in above sections, both mtDNA mutation and defects in nuclear control of mitochondrial biogenesis and function have been implication in mitochondrial dysfunction. The altered metabolism resulted from mitochondrial dysfunction could further manifest into tissue-specific phenotypes under different bioenergetical and physiological conditions during aging (Figure 2).
Figure 2.
The mtDNA mutations accumulation and transcriptional dysregulation both contribute to the process of tissue-specific aging. As there is no synchronization between cell division and mtDNA replication, a mutated mtDNA of different tissue cells may be replicated many times and lead to accumulation of mtDNA mutations and impaired respiratory chain function. On the other side, there is a finely-tuned transcriptional network regulating mitochondrial biogenesis and function. PGC-1α can coactivate several important transcriptional factors such as NRF-1/-2, ERRα and PPARα. These factors play vital role in mitochondrial biogenesis and function. As the expression and regulation of these factors and the mtDNA mutation accumulation are all tissue-specific, there are tissue-specific physiological consequences when people age.
Over the past two decades, enormous attention has been focused on mutations in mitochondrial genomes during aging in various organisms. Though still controversial, a tremendous amount of useful information has been generated. With the increasing understanding about regulation of mitochondrial biogenesis and function, and the availability of new technology and animal models involving nDNA encoded factors regulating mitochondrial biogenesis and function, we can now investigate the implication of mitochondrial dysfunction in different tissues during aging within a new context.
At the same time, the emerging fields of mitochondrial proteomics and mitochondrial metabolomics provide an opportunity to integrate the information on mtDNA to energetics and physiology. As a result, we should expect to identify better tissue-specific biomarkers for aging. Finally, last but certainly not least, we are entering a stage when targeting mitochondrial dysfunction with the purpose of either curing age-related diseases or mitigating the aging process itself will attract more and more attention. Approaches that build on preventing oxidative damage, enhancing mitochondrial biogenesis and function, eliminating dysfunctional mitochondrial components or the mitochondrion itself, and possibly other novel ideas, will be extensively explored.
Acknowledgments
The work in the laboratory of the authors has been supported by Morrison Trust and grants from National Science Foundation of China (31070765/C050605) and Zhejiang provincial natural science foundation of China (Y2090378).
Abbreviations
- OXPHOS
oxidative phosphorylation
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
- NRF
nuclear respiratory factor
- PGC-1α
PPARγ coactivator-1α
- SIRT1
Silent Information Regulator
- AD
Alzheimer’s disease
References
- 1.Boss GR, Seegmiller JE. Age-related physiological changes and their clinical significance. West J Med. 1981;135(6):434–40. [PMC free article] [PubMed] [Google Scholar]
- 2.Anderton BH. Ageing of the brain. Mech Ageing Dev. 2002;123(7):811–7. doi: 10.1016/s0047-6374(01)00426-2. [DOI] [PubMed] [Google Scholar]
- 3.Anderton BH. Changes in the ageing brain in health and disease. Philos Trans R Soc Lond B Biol Sci. 1997;352(1363):1781–92. doi: 10.1098/rstb.1997.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes VA, Frontera WR, Roubenoff R, Evans WJ, Singh MA. Longitudinal changes in body composition in older men and women: role of body weight change and physical activity. Am J Clin Nutr. 2002;76(2):473–81. doi: 10.1093/ajcn/76.2.473. [DOI] [PubMed] [Google Scholar]
- 5.Nair KS. Aging muscle. Am J Clin Nutr. 2005;81(5):953–63. doi: 10.1093/ajcn/81.5.953. [DOI] [PubMed] [Google Scholar]
- 6.Rossi P, Marzani B, Giardina S, Negro M, Marzatico F. Human skeletal muscle aging and the oxidative system: cellular events. Curr Aging Sci. 2008;1(3):182–91. doi: 10.2174/1874609810801030182. [DOI] [PubMed] [Google Scholar]
- 7.Macieira-Coelho A, Diatloff C, Malaise E. Concept of fibroblast aging in vitro: implications for cell biology. Gerontology. 1977;23(4):290–305. doi: 10.1159/000212199. [DOI] [PubMed] [Google Scholar]
- 8.Martin GM, Sprague CA, Epstein CJ. Replicative life-span of cultivated human cells. Effects of donor’s age, tissue, and genotype. Lab Invest. 1970;23(1):86–92. [PubMed] [Google Scholar]
- 9.Makrantonaki E, Zouboulis CC. Molecular mechanisms of skin aging: state of the art. Ann N Y Acad Sci. 2007;1119:40–50. doi: 10.1196/annals.1404.027. [DOI] [PubMed] [Google Scholar]
- 10.Timchenko NA. Aging and liver regeneration. Trends Endocrinol Metab. 2009;20(4):171–6. doi: 10.1016/j.tem.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Anantharaju A, Feller A, Chedid A. Aging Liver. A review. Gerontology. 2002;48(6):343–53. doi: 10.1159/000065506. [DOI] [PubMed] [Google Scholar]
- 12.Costantini LC, Barr LJ, Vogel JL, Henderson ST. Hypometabolism as a therapeutic target in Alzheimer’s disease. BMC Neurosci. 2008;9(Suppl 2):S16. doi: 10.1186/1471-2202-9-S2-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillon LM, Rebelo AP, Moraes CT. The role of PGC-1 coactivators in aging skeletal muscle and heart. IUBMB Life. 2012;64(3):231–41. doi: 10.1002/iub.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hultman E, Chasiotis D, Sjoholm H. Energy metabolism in muscle. Prog Clin Biol Res. 1983;136:257–72. [PubMed] [Google Scholar]
- 15.Soboll S. Regulation of energy metabolism in liver. J Bioenerg Biomembr. 1995;27(6):571–82. doi: 10.1007/BF02111655. [DOI] [PubMed] [Google Scholar]
- 16.Wallace DC. Mitochondrial DNA mutations in diseases of energy metabolism. J Bioenerg Biomembr. 1994;26(3):241–50. doi: 10.1007/BF00763096. [DOI] [PubMed] [Google Scholar]
- 17.Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Oxidative stress, mitochondrial bioenergetics, and cardiolipin in aging. Free Radic Biol Med. 2010;48(10):1286–95. doi: 10.1016/j.freeradbiomed.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Park CB, Larsson NG. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193(5):809–18. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebelo AP, Dillon LM, Moraes CT. Mitochondrial DNA transcription regulation and nucleoid organization. J Inherit Metab Dis. 2011;34(4):941–51. doi: 10.1007/s10545-011-9330-8. [DOI] [PubMed] [Google Scholar]
- 20.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 21.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–40. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 22.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88(2):611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 23.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813(7):1269–78. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JS, Huang PH, Wang CH, Lin FY, Tsai HY, Wu TC, Lin SJ, Chen JW. Nrf-2 mediated heme oxygenase-1 expression, an antioxidant-independent mechanism, contributes to anti-atherogenesis and vascular protective effects of Ginkgo biloba extract. Atherosclerosis. 2011;214(2):301–9. doi: 10.1016/j.atherosclerosis.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, Choi YB, Lee AH, Lee KH, Park C, Obin MS, Park SK, Seo YJ, Oh GT, Lee HW, Shin J. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13(2):150–6. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010;244(1):16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 28.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18(4):357–68. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 29.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277(3):1645–8. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 30.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 31.Andersson U, Scarpulla RC. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21(11):3738–49. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci U S A. 2006;103(26):10086–91. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, Zhang D, Litwin SE, Zaha VG, Fountain KT, Boudina S, Jimenez-Linan M, Blount M, Lopez M, Meirhaeghe A, Bohlooly YM, Storlien L, Stromstedt M, Snaith M, Oresic M, Abel ED, Cannon B, Vidal-Puig A. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4(11):e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22(14):1948–61. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson R, Prolla T. PGC-1alpha in aging and anti-aging interventions. Biochim Biophys Acta. 2009;1790(10):1059–66. doi: 10.1016/j.bbagen.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanazawa A, Nishio Y, Kashiwagi A, Inagaki H, Kikkawa R, Horiike K. Reduced activity of mtTFA decreases the transcription in mitochondria isolated from diabetic rat heart. Am J Physiol Endocrinol Metab. 2002;282(4):E778–85. doi: 10.1152/ajpendo.00255.2001. [DOI] [PubMed] [Google Scholar]
- 37.Nishio Y, Kanazawa A, Nagai Y, Inagaki H, Kashiwagi A. Regulation and role of the mitochondrial transcription factor in the diabetic rat heart. Ann N Y Acad Sci. 2004;1011:78–85. doi: 10.1007/978-3-662-41088-2_9. [DOI] [PubMed] [Google Scholar]
- 38.Yoon JC, Xu G, Deeney JT, Yang SN, Rhee J, Puigserver P, Levens AR, Yang R, Zhang CY, Lowell BB, Berggren PO, Newgard CB, Bonner-Weir S, Weir G, Spiegelman BM. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev Cell. 2003;5(1):73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 39.Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117(11):3463–74. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10(5):530–4. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 41.Lee HK, Cho YM, Kwak SH, Lim S, Park KS, Shim EB. Mitochondrial dysfunction and metabolic syndrome-looking for environmental factors. Biochim Biophys Acta. 2010;1800(3):282–9. doi: 10.1016/j.bbagen.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Ha MH, Lee DH, Son HK, Park SK, Jacobs DR., Jr Association between serum concentrations of persistent organic pollutants and prevalence of newly diagnosed hypertension: results from the National Health and Nutrition Examination Survey 1999–2002. J Hum Hypertens. 2009;23(4):274–86. doi: 10.1038/jhh.2008.124. [DOI] [PubMed] [Google Scholar]
- 43.Lee DH, Lee IK, Porta M, Steffes M, Jacobs DR., Jr Relationship between serum concentrations of persistent organic pollutants and the prevalence of metabolic syndrome among non-diabetic adults: results from the National Health and Nutrition Examination Survey 1999–2002. Diabetologia. 2007;50(9):1841–51. doi: 10.1007/s00125-007-0755-4. [DOI] [PubMed] [Google Scholar]
- 44.Mokdad AH, Bowman BA, Ford ES, Vinicor F, Marks JS, Koplan JP. The continuing epidemics of obesity and diabetes in the United States. JAMA. 2001;286(10):1195–200. doi: 10.1001/jama.286.10.1195. [DOI] [PubMed] [Google Scholar]
- 45.Lim S, Ahn SY, Song IC, Chung MH, Jang HC, Park KS, Lee KU, Pak YK, Lee HK. Chronic exposure to the herbicide, atrazine, causes mitochondrial dysfunction and insulin resistance. PLoS One. 2009;4(4):e5186. doi: 10.1371/journal.pone.0005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shertzer HG, Genter MB, Shen D, Nebert DW, Chen Y, Dalton TP. TCDD decreases ATP levels and increases reactive oxygen production through changes in mitochondrial F(0)F(1)-ATP synthase and ubiquinone. Toxicol Appl Pharmacol. 2006;217(3):363–74. doi: 10.1016/j.taap.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 48.Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–78. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 49.Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci U S A. 2002;99(8):5521–6. doi: 10.1073/pnas.072670199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker WD, Jr, Parks JK. Mitochondrial ND5 mutations in idiopathic Parkinson’s disease. Biochem Biophys Res Commun. 2005;326(3):667–9. doi: 10.1016/j.bbrc.2004.11.093. [DOI] [PubMed] [Google Scholar]
- 51.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19(7):802–15. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kukat A, Edgar D, Bratic I, Maiti P, Trifunovic A. Random mtDNA mutations modulate proliferation capacity in mouse embryonic fibroblasts. Biochem Biophys Res Commun. 2011;409(3):394–9. doi: 10.1016/j.bbrc.2011.04.145. [DOI] [PubMed] [Google Scholar]
- 54.Sharma LK, Fang H, Liu J, Vartak R, Deng J, Bai Y. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20(23):4605–16. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler JA, Ventura N, Johnson TE, Rea SL. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB J. 2010;24(12):4977–88. doi: 10.1096/fj.10-162941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20(4):145–7. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 57.Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2010;67(1):1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23(15):1714–36. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–4. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 60.Hutter E, Skovbro M, Lener B, Prats C, Rabol R, Dela F, Jansen-Durr P. Oxidative stress and mitochondrial impairment can be separated from lipofuscin accumulation in aged human skeletal muscle. Aging Cell. 2007;6(2):245–56. doi: 10.1111/j.1474-9726.2007.00282.x. [DOI] [PubMed] [Google Scholar]
- 61.Hebert SL, Lanza IR, Nair KS. Mitochondrial DNA alterations and reduced mitochondrial function in aging. Mech Ageing Dev. 2010;131(7–8):451–62. doi: 10.1016/j.mad.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu VW, Zhang C, Nagley P. Mutations in mitochondrial DNA accumulate differentially in three different human tissues during ageing. Nucleic Acids Res. 1998;26(5):1268–75. doi: 10.1093/nar/26.5.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McInerny SC, Brown AL, Smith DW. Region-specific changes in mitochondrial D-loop in aged rat CNS. Mech Ageing Dev. 2009;130(5):343–9. doi: 10.1016/j.mad.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 64.Navarro A, Boveris A. Brain mitochondrial dysfunction in aging: conditions that improve survival, neurological performance and mitochondrial function. Front Biosci. 2007;12:1154–63. doi: 10.2741/2133. [DOI] [PubMed] [Google Scholar]
- 65.Navarro A, Boveris A. Brain mitochondrial dysfunction in aging, neurodegeneration, and Parkinson’s disease. Front Aging Neurosci. 2010;2 doi: 10.3389/fnagi.2010.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Navarro A, Boveris A. Brain mitochondrial dysfunction and oxidative damage in Parkinson’s disease. J Bioenerg Biomembr. 2009;41(6):517–21. doi: 10.1007/s10863-009-9250-6. [DOI] [PubMed] [Google Scholar]
- 67.Gomez C, Bandez MJ, Navarro A. Pesticides and impairment of mitochondrial function in relation with the parkinsonian syndrome. Front Biosci. 2007;12:1079–93. doi: 10.2741/2128. [DOI] [PubMed] [Google Scholar]
- 68.Cardoso SM, Proenca MT, Santos S, Santana I, Oliveira CR. Cytochrome c oxidase is decreased in Alzheimer’s disease platelets. Neurobiol Aging. 2004;25(1):105–10. doi: 10.1016/s0197-4580(03)00033-2. [DOI] [PubMed] [Google Scholar]
- 69.Kish SJ, Bergeron C, Rajput A, Dozic S, Mastrogiacomo F, Chang LJ, Wilson JM, DiStefano LM, Nobrega JN. Brain cytochrome oxidase in Alzheimer’s disease. J Neurochem. 1992;59(2):776–9. doi: 10.1111/j.1471-4159.1992.tb09439.x. [DOI] [PubMed] [Google Scholar]
- 70.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429(6994):883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 71.Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res Rev. 2009;8(3):173–88. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chong ZZ, Shang YC, Wang S, Maiese K. SIRT1: new avenues of discovery for disorders of oxidative stress. Expert Opin Ther Targets. 2012;16(2):167–78. doi: 10.1517/14728222.2012.648926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135(5):907–18. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu F, Benashski SE, Persky R, Xu Y, Li J, McCullough LD. Age-related changes in AMP-activated protein kinase after stroke. Age (Dordr) 2012;34(1):157–68. doi: 10.1007/s11357-011-9214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127(1):59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 76.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 77.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331(6158):717–9. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 78.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992;2(4):318–23. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 79.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–20. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 80.Ochoa JJ, Pamplona R, Ramirez-Tortosa MC, Granados-Principal S, Perez-Lopez P, Naudi A, Portero-Otin M, Lopez-Frias M, Battino M, Quiles JL. Age-related changes in brain mitochondrial DNA deletion and oxidative stress are differentially modulated by dietary fat type and coenzyme Q. Free Radic Biol Med. 2011;50(9):1053–64. doi: 10.1016/j.freeradbiomed.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 81.Simon DK, Lin MT, Zheng L, Liu GJ, Ahn CH, Kim LM, Mauck WM, Twu F, Beal MF, Johns DR. Somatic mitochondrial DNA mutations in cortex and substantia nigra in aging and Parkinson’s disease. Neurobiol Aging. 2004;25(1):71–81. doi: 10.1016/s0197-4580(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 82.Horvath R, Kley RA, Lochmuller H, Vorgerd M. Parkinson syndrome, neuropathy, and myopathy caused by the mutation A8344G (MERRF)in tRNALys. Neurology. 2007;68(1):56–8. doi: 10.1212/01.wnl.0000250334.48038.7a. [DOI] [PubMed] [Google Scholar]
- 83.Mancuso M, Nesti C, Petrozzi L, Orsucci D, Frosini D, Kiferle L, Bonuccelli U, Ceravolo R, Murri L, Siciliano G. The mtDNA A8344G “MERRF” mutation is not a common cause of sporadic Parkinson disease in Italian population. Parkinsonism Relat Disord. 2008;14(4):381–2. doi: 10.1016/j.parkreldis.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 84.Richter G, Sonnenschein A, Grunewald T, Reichmann H, Janetzky B. Novel mitochondrial DNA mutations in Parkinson’s disease. J Neural Transm. 2002;109(5–6):721–9. doi: 10.1007/s007020200060. [DOI] [PubMed] [Google Scholar]
- 85.Mancuso M, Coppede F, Migliore L, Siciliano G, Murri L. Mitochondrial dysfunction, oxidative stress and neurodegeneration. J Alzheimers Dis. 2006;10(1):59–73. doi: 10.3233/jad-2006-10110. [DOI] [PubMed] [Google Scholar]
- 86.Azari NP, Pettigrew KD, Schapiro MB, Haxby JV, Grady CL, Pietrini P, Salerno JA, Heston LL, Rapoport SI, Horwitz B. Early detection of Alzheimer’s disease: a statistical approach using positron emission tomographic data. J Cereb Blood Flow Metab. 1993;13(3):438–47. doi: 10.1038/jcbfm.1993.58. [DOI] [PubMed] [Google Scholar]
- 87.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105(11):4441–6. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trimmer PA, Borland MK. Differentiated Alzheimer’s disease transmitochondrial cybrid cell lines exhibit reduced organelle movement. Antioxid Redox Signal. 2005;7(9–10):1101–9. doi: 10.1089/ars.2005.7.1101. [DOI] [PubMed] [Google Scholar]
- 89.Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, Head E, Cotman CW, Wallace DC. Systemic mitochondrial dysfunction and the etiology of Alzheimer’s disease and down syndrome dementia. J Alzheimers Dis. 2010;20(Suppl 2):S293–310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101(29):10726–31. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Elson JL, Herrnstadt C, Preston G, Thal L, Morris CM, Edwardson JA, Beal MF, Turnbull DM, Howell N. Does the mitochondrial genome play a role in the etiology of Alzheimer’s disease? Hum Genet. 2006;119(3):241–54. doi: 10.1007/s00439-005-0123-8. [DOI] [PubMed] [Google Scholar]
- 92.Conley KE, Jubrias SA, Esselman PC. Oxidative capacity and ageing in human muscle. J Physiol. 2000;526(Pt 1):203–10. doi: 10.1111/j.1469-7793.2000.t01-1-00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci U S A. 2005;102(15):5618–23. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282(41):30014–21. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 95.Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A. 2009;106(48):20405–10. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Wenz T, Diaz F, Spiegelman BM, Moraes CT. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8(3):249–56. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, Schon EA, Lamperti C, Zeviani M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab. 2011;14(1):80–90. doi: 10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee CM, Chung SS, Kaczkowski JM, Weindruch R, Aiken JM. Multiple mitochondrial DNA deletions associated with age in skeletal muscle of rhesus monkeys. J Gerontol. 1993;48(6):B201–5. doi: 10.1093/geronj/48.6.b201. [DOI] [PubMed] [Google Scholar]
- 99.Chung SS, Weindruch R, Schwarze SR, McKenzie DI, Aiken JM. Multiple age-associated mitochondrial DNA deletions in skeletal muscle of mice. Aging (Milano) 1994;6(3):193–200. doi: 10.1007/BF03324239. [DOI] [PubMed] [Google Scholar]
- 100.Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, Oehmichen M. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol. 2008;43(7):645–52. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 101.Yu-Wai-Man P, Lai-Cheong J, Borthwick GM, He L, Taylor GA, Greaves LC, Taylor RW, Griffiths PG, Turnbull DM. Somatic mitochondrial DNA deletions accumulate to high levels in aging human extraocular muscles. Invest Ophthalmol Vis Sci. 2010;51(7):3347–53. doi: 10.1167/iovs.09-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kadenbach B, Munscher C, Frank V, Muller-Hocker J, Napiwotzki J. Human aging is associated with stochastic somatic mutations of mitochondrial DNA. Mutat Res. 1995;338(1–6):161–72. doi: 10.1016/0921-8734(95)00021-w. [DOI] [PubMed] [Google Scholar]
- 103.Fayet G, Jansson M, Sternberg D, Moslemi AR, Blondy P, Lombes A, Fardeau M, Oldfors A. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul Disord. 2002;12(5):484–93. doi: 10.1016/s0960-8966(01)00332-7. [DOI] [PubMed] [Google Scholar]
- 104.Rommelaere G, Michel S, Malaisse J, Charlier S, Arnould T, Renard P. Hypersensitivity of A8344G MERRF mutated cybrid cells to staurosporine-induced cell death is mediated by calcium-dependent activation of calpains. Int J Biochem Cell Biol. 2012;44(1):139–49. doi: 10.1016/j.biocel.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 105.Li Y, Li HZ, Hu P, Deng J, Banoei MM, Sharma LK, Bai Y. Generation and bioenergetic analysis of cybrids containing mitochondrial DNA from mouse skeletal muscle during aging. Nucleic Acids Res. 2010;38(6):1913–21. doi: 10.1093/nar/gkp1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Greco M, Villani G, Mazzucchelli F, Bresolin N, Papa S, Attardi G. Marked aging-related decline in efficiency of oxidative phosphorylation in human skin fibroblasts. FASEB J. 2003;17(12):1706–8. doi: 10.1096/fj.02-1009fje. [DOI] [PubMed] [Google Scholar]
- 107.Liang H, Bai Y, Li Y, Richardson A, Ward WF. PGC-1alpha-induced mitochondrial alterations in 3T3 fibroblast cells. Ann N Y Acad Sci. 2007;1100:264–79. doi: 10.1196/annals.1395.028. [DOI] [PubMed] [Google Scholar]
- 108.Gerhard GS, Benko FA, Allen RG, Tresini M, Kalbach A, Cristofalo VJ, Gocke CD. Mitochondrial DNA mutation analysis in human skin fibroblasts from fetal, young, and old donors. Mech Ageing Dev. 2002;123(2–3):155–66. doi: 10.1016/s0047-6374(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 109.Schroeder P, Gremmel T, Berneburg M, Krutmann J. Partial depletion of mitochondrial DNA from human skin fibroblasts induces a gene expression profile reminiscent of photoaged skin. J Invest Dermatol. 2008;128(9):2297–303. doi: 10.1038/jid.2008.57. [DOI] [PubMed] [Google Scholar]
- 110.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286(5440):774–9. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 111.Birket MJ, Birch-Machin MA. Ultraviolet radiation exposure accelerates the accumulation of the aging-dependent T414G mitochondrial DNA mutation in human skin. Aging Cell. 2007;6(4):557–64. doi: 10.1111/j.1474-9726.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 112.Birket MJ, Passos JF, von Zglinicki T, Birch-Machin MA. The relationship between the aging- and photo-dependent T414G mitochondrial DNA mutation with cellular senescence and reactive oxygen species production in cultured skin fibroblasts. J Invest Dermatol. 2009;129(6):1361–6. doi: 10.1038/jid.2008.373. [DOI] [PubMed] [Google Scholar]
- 113.Muller-Hocker J, Aust D, Rohrbach H, Napiwotzky J, Reith A, Link TA, Seibel P, Holzel D, Kadenbach B. Defects of the respiratory chain in the normal human liver and in cirrhosis during aging. Hepatology. 1997;26(3):709–19. doi: 10.1002/hep.510260324. [DOI] [PubMed] [Google Scholar]
- 114.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 115.Gupte AA, Liu JZ, Ren Y, Minze LJ, Wiles JR, Collins AR, Lyon CJ, Pratico D, Finegold MJ, Wong ST, Webb P, Baxter JD, Moore DD, Hsueh WA. Rosiglitazone attenuates age- and diet-associated nonalcoholic steatohepatitis in male low-density lipoprotein receptor knockout mice. Hepatology. 2010;52(6):2001–11. doi: 10.1002/hep.23941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85(17):6465–7. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yen TC, Su JH, King KL, Wei YH. Ageing-associated 5 kb deletion in human liver mitochondrial DNA. Biochem Biophys Res Commun. 1991;178(1):124–31. doi: 10.1016/0006-291x(91)91788-e. [DOI] [PubMed] [Google Scholar]
- 118.Quiles JL, Ochoa JJ, Ramirez-Tortosa MC, Huertas JR, Mataix J. Age-related mitochondrial DNA deletion in rat liver depends on dietary fat unsaturation. J Gerontol A Biol Sci Med Sci. 2006;61(2):107–14. doi: 10.1093/gerona/61.2.107. [DOI] [PubMed] [Google Scholar]
- 119.Rotskaya UN, Rogozin IB, Vasyunina EA, Malyarchuk BA, Nevinsky GA, Sinitsyna OI. High frequency of somatic mutations in rat liver mitochondrial DNA. Mutat Res. 2010;685(1–2):97–102. doi: 10.1016/j.mrfmmm.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 120.Liu VW, Zhang C, Linnane AW, Nagley P. Quantitative allele-specific PCR: demonstration of age-associated accumulation in human tissues of the A-->G mutation at nucleotide 3243 in mitochondrial DNA. Hum Mutat. 1997;9(3):265–71. doi: 10.1002/(SICI)1098-1004(1997)9:3<265::AID-HUMU8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 121.Beregi E, Regius O. Relationship of mitochondrial damage in human lymphocytes and age. Aktuelle Gerontol. 1983;13(6):226–8. [PubMed] [Google Scholar]
- 122.Drouet M, Lauthier F, Charmes JP, Sauvage P, Ratinaud MH. Age-associated changes in mitochondrial parameters on peripheral human lymphocytes. Exp Gerontol. 1999;34(7):843–52. doi: 10.1016/s0531-5565(99)00058-3. [DOI] [PubMed] [Google Scholar]
- 123.Li CT, Li L, Liu Y. Relationship between human age and mtDNA 4977 deletions in human blood cells. Fa Yi Xue Za Zhi. 2006;22(5):346–8. [PubMed] [Google Scholar]
- 124.Meissner C, Mohamed SA, Klueter H, Hamann K, von Wurmb N, Oehmichen M. Quantification of mitochondrial DNA in human blood cells using an automated detection system. Forensic Sci Int. 2000;113(1–3):109–12. doi: 10.1016/s0379-0738(00)00249-8. [DOI] [PubMed] [Google Scholar]
- 125.Ross OA, Hyland P, Curran MD, McIlhatton BP, Wikby A, Johansson B, Tompa A, Pawelec G, Barnett CR, Middleton D, Barnett YA. Mitochondrial DNA damage in lymphocytes. a role in immunosenescence? Exp Gerontol. 2002;37(2–3):329–40. doi: 10.1016/s0531-5565(01)00200-5. [DOI] [PubMed] [Google Scholar]
- 126.von Wurmb-Schwark N, Schwark T, Caliebe A, Drenske C, Nikolaus S, Schreiber S, Nebel A. Low level of the mtDNA(4977) deletion in blood of exceptionally old individuals. Mech Ageing Dev. 2010;131(3):179–84. doi: 10.1016/j.mad.2010.01.005. [DOI] [PubMed] [Google Scholar]