Abstract
If fully stretched out, a typical bacterial chromosome would be nearly one millimeter long, or approximately 1000 times the length of a cell. Not only must cells massively compact their genetic material, but they must also organize their DNA in a manner that is compatible with a range of cellular processes, including DNA replication, DNA repair, homologous recombination, and horizontal gene transfer. Recent work, driven in part by technological advances, has begun to reveal the general principles of chromosome organization in bacteria. Here, drawing on studies of many different organisms, we review the emerging picture of how bacterial chromosomes are structured at multiple length-scales, highlighting the functions of various DNA-binding proteins and impact of physical forces. Additionally, we discuss the spatial dynamics of chromosomes, particularly during their segregation to daughter cells. Although there has been tremendous progress, we also highlight gaps that remain in understanding chromosome organization and segregation.
Keywords: macrodomains, supercoiling, transcription, ParA-ParB-parS, nucleoid-associated proteins, Hi-C
Introduction
The chromosomes of all organisms must be compacted nearly three orders of magnitude to fit within cells. Moreover, DNA must be packaged in a way that is compatible with a myriad of DNA-based processes including replication, transcription, repair, recombination, and integration. This challenge is particularly acute in bacteria as chromosome segregation occurs concomitantly with DNA replication, rather than being separated temporally as in eukaryotes. Efforts to understand the structure and organization of bacterial chromosomes have been greatly enhanced in recent years with major technical developments and innovations, including microscopy-based methods for accurately probing the spatial and temporal dynamics of individual DNA loci and genomic methods for investigating the global conformation and folding properties of chromosomes. These new techniques, in combination with the tried-and-true approaches of genetics, biochemistry, biophysics, and cell biology, have begun to reveal the remarkable mechanisms used by bacterial cells to compact, organize, and segregate their chromosomes. Here, we review these mechanisms and the organizing principles of chromosomes in a top-down manner, from the micron to nanometer scale, before discussing recent work on understanding chromosome segregation.
Bacterial Chromosome Compaction and Organization
Global organization
Bacterial chromosomes were originally thought to fit randomly within cells with no stereotypical or reproducible organization. This assumption was initially dispelled by light microscopy studies of Escherichia coli cells stained with DNA-specific dyes which revealed a discrete body of DNA named the nucleoid (reviewed in Robinow & Kellenberger 1994). Early electron microscopy (EM) and subsequent cryo-EM images of vitreous sections of E. coli suggested that the nucleoid forms an irregular structure with extensions projecting into the cytoplasm. In rich growth media, the E. coli nucleoid occupies about half of the cytoplasmic area and seems to exclude most ribosomes. This overall arrangement was seen in living E. coli and B. subtilis cells using fluorescently-tagged nucleoid-associated proteins (NAPs), which bind nonspecifically to DNA, and ribosome subunits (Azam et al. 2000, Bakshi et al. 2012, Lewis et al. 2000). The separation of chromosomes from the bulk ribosomes was subsequently observed in other organisms, including Myxococcus xanthus and Streptomyces coelicolor (Dyson et al. 2011, Harms et al. 2013).
Imaging of fluorescently-tagged NAPs in E. coli has further suggested that the nucleoid assumes a loosely twisted overall conformation, with no particular handedness (Fisher et al. 2013, Hadizadeh Yazdi et al. 2012). A helical-like conformation was also observed in replicating B. subtilis when newly-replicated chromosomes were followed microscopically using fluorescent dNTP derivatives that incorporate into DNA as replication proceeds (Berlatzky et al. 2008). In addition, whole cell cryo-tomography of Bdellovibrio bacteriovorous revealed an apparent helical-like structure of the chromosome (Butan et al. 2011). The biological significance of a helical fold is unknown but may represent an energy-minimal configuration for fitting chromosomes within rod-shaped cells (Fisher et al. 2013).
Time-lapse microscopy using fluorescently-tagged NAPs has also revealed the temporal dynamics of chromosomes. In E. coli, waves of nucleoid density flux along the long axis of the cell; the function of this nucleoid mobility is not clear, but may impact chromosome segregation (discussed later) (Fisher et al. 2013).
The spatial arrangement of chromosomes has also been inferred by tracking the subcellular positions of individual loci using fluorescence in situ hybridization (FISH), fluorescent repressor-operator systems (FROS), and ParB/parS systems (Le & Laub 2014). In C. crescentus, 112 loci were examined by FROS in cells containing a single chromosome. The spatial positions of loci within the cell recapitulated the genetic map with the origin of replication (oriC) at one cell pole, the replication terminus (ter) at the opposite pole, and the left and right chromosomal arms likely running in parallel down the long axis of the cell, a pattern referred to as the ori-ter configuration (Viollier et al. 2004) (Fig. 1a). Recent data from large-scale chromosome conformation capture assays (5C and Hi-C) performed on C. crescentus are consistent with this pattern (Le et al. 2013, Umbarger et al. 2011). Those studies revealed high frequency interactions between loci nearby on the same chromosomal arms and slightly lower frequency interactions between loci at similar positions on opposite arms (also see Fig. 3b). In replicating C. crescentus cells, one new copy of oriC is rapidly segregated to the opposite pole. As replication proceeds, newly-generated DNA moves to its respective position, again with loci arranged relative to the origin in a manner that reflects the genetic map (Viollier et al. 2004). Ultimately the two termini end up at mid-cell, thereby recreating the ori-ter pattern in each daughter cell.
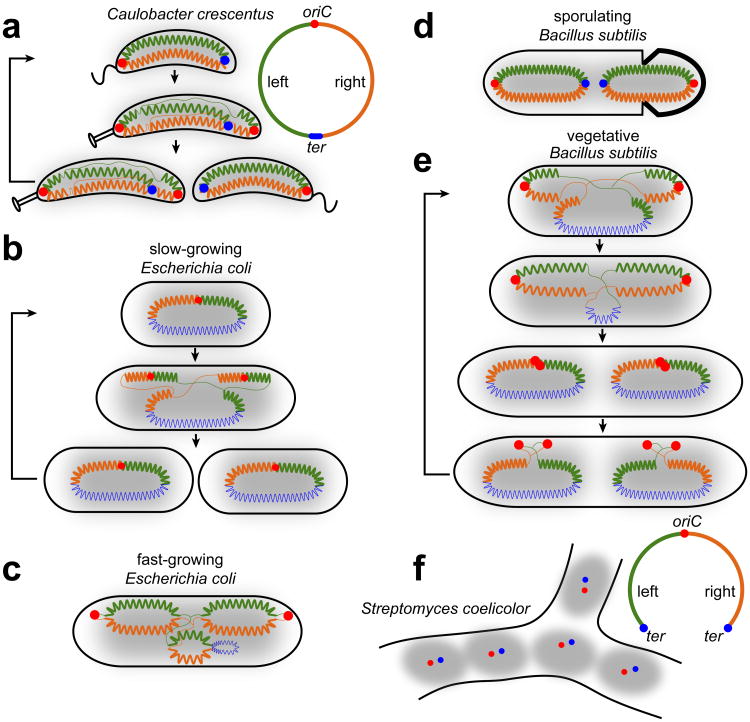
Figure 1. The global organization of bacterial chromosomes.
For the organism indicated in each panel, the schematics represent the origin of replication (oriC) as a red dot and terminus (ter) as blue dot or line. The left and right arms of the chromosome are colored green and orange, respectively. Thick zigzag lines denote compacted parts of the chromosome, while newly-synthesized DNA and hypothetically less-organized DNA are illustrated as thin lines. Overall nucleoid distribution is illustrated by grey shading. Black arrows indicate the progression of the global chromosome organization through a cell cycle.
Figure 3. Macrodomains and chromosomal-interaction domains.
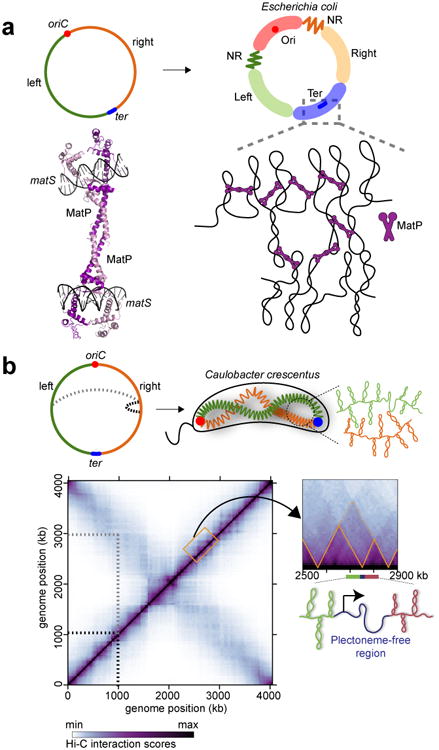
(a) Macrodomain-organization of E. coli chromosome, shown as in Figure 1 (left) or with the four macrodomains, Ori, Ter, Left and Right, and the two non-structured regions (NR) (right). MatP (colored purple) organizes the Ter macrodomain. The crystal structure of two MatP dimers, each bound to a matS recognition site is shown (PDB: 4D8J). (b) Chromosome conformation capture assay (5C and Hi-C) and computational modeling have revealed the organization of the C. crescentus chromosome. The Hi-C heat map (Le et al. 2013) indicates frequency of DNA-DNA interactions across the genome using the color scale shown. The most prominent diagonal indicates frequent interactions within a chromosomal arm (black dotted lines), while the other, less prominent diagonal shows interactions between the two arms (grey dotted lines). Orange triangles in the inset (right) indicate CIDs, or chromosomal-interaction domains (see text for details). Highly-transcribed genes are thought to create a less compacted, plectoneme-free region (blue) that serves to spatially insulate DNA (green and red) in adjacent domains, creating a CID boundary.
The chromosome configuration is substantially different in slow growing E. coli. The origin resides near mid-cell with the two chromosomal arms on opposite sides of the cell, and the terminus variably around mid-cell, a so-called left-ori-right configuration (Nielsen et al. 2006b, Wang et al. 2006) (Fig. 1b). DNA replication and segregation of the origins to cell quarter positions regenerates a left-ori-right organization for each chromosome. In contrast, fast growing E. coli cells adopt an ori-ter configuration of the chromosome with polarly-localized origins (Youngren et al. 2014) (Fig. 1c). Studies of chromosome organization in fast growing E. coli also demonstrated that in a cross-section of the cell, the chromosomal arms occupy the outer shell with the origin and terminus regions within the nucleoid core (Youngren et al. 2014).
In B. subtilis, global chromosome organization depends on its cell cycle and developmental stage. In sporulating B. subtilis, the two chromosomes adopt ori-ter/ter-ori configurations with an asymmetric septum trapping a quarter of one chromosome in the pre-spore compartment (Wang et al. 2014a, Wang & Rudner 2014) (Fig. 1d). During vegetative growth, the chromosome alternates between an ori-ter and E. coli-like left-ori-right pattern (Wang et al. 2014a, Wang & Rudner 2014) (Fig. 1e). Template DNA initially adopts a left-ori-right configuration, with replicated chromosomes then adopting an ori-ter pattern prior to cell division. Why B. subtilis employs both configurations is not clear, but it may allow replisomes to move independently on the two arms, while also ensuring segregation of newly replicated chromosomes to opposite sides of the cell (Wang & Rudner 2014).
Although the majority of bacterial chromosomes are circular, some are linear, including the multiple ∼1 Mb chromosomes in Borrelia species and the ∼8 Mb chromosomes of Streptomyces species (Chaconas & Kobryn 2010, Dyson 2011). Streptomyces oriC is flanked by the two chromosomal arms whose ends are spatially close, suggesting that the linear chromosome folds back on itself (Yang & Losick 2001) (Fig. 1f). Telomere-binding proteins that cap the chromosome ends may interact, effectively forming a topologically-closed chromosome (Tsai et al. 2011) although it remains unknown whether the Streptomyces chromosome adopts an ori-ter or left-ori-right configuration.
There is still relatively little known about chromosome organization in coccoid or other non-rod shaped bacteria. Finally, it is important to emphasize that the spatial positioning of a given locus typically varies up to 10% of the cell length in a population of cells and within a cell over time (Viollier et al. 2004, Wang et al. 2006, Wiggins et al. 2010). DNA typically moves in a sub-diffusive manner, i.e. more constrained than expected for Brownian motion. This tight positional variation may result from the crowded, viscoelastic environment of the nucleoid, from intranucleoid linkages that restrict DNA movement, supercoiling, or protein binding. The movement of DNA likely also depends on ATP-dependent mechanical processes as inhibiting ATP synthesis significantly reduces the diffusion coefficient of individual loci (Weber et al. 2012). Nevertheless, certain DNA loci occasionally exhibit super-diffusive or ‘near-ballistic’ motion, implying active segregation mechanisms or relaxations back to a “home” position (Bates & Kleckner 2005, Javer et al. 2014, Joshi et al. 2011). Advances in time-lapse fluorescence microscopy promise to reveal much more about DNA dynamics in the coming years.
Proteins that anchor specific DNA regions
In general, the ori-ter chromosome pattern appears most common in rod-shaped bacterial species. Whether this configuration affords an advantage is unclear, but the polar anchoring of origins, which likely enforces the ori-ter pattern, may help ensure that each daughter cell inherits a full copy of the genome. Studies in several organisms have identified proteins that localize to the cell poles and that bind oriC-proximal regions.
In C. crescentus, a parS site, critical for chromosome segregation (discussed later) is located ∼13 kb from the origin and is bound by ParB (Mohl et al. 2001, Toro et al. 2008), which also binds PopZ, a cytoplasmic protein that self-aggregates into a proteinaceous matrix at cell poles (Bowman et al. 2008, Ebersbach et al. 2008) (Fig. 2a). Moving parS away from the origin leads to a global rotation of the chromosome such that the relocated parS sites are still polar but the origins are not (Umbarger et al. 2011). The fact that parS position dictates global orientation of the chromosome implies that most loci are not actively positioned and instead effectively fall into place based on the position of parS and lengthwise compaction of the nucleoid.
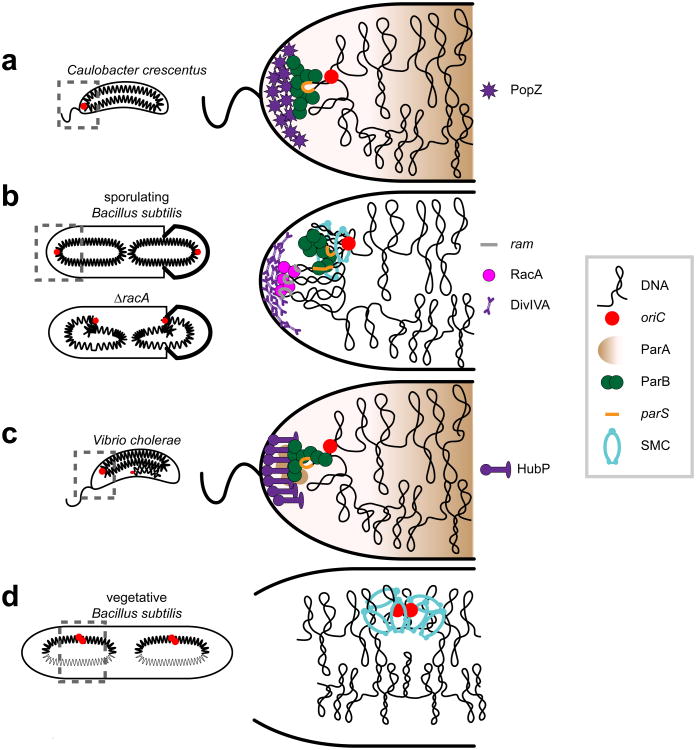
Figure 2. Protein-based systems that anchor specific DNA regions.
Schematics of polar anchoring complexes are shown for (a) C. crescentus, (b) sporulating B. subtilis, (c) V. cholerae and (d) vegetative B. subtilis. The likely global chromosome organization defect of a B. subtilis strain lacking RacA is shown in panel b. Specific DNA elements and proteins common to each organism are represented as shown in the legend (right), with species-specific factors indicated adjacent to each panel. Note that Soj/ParA is not represented in B. subtilis; although Soj/ParA is required for the bipolar localization of origins, its own localization is complex (Murray & Errington 2008), and precisely how localization impacts function is unclear. Schematics derived from similar drawings in (Wang & Rudner 2014).
In B. subtilis, a protein called RacA accumulates prior to sporulation and concentrates near the cell pole (Ben-Yehuda et al. 2003, Wu & Errington 2003). RacA binds 25 ram sites near oriC helping to tether ori-proximal regions of the chromosome to the pole (Ben-Yehuda et al. 2005) (Fig. 2b). Polar localization of RacA requires a small peripheral membrane protein called DivIVA, which recognizes the concave curvature of the polar membrane (Lenarcic et al. 2009, Oliva et al. 2010, Ramamurthi & Losick 2009). Cells lacking either RacA or DivIVA have disoriented chromosomes with oriC positioned near mid-cell rather than at the poles, and they frequently form empty pre-spore compartments (Ben-Yehuda et al. 2003).
In V. cholerae, a membrane-associated protein called HubP anchors the origin of the large chromosome, ChrI, to the pole (Yamaichi et al. 2012) (Fig. 2c). HubP interacts with ParAI, which likely interacts with ParBI, which in turn binds a parS site near the ChrI origin. HubP has a peptidoglycan-binding LysM domain, which is required for polar localization.
Although PopZ, RacA, and HubP each anchor chromosomes to a cell pole, these proteins bear no sequence similarity suggesting they arose independently, further supporting the notion that an ori-ter chromsome configuration may be selectively advantageous. However, pole-anchoring proteins may not be strictly necessary for the ori-ter arrangement as some organisms, such as M. xanthus and P. aeruginosa, adopt an ori-ter pattern, but have a large cytoplasmic gap between oriC and the cell pole, suggesting that the origin is not anchored (Harms et al. 2013, Vallet-Gely & Boccard 2013).
In E. coli, no polar anchoring complex has been identified, although if one exists, it may function only during fast growth when chromosomes exhibit an ori-ter pattern. However, the structural maintenance of chromosomes (SMC) complex (discussed later) and called MukBEF in E. coli, is required to maintain the left-ori-right configuration; cells lacking mukB adopt an ori-ter configuration even in slow growth conditions (Danilova et al. 2007). Similarly, in vegetative B. subtilis cells, where the chromosome alternates between left-ori-right and ori-ter configurations, SMC is also required for the left-ori-right pattern (Wang et al. 2014a) (Fig. 2d). Whether SMC promotes a traverse left-ori-right pattern by anchoring origin-proximal regions to mid-cell or non-polar regions is unclear. B. subtilis SMC and E. coli MukBEF do associate with origin-proximal regions (Danilova et al. 2007, Gruber & Errington 2009, Sullivan et al. 2009), but there is no evidence that SMC associates with the cell membrane.
Macrodomains and chromosomal-interaction domains
Bacterial chromosomes are further organized into Mb-sized domains called macrodomains, which were first suggested from FISH studies in E. coli demonstrating that certain loci frequently co-occupy the same restricted cytoplasmic space (Niki et al. 2000). Subsequent use of a λ recombination-based assay found that loci within a given macrodomain interact, and hence recombine, more frequently than loci in different macrodomains (Valens et al. 2004). Collectively, E. coli has four macrodomains, called Ori, Ter, Left, and Right, with two less-structured DNA regions flanking the Ori macrodomain (Fig. 3a). DNA within macrodomains is more restricted in its movement than DNA in unstructured regions (Espeli et al. 2008). DNA inversions are also more easily tolerated if occurring within a macrodomain suggesting that macrodomains are a critical level of chromosome organization (Thiel et al. 2012).
A breakthrough in understanding macrodomain organization came from the discovery of E. coli MatP, which binds to 13 bp matS sites present exclusively in the ∼800 kb Ter macrodomain (Mercier et al. 2008). A MatP dimer bound to one matS site can form a tetramer with a MatP dimer bound at another site, bringing distal MatP-matS complexes together, helping to compact the Ter macrodomain in space and looping the intervening DNA (Dupaigne et al. 2012) (Fig. 3a). Cells lacking MatP exhibit chromosome segregation and terminus resolution problems (Dupaigne et al. 2012, Mercier et al. 2008). Similar proteins may structure the other E. coli macrodomains but have not been identified yet, and MatP homologs appear restricted to enteric bacteria.
Macrodomains per se have not been documented in C. crescentus. However, recent Hi-C analyses revealed that the C. crescentus chromosome is divided into ∼23 chromosomal interaction domains (CIDs), each ∼166 kb on average (Le et al. 2013). Loci within a domain interact preferentially with other loci in the same domain (Le et al. 2013) (Fig. 3b). Notably, these domains are often nested, with several adjacent domains forming larger entities potentially similar to macrodomains. Whether chromosomal domains akin to those in Caulobacter are also present within E. coli macrodomains cannot be resolved by FROS and awaits high-resolution Hi-C studies.
The chromosomal interaction domains in Caulobacter are created in part by highly expressed genes (Le et al. 2013) (Fig. 3b). Domain boundaries often coincide with the most highly expressed genes, such as those encoding ribosomal proteins. Inhibiting transcription elongation by adding rifampicin to cells causes an almost complete loss of domains (Le et al. 2013). Additionally, relocating a highly expressed gene, rsaA, to an ectopic location was sufficient to induce a new domain boundary (Le et al. 2013). High rates of transcription and the frequent unwinding of DNA likely creates local, plectoneme-free regions in the chromosome. These plectoneme-free regions may prevent the diffusion of supercoils and physically separate the flanking domains, thereby decreasing the contact probabilities between loci in neighboring domains. The boundaries between chromosomal domains vary in sharpness, which might reflect variation in the rate of transcription of highly expressed genes and differences in transcript length, as well as variability in the local density of DNA-binding proteins. Supercoiling (discussed below) is also important for domain formation and/or maintenance as the addition of novobiocin largely eliminated domains. This finding may reflect the fact that negative supercoils must be introduced to offset positive supercoils, which can impede RNA polymerase during transcription. Finally, it should be noted that Hi-C reflects the DNA-DNA interactions in a population of cells. Thus, domains identified by Hi-C must be present in most cells; individual cells may have additional, transient domain boundaries.
Supercoil domains
Within macrodomains and chromosomal interaction domains, loops of genomic DNA are supercoiled, likely forming plectonemes that coil up around them selves while attached at their base to proteins that help to topologically isolate the looped DNA. These plectonemic loops, also called supercoil domains or topological domains, were first seen in electron micrographs of gently-lysed E. coli cells (Kavenoff & Ryder 1976). Subsequent studies have tried to estimate the number of supercoil domains in E. coli by assessing the number of nicks required to fully relax the chromosome, assuming that an individual nick relaxes only the DNA within a given supercoil domain. Initially, relaxation was assessed by measuring the incorporation of trimethyl-psoralen, an intercalating dye that preferentially interacts with negatively supercoiled DNA; these assays suggested that E. coli harbors ∼40 topologically isolated domains during exponential growth (Sinden & Pettijohn 1981). A subsequent study examined supercoil domains by assessing the transcriptional response to double-strand breaks (Postow et al. 2004). The idea was that only DNA within a single, topologically isolated domain will be relaxed after a double-strand break, leading to changes in the transcription of supercoiling-sensitive genes only within that domain. This method estimated the average supercoil domain at ∼10 kb, implying ∼400 domains in the E. coli chromosome, a number that agrees well with the number of loops in chromosomes from lysed cells imaged by EM (Postow et al. 2004).
Supercoil domains have also been probed using recombination as an indirect readout. Unlike λ-Int, the γδ and Tn3 resolvases only recombine if two res sites are brought into precise alignment through the slithering of plectonemic DNA. The rate of recombination thus depends on the genomic distances between res sites, being almost undetectable if separated ∼100 kb, thereby setting a likely upper limit on the size of plectonemes in vivo (Higgins et al. 1996). Studies with the γδ and Tn3 resolvases indicate that the average plectoneme size in wild-type S. typhimurium is ∼10 kb, again implying ∼400 supercoil domains per chromosome (Stein et al. 2005). Certain gyrase mutants can, however, harbor double the number of supercoil domains.
Hi-C data has also contributed to our understanding of plectonemes in vivo. A polymer model of the Caulobacter chromosome comprised of plectonemes was recently constructed with parameters corresponding to plectoneme length, width, diameter, flexibility, and radius of collisions (Le et al. 2013). A search for parameter values that reproduced Hi-C data suggested an average length for plectonemes of ∼8 kb, similar to that measured by recombination and relaxation assays (Stein et al. 2005).
The boundaries between supercoil domains are often dynamic, and may depend on both DNA-binding proteins and gene expression. DNA-binding proteins (discussed below) can bridge distant loci, topologically isolating the intervening DNA and preventing the spread of supercoils between adjacent domains. Gene expression also plays a major role in establishing supercoil domains. As noted for chromosomal interaction domains, loci undergoing high rates of transcription can be boundary elements that prevent plectoneme diffusion, although the precise underlying mechanism is not clear. Additionally, transcription contributes to the supercoiling structure of the genome as RNA polymerase introduces negative supercoils behind it and positive supercoils in front.
Some supercoil domains likely vary significantly between cells in a population and within a given cell over time. This variability in supercoil location may, in turn, impact the expression of genes whose promoters are sensitive to supercoiling status. However, the domain boundaries associated with very highly expressed genes, and observed by Hi-C, are static. Notably, these domain boundaries are relatively well distributed across the genome and bioinformatic analyses indicate that such a distributed pattern of highly expressed genes is common (Wright et al. 2007). The advantage, if any, of distributing domains across a genome is not known, but domain boundaries could help periodically pause DNA replication to promote compaction of recently replicated domains and the decatenation of sister chromosomes. Alternatively, or in addition, dividing the genome into domains may help limit how much of the chromosome relaxes following a nick or double-strand break.
In sum, the relationship between various domains - macrodomains, chromosomal interaction domains, and supercoil domains - is not fully clear yet, but we envision a hierarchical organization. Megabase-sized macrodomains are likely comprised of multiple chromosomal interaction domains, each ∼100-200 kb and containing multiple, diffusible supercoil domains, each ∼10 kb in size. Very highly expressed genes appear to play a critical role in establishing chromosomal interaction domain boundaries and are relatively fixed in a population of cells. The expression of other genes may form transient domains and transient boundaries. The position of genes within domains (at every level) may influence their expression, but the precise relationship between chromosome structure and gene expression remains to be defined.
Nucleoid-associated proteins (NAPs)
The organization of bacterial chromosomes is profoundly influenced by DNA-binding proteins, and in particular by a heterogeneous class of abundant proteins called nucleoid-associated proteins (NAPs). NAPs typically bind relatively non-specifically across bacterial genomes, wrapping, bending, or bridging DNA (Fig. 4). The local action of NAPs ultimately influences global chromosome organization and, in many cases, transcriptional patterns (reviewed in Dillon & Dorman 2010).
Figure 4. Nucleoid-associated proteins with DNA bridging, wrapping, or bending activities contribute to the organization of the chromosome.
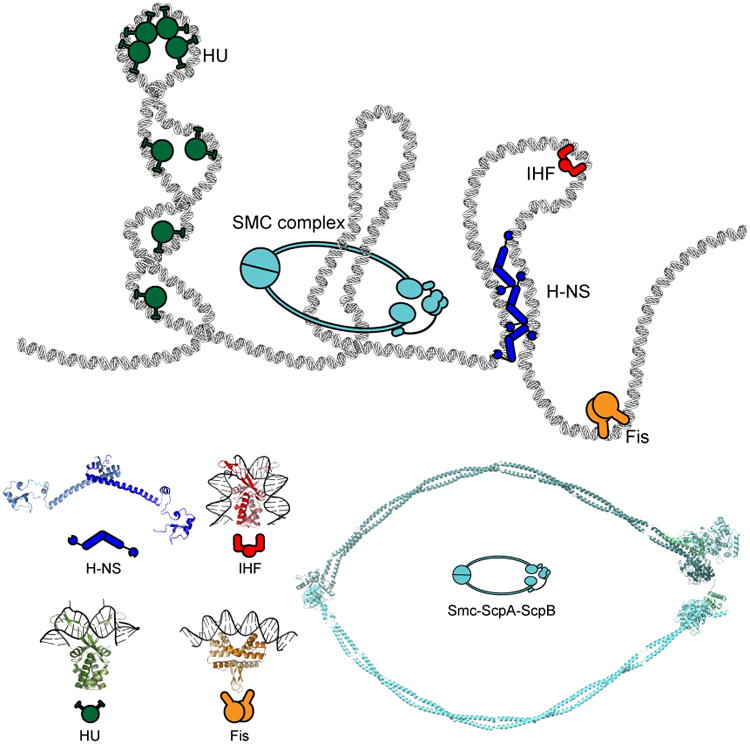
The functions of well-studied NAPs are schematized at the top, with the corresponding crystal structures below. H-NS dimers of dimers (blue) bridge DNA. The abundant HU (green) introduces ∼90° bending to DNA and may wrap DNA around itself, thereby promoting short-range DNA interactions. IHF (red) binding to DNA induces a dramatic U-turn on DNA that drastically changes the trajectory of the DNA backbone. Fis (orange) is another NAP with DNA-bending activity. SMC complexes (cyan) likely form a ring structure that can bring together and handcuff loci that are distal in primary sequence. The protein and protein:DNA complexes shown have PDB IDs: 1P78, 1IHF, and 3JRA for HU, IHF, and Fis, respectively. A hypothetical model of H-NS was constructed from PDB structures 3NR7 and 1HNR. A model of SMC-ScpA-ScpA was derived from PDB structures 4I98, 4I99 and 3ZGX.
E. coli H-NS is a small (15.5 kDa) protein that can bridge DNA, bringing loci separated on the primary sequence level into close physical proximity (Fig. 4). H-NS has an N-terminal domain, which drives oligomerization that is connected by a flexible linker to a C-terminal DNA-binding domain. ChIP studies indicate that E. coli H-NS binds hundreds of sites in the genome, with a preference for AT-rich or curved DNA (Grainger et al. 2006, Kahramanoglou et al. 2011). Bridging of different segments of DNA by H-NS has been directly demonstrated by both single-molecule and atomic force microscopy (Dame et al. 2000, 2006).
DNA bridging by H-NS likely enables it to constrain negative supercoils, by effectively isolating the intervening, looped region of the chromosome. H-NS binding sites also often coincide with supercoiling-sensitive promoters further suggesting a tight relationship between H-NS and supercoiling (Higgins et al. 1988); in fact, H-NS was originally discovered in a screen for E. coli mutants with reduced negative supercoiling (Hardy & Cozzarelli 2005). H-NS has also been examined in vivo by super-resolution microscopy, forming two discrete foci within the cytoplasm (Wang et al. 2011), although the functional significance of these foci and the DNA loci associated with them are unknown. In addition to bridging distant DNA segments, H-NS can also oligomerize and spread along DNA. Such oligomers can occlude binding sites for RNA polymerase or transcription activators, thereby enabling H-NS to regulate gene expression. This oligomerization of H-NS also enables it to silence spurious transcriptional promoters and horizontally acquired DNA, which is often more AT-rich than host chromosomal DNA (Lucchini et al. 2006, Navarre et al. 2006, Singh et al. 2014). H-NS orthologs, and paralogs called StpA, are found in many species, although H-NS is not universal. However, other, unrelated proteins, such as Rok in B. subtilis, may similarly bridge DNA or oligomerize along AT-rich DNA (Smits & Grossman 2010).
HU is another small (18 kDa), abundant (∼30,000 copies/cell) NAP found in many bacteria that coats and wraps chromosomal DNA around itself, grossly similar to histones (Azam et al. 1999) (Fig. 4). There are two HU subunits, alpha and beta, and both homo- and hetero-dimers exist, depending on growth phase in E. coli (Claret & Rouviere-Yaniv 1997). HU inserts conserved proline residues into the minor groove of DNA, inducing a sharp bend in the DNA (Swinger et al. 2003). Structural studies also suggest that HU can form an octameric structure with DNA coiled around it (Guo & Adhya 2007, Swinger et al. 2003).
HU has little or no DNA-binding specificity based on ChIP-Seq analyses, and given its abundance in E. coli, HU may coat ∼10% of the chromosome (Prieto et al. 2012). Consistent with widespread genomic binding, strains lacking HU often produce anucleate cells suggestive of a general chromosome compaction or segregation defect (Huisman et al. 1989), and strains harboring HU variants with higher DNA binding affinity have over-compacted nucleoids (Kar et al. 2005). Additionally, Hi-C studies of an HU mutant in Caulobacter revealed a significant decrease in short-range interactions, supporting the notion that HU helps to broadly compact the chromosome, possibly by stabilizing plectonemes (Le et al. 2013). HU binding to DNA may also affect supercoiling status of the chromosome as HU mutants in E. coli show decreased supercoiling, are rescued by mutations in gyrase, and are synthetically lethal with mutations in topoisomerase I (Bensaid et al. 1996, Malik et al. 1996). Additionally, the variants of HU with increased affinity for DNA also increase global supercoiling levels (Kar et al. 2005).
Some organisms encode divergent HU paralogs. For example, S. coelicolor and Mycobacteria encode an HU paralog called HupS/Hlp that has an extensive C-terminal extension with homology to eukaryotic histone H1 (Mukherjee et al. 2009, Salerno et al. 2009). In M. tuberculosis, the phosphorylation of HupS decreases its interaction with DNA highlighting the possibility of post-translational regulation of DNA compaction (Gupta et al. 2014).
Two proteins that can sharply bend DNA are integration host factor, IHF, and factor for inversion stimulation, Fis. IHF bears some sequence similarity to HU and is similarly composed of two subunits, although IHF binds DNA more specifically and introduces dramatic ∼160° bends (Rice et al. 1996) (Fig. 4). Consequently IHF can dramatically alter DNA shape and facilitate the formation of loops, frequently bringing RNA polymerase together with distant regulatory proteins. IHF also impacts a range of other DNA-based processes, including replication initiation and recombination (Leonard & Grimwade 2005, Mumm et al. 2006).
Like IHF, Fis can bend DNA. It is among the most highly expressed genes during fast growth in E. coli, especially following nutrient upshifts (Azam et al. 1999). Fis homodimers bind to AT-rich DNA sequences with narrow minor grooves, bending the DNA by ∼50-90° and forming very stable, long-lived nucleoprotein complexes (Stella et al. 2010) (Fig. 4). Fis binds throughout the genome (Kahramanoglou et al. 2011), impacting transcription, replication and recombination. Given its genome-wide distribution, Fis probably also influences chromosome compaction and organization in significant ways. Fis-mediated bending of DNA can displace nearby supercoils and it can preserve the writhe of DNA, potentially maintaining supercoiled plectonemic loops (Auner et al. 2003). Indirectly, Fis also influences global supercoiling levels by modulating the expression of gyrase (Schneider et al. 1999).
Although NAPs are generally small proteins, some large proteins also stably associate with and influence the structure of chromosomes. Most prominent in this category is the widely-conserved protein SMC, homologous to eukaryotic condensin (reviewed in Nolivos & Sherratt 2014). The >125 kDa SMC forms an extended, antiparallel coiled coil with a so-called hinge domain at one end and an ATPase domain at the other (Fig. 4). Homodimerization via the hinge domains creates a ring-like structure that may encircle DNA. SMC associates with two regulatory proteins, ScpA and ScpB, that likely modulate the ATPase activity of SMC, thereby affecting the opening and closing of the homodimeric ring. E. coli and other γ-proteobacteria do not encode SMC/ScpA/ScpB, and instead produce an analogous complex MukB/MukE/MukF (Nolivos & Sherratt 2014).
Mutations in SMC produce a range of chromosomal defects in different bacteria, often including an increase in anucleate cells. SMC likely contributes to both chromosome segregation (discussed in detail below) and chromosome compaction. In B. subtilis and E. coli, mutations in smc and mukB, respectively, lead to chromosome decondensation visible by DAPI staining (Tadesse et al. 2005, Weitao et al. 1999). Additionally, in E. coli, mukB mutants display altered supercoiling levels, and these mutants can be partially rescued by other mutations that increase DNA gyrase activity and negative supercoiling (Sawitzke & Austin 2000).
Precisely how SMC proteins affect chromosome compaction is not yet clear. By virtue of its extended, ring-like structure, SMC may bridge different loci in the chromosome. This bridging could help compact the DNA and it may also constrain supercoils by producing topologically isolated DNA loops. Notably, in both E. coli and B. subtilis, SMC proteins associate with origin-proximal regions and are required for the proper positioning of origins (Danilova et al. 2007, Gruber & Errington 2009, Sullivan et al. 2009). Whether the origin-proximal regions are preferentially compacted by the associated SMC proteins is not yet clear. ChIP-chip analysis in B. subtilis also indicated enrichment of SMC at regions of high transcription, a pattern also seen with eukaryotic SMC, but the functional significance of this localization is unknown (Gruber & Errington 2009).
In Caulobacter, cells lacking SMC do not exhibit major defects in chromosome organization (Le et al. 2013), as originally suggested (Jensen & Shapiro 1999), although an ATPase defective mutant shows a severe defect in sister chromosome separation (Schwartz & Shapiro 2011). Additionally, Hi-C studies of a Δsmc strain indicated lower frequencies of interactions between loci at approximately equivalent positions on opposite arms of the chromosome down nearly the entire long axis of the cell (Le et al. 2013). This could indicate that SMC tethers the arms together. Alternatively, SMC could promote the colinearity of the two chromosomal arms by promoting the compaction of each arm along the long axis of the cell. Cells lacking SMC may then end up with irregularities in the relative positions of loci in each chromosomal arm, disrupting the colinearity of loci observed by Hi-C.
Some traditional transcription factors also have NAP-like properties. The cyclic AMP regulatory protein CRP, which can bend DNA ∼90°, binds several hundred sites in the E. coli chromosome (Grainger et al. 2005). The leucine-responsive regulatory protein, Lrp, which may influence the expression of ∼10% of E. coli genes, can form a dimer, octamer, and hexadecamer, with its DNA-binding domain exposed, potentially enabling Lrp to bend or wrap DNA (Chen & Calvo 2002).
The abundance of many NAPs varies significantly depending on growth phase and environmental conditions. Indeed, nucleoid-associated proteins likely play a critical role in shaping or adjusting chromosome organization in response to different growth conditions. For example, in stationary phase, the E. coli nucleoid contains fewer loops than in exponential phase and each loop has more relaxed DNA; however, on the cellular level, the nucleoid becomes significantly more compact in stationary phase. These changes in structure result in part from the elimination of Fis as cells enter stationary phase and the massive upregulation of Dps (DNA-binding protein from starved cells). Dps binds throughout the chromosome, inducing a stable, crystalline state for the DNA that persists even if cells are lysed (Wolf et al. 1999). Dps physically protects the chromosome from damage during stationary phase. Additionally, Dps chelates Fe2+, helping to prevent it from producing hydroxyl radicals via a Fenton reaction that could damage the DNA (Frenkiel-Krispin & Minsky 2006). How Dps is released from DNA as cells exit stationary phase is unknown. SASP (small acid soluble protein) has a similar role as Dps in protecting the chromosome of B. subtilis and Clostridium difficile during sporulation (Nicholson et al. 2000). SASP non-specifically coats the chromosome and induces a ring-like structure that likely physically shields the chromosome and that may promote non-homologous end-joining repair following double-strand breaks by preventing the cut ends from diffusing apart (Frenkiel-Krispin et al. 2004).
In sum, NAPs and other chromosome-associated proteins are clearly central players in chromosome organization. Although the local, biophysical properties of many of these proteins have been well-studied, much remains to be learned about their in vivo functions and how, on a global level, they combine to compact, shape, and organize the genome, and, in turn, how they affect DNA-based transactions within cells.
Non-proteinaceous factors that contribute to chromosome organization
Other factors, beyond DNA-binding proteins, also contribute significantly to chromosome organization. Macromolecular crowding in the viscous bacterial cytoplasm may help with compaction (de Vries 2010). As noted, the movement of chromosomal loci is generally subdiffusive, implying that the viscoelastic cellular environment influences motion and compaction. Occasional super diffusive motions, which may reflect stress-relaxation mechanisms, further suggests that the chromosome is subject to strong mechanical forces that ultimately impact its compaction and organization.
The physical properties of the chromosome as a large polymer may also influence its organization. One model suggested that the chromosome is a self-avoiding polymer, and argued that entropic forces may significantly influence chromosome organization, favoring the separation of supercoil domains (Jun & Mulder 2006). Indeed, in some bacteria, such as Caulobacter, the chromosome occupies nearly the entire cytoplasmic space, further suggesting that the inner membrane influences chromosome organization through physical confinement. Additionally, Hi-C studies of Caulobacter chromosomes during DNA replication indicated little interaction between sister chromosomes (Le et al. 2013), possibly consistent with DNA supercoiling loops repelling each other. However, in many species, the chromosome does not fill the entire cell, and nucleoid-associated proteins that decorate bacterial DNA likely render chromosomes self-adherent filaments (Hadizadeh Yazdi et al. 2012).
Another factor that may affect chromosome organization is transertion, the coupled translation and insertion of proteins into the membrane by the signal recognition particle SRP and the Sec translocase. Transertion may tether DNA to the cell membrane, pulling some DNA out of the nucleoid, which could affect chromosome compaction and segregation. Evidence for transertion has been scant, but a recent FROS study tracking the intracellular position of tetA, which encodes a membrane efflux pump, showed that this locus, and nearly 90 kb around it, moves toward the membrane shortly after inducing tetA expression (Libby et al. 2012). Additionally, treating cells with either a transcription inhibitor (rifampicin) or translation inhibitor (chloramphenicol) causes radial shrinkage of the E. coli nucleoid, further supporting the notion that transertion represents an expansion force for the chromosome (Bakshi et al. 2012).
Bacterial Chromosome Segregation
Chromosome segregation is essential for daughter cells to each inherit a full copy of the genome. Unlike in eukaryotes, chromosome replication and segregation occur concomitantly in bacteria and, apparently, without a dedicated, spindle-like apparatus (Nielsen et al. 2006a, Viollier et al. 2004, Wang et al. 2006). The molecular mechanisms responsible for bacterial chromosome segregation are only just beginning to emerge, and involve both specific protein components as well as non-protein, mechanical-based mechanisms.
One of the earliest models for bacterial chromosome segregation was proposed by François Jacob who suggested that newly replicated origins may get anchored to the cell membrane and segregated passively by cell growth/elongation between them (Jacob et al. 1963). However, subsequent studies tracking origins have shown that they segregate much faster than the rate of cell elongation (Fiebig et al. 2006, Viollier et al. 2004, Wang & Sherratt 2010).
DNA replication has also been implicated in chromosome segregation. Early work in B. subtilis suggested that the replisomes formed a factory at midcell, pulling DNA toward it for replication and extruding replicated DNA to either side of it (Lemon & Grossman 1998). Although this “capture-extrusion” model could explain bulk, symmetric segregation of chromosomal regions after replication, it cannot apply to bacteria where the chromosome is asymmetrically replicated and segregated. Additionally, recent studies using fluorescence time-lapse microscopy in E. coli, C. crescentus, and B. subtilis have indicated that the replisomes are mobile, tracking independently along the chromosome (Bates & Kleckner 2005, Jensen et al. 2001, Reyes-Lamothe et al. 2008, Wang et al. 2014a). Thus, while the replisome and act of DNA replication per se could aid chromosome segregation, it likely cannot provide all of the force necessary.
The ParAB system for origin segregation
The first section of the chromosome segregated is usually the origin-proximal region. In many bacteria, origins are segregated actively via the parABS partitioning system (Fogel & Waldor 2006, Ireton et al. 1994, Lin & Grossman 1998, Mohl et al. 2001), first discovered in plasmids where they are often essential for plasmid maintenance (Austin et al. 1985, Gerdes et al. 2010). Homologs of parABS were subsequently found to facilitate chromosome segregation in some bacteria, with nearly 65% of bacteria harboring this system (Livny et al. 2007). In some cases, the parABS system is essential for viability; even when not formally essential, deletion of the system often leads to a significant increase in anucleate cells, demonstrating its importance in chromosome segregation.
In most bacteria, parS sites are located near the origin. ParB specifically recognizes and binds to these parS sites, often spreading in that region and perhaps bridging more distant DNA to form a large nucleoprotein complex (Graham et al. 2014, Lin & Grossman 1998, Murray et al. 2006). On its own, ParA has weak ATPase activity and binds DNA non-specifically; its ATPase activity is directly stimulated by ParB (Easter & Gober 2002, Leonard et al. 2005). It was originally proposed, based on studies of plasmids (Ebersbach et al. 2006, Gerdes et al. 2010, Ringgaard et al. 2009), that ParA forms dynamic filaments that segregate the ParB:parS complexes that form on sister chromosomes after replication by either a pulling or pushing mechanism. According to the pulling model, a ParA filament forms away from the partition complex; the edge of this filament captures a ParB:parS complex and the filament retracts, pulling the DNA with it. According to the pushing model, a ParA filament forms between duplicated ParB:parS complexes and grows between them, thus pushing them apart.
Early evidence for a pulling mechanism came from studies of origin segregation in V. cholerae, which encodes two par systems, one for each chromosome (Fogel & Waldor 2006). ParAI was proposed to segregate origins by pulling as ParAI-YFP does not localize between ParBI:parSI complexes and instead localizes between the new cell pole and the segregating ParBI:parSI complex. ParAI appeared, based on epi-fluorescence microscopy, to form dynamic filaments that retract toward the cell pole in concert with the movement of the ParBI:parSI complex in the same direction, implying a pulling mechanism. However, the precise mechanism of pulling and whether ParAI forms a continuous filament are not clear.
ParAB-dependent origin segregation has also been studied in C. crescentus and initial studies also proposed a pulling mechanism (Ptacin et al. 2010, Shebelut et al. 2010). Origin segregation in C. crescentus is a two-step process. After replication, the duplicated origins are first released from the pole and separate slightly from one another before one of the origins is translocated to the opposite cell pole, effectively unidirectionally (Shebelut et al. 2010). The initial separation does not require ParA, but the subsequent step does (Shebelut et al. 2010, Toro et al. 2008).
ATP-bound ParA was postulated to form a filamentous structure across the cell (Ptacin et al. 2010). ParB bound to parS sites would contact the edge of this filament and stimulate ParA ATPase activity, resulting in dissociation of ParA molecules from the edge of the filament and a net retraction of the filament away from ParB. Brownian movement of the ParB:parS complex would then renew contact with the ParA filament, and the ATP hydrolysis and dissociation cycle would repeat. The higher affinity of ParB for ATP-bound ParA would ensure that a ParB:parS complex moves with the retracting ATP-bound ParA filament toward the opposite cell pole.
Although attractive, this pulling model initially assumed that ParA forms a single, continuous filament. Whether such filaments actually occur in vivo is uncertain, and recent studies suggest that an extended filament is not necessary for directional movement; instead, the Mizuuchi group has proposed a “diffusion-ratchet” model (Hwang et al. 2013, Vecchiarelli et al. 2010, 2012, 2014). By reconstituting a plasmid parABS system in vitro, they showed that ParA-ATP binds DNA non-specifically. ParB bound to parS on a plasmid stimulated ParA ATPase activity, resulting in the release of ParA and local depletion of ATP-bound ParA. The ParB:parS complex then diffused up the ParA-ATP gradient, resulting in net directional movement of the ParB-bound plasmid.
The “diffusion-ratchet” model was derived from studies of plasmid partitioning, and a subsequent study suggested that it may also apply to chromosome partitioning in Caulobacter (Lim et al. 2014). However, that study argued, based on mathematical modeling, that the diffusion of ParB:parS up short-range ParA-ATP gradients was insufficient to provide the observed directionality of chromosome segregation. Instead, it was suggested that the elasticity and dynamic motion of the chromosome helps relay, or drive translocation of, the chromosome short distances. This ‘DNA relay’ model essentially extends the ‘diffusion-ratchet’ model, providing a plausible mechanism for the directional segregation of chromosomal loci via the parABS system, without invoking or requiring large ParA filaments (Fig. 5a).
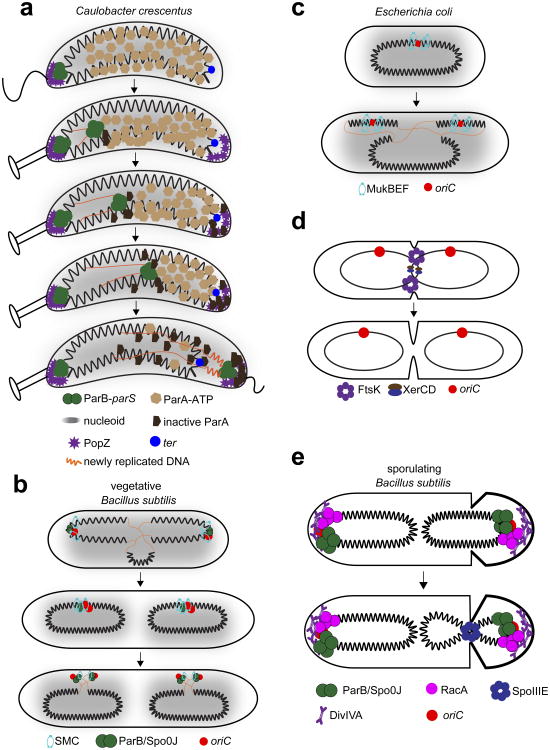
Figure 5. Chromosome segregation.
(a) Origin segregation in C. crescentus relies on the parABS system. ParB (green) binds parS sites located near the origin. Shortly after replication, one ParB:parS complex remains polarly localized while the second complex comes in contact with ATP-bound ParA (light brown). ParB stimulates ParA ATPase activity, resulting in the release of ParA from DNA (dark brown) and contraction of the cloud of ParA-ATP. The migrating ParB:parS complex can then move toward the retracting ParA-ATP and thus toward the opposite pole, eventually resulting in full segregation of the origin. PopZ (purple) influences ParAB activity directly or indirectly to promote origin segregation. (b) In vegetatively growing B. subtilis, chromosome organization oscillates between ori-ter and left-ori-right patterns. While ParA/Soj and ParB/Spo0J (green) ensure origin movement towards opposite poles, the SMC complex (cyan) relocates the origins to mid cell during the initial phase of DNA replication. This oscillation in chromosome organization may promote chromosome segregation by preventing entanglement of the chromosomes. (c) Origin segregation in E. coli. Unlike Caulobacter and B. subtilis, E. coli does not have a ParAB-like system for origin segregation. A distant relative of the SMC complex, MukBEF (cyan), localizes around the origin region and is thought to promote origin segregation and origin-proximal chromosome organization. MukBEF may also promote bulk chromosome segregation. (d) Circular chromosome replication can result in dimeric or catenated chromosomes, whose resolution requires the action of the DNA translocase FtsK (purple) and the tyrosine recombinase XerCD (blue-brown). Schematic shown is for E. coli. (e) Chromosome segregation in sporulating B. subtilis. Segregation of the origin region depends on RacA (pink) and Spo0J (green), with the rest of the chromosome pumped into the forespore by the DNA translocase SpoIIIE (blue). The origin is anchored to the cell pole by RacA and DivIVA (purple sticks).
The polar anchoring protein PopZ, discussed above, may also help ensure directional movement of one origin toward the new cell pole in Caulobacter by anchoring the origin region to the cell pole (Bowman et al. 2008, Ebersbach et al. 2008, Laloux & Jacobs-Wagner 2013). PopZ may also regulate ParA activity (Ptacin et al. 2014, Schofield et al. 2010), possibly by sequestering ATP-hydrolysed ParA generated near the translocating origin away from the nucleoid, and by regenerating ParA-ATP that can bind the nucleoid again near the pole (Ptacin et al. 2014). This PopZ-dependent regulation of ParA may help ensure unidirectional movement of the translocating origin.
Although many organisms segregate replicated origins to opposite cell poles, some species produce multiple chromosomes that must be spaced out evenly across the cell, such as Synechococcus elongatus and S. coelicolor during sporulation (Jain et al. 2012, Jakimowicz et al. 2007). In the latter case, ParAB is required to space out chromosomes, and ParB itself is regularly distributed across the cell. ParA ATPase activity is essential for segregation and ParA forms an apparent filament during segregation that disassembles prior to septation (Jakimowicz et al. 2007).
Although widespread, parABS is absent from some species, including E. coli. However, E. coli does harbor a parS-like site called migS that helps promote the bipolar segregation of origins in E. coli, although migS is not formally essential for successful chromosome segregation (Fekete & Chattoraj 2005, Wang & Sherratt 2010, Yamaichi & Niki 2004). Interestingly, a recent study in E. coli suggested that the MinDE system, which directly regulates cell division, could also promote chromosome segregation, albeit not by binding a specific site as ParAB does (Di Ventura et al. 2013). MinD and MinE normally oscillate back and forth across cells, inhibiting polymerization of the cytokinetic ring protein FtsZ at the poles and thereby helping force cell division to occur at mid-cell. MinD was proposed to also simultaneously bind the membrane and DNA non-specifically, with MinDE oscillations biasing the movement of replicated DNA regions towards cell poles. However, the role of the Min system in chromosome segregation is difficult to discern given that MinDE may indirectly affect chromosome organization and segregation through their effect on cell division.
In addition to promoting the segregation of origins, ParA, ParB, and parS sometimes have additional functions and interaction partners. In B. subtilis, ParA (Soj) can regulate DNA replication initiation by interacting with DnaA (Murray & Errington 2008, Scholefield et al. 2011). Monomeric ParA/Soj inhibits DnaA from forming an oligomeric helix on DNA, thereby preventing replication initiation. In contrast, dimeric ParA/Soj, which binds DNA, appears to promote replication initiation through DnaA, although the precise mechanism of activation is still unclear. Other work in B. subtilis has shown that ParB (Spo0J) interacts with SMC, recruiting it to the origin region (Gruber & Errington 2009, Sullivan et al. 2009). This origin-localized SMC helps promote chromosome segregation (see next section) and somehow promotes the transient left-ori-right configuration of B. subtilis chromosomes noted above (Wang et al. 2014a) (Fig. 5b).
In Caulobacter, ParB interacts directly with MipZ, a protein similar to MinD, that helps determine the mid-cell placement of FtsZ (Thanbichler & Shapiro 2006). MipZ inhibits FtsZ polymerization; hence, by associating with ParB, MipZ ends up localized primarily to the polar regions of the Caulobacter, leaving the mid-cell region free for FtsZ polymerization. A recent study suggested that the parS site in Caulobacter interacts not only with ParB, but potentially also with DnaA, providing a link between DNA replication initiation and origin segregation (Mera et al. 2014). The model proposed posits that DnaA binds parS sites, somehow altering the structure of the DNA around it in a manner that promotes ParB binding and, consequently, proper segregation.
Bulk chromosome segregation and SMC proteins
The faithful segregation of two recently replicated origins by parABS, and the subsequent anchoring of these origins to opposite cell poles, may dictate the organization and segregation of the rest of the replicated chromosome (Umbarger et al. 2011). In other words, once the global orientation of chromosomes is set by polar anchoring of the origins, purely physical forces could then drive the rest of segregation. As noted, the extrusion of DNA from replication forks may help push DNA toward opposite poles. One study suggested that segregation might result largely from entropic forces, arguing that, if the chromosome is a self-avoiding polymer, the maximization of entropy will intrinsically separate two chromosomes (Jun & Mulder 2006, Jun & Wright 2010). However, entropy alone may not explain the speed of bulk chromosome segregation in E. coli. Additionally, chromosomes are thought to be self-adherent rather than self-avoiding polymers (Fisher et al. 2013, Hadizadeh Yazdi et al. 2012, Kleckner et al. 2014).
Studies in E. coli have shown that replicated sisters are initially cohesed together for ∼7-10 minutes before being rapidly segregated apart, with some origin-proximal loci remaining cohesed even longer (Joshi et al. 2011). The cohesion of DNA is modulated, at least part in E. coli, by SeqA, which binds recently duplicated, hemi-methylated DNA (Joshi et al. 2013, Sánchez-Romero et al. 2010). Additionally, sister chromosomes likely form precatenanes, structures in which the DNA from sister chromosomes effectively becomes topologically entangled (Joshi et al. 2011, 2013, Wang et al. 2008). Thus, recently replicated regions cannot separate until sisters are disentangled via topoisomerase IV. Once free of topological constraints and protein-based tethers, duplicated DNA moves bidirectionally, likely producing a more relaxed state of the nascent, sister chromosomes. In this way, the periodic build up and release of mechanical stress may ultimately drive bulk chromosome segregation (Fisher et al. 2013, Kleckner et al. 2014). Some sister loci with particularly long periods of cohesion separate very rapidly and abruptly, consistent with the study noted earlier in which certain regions of the nucleoid displayed near-ballistic movements, sometimes during chromosome segregation (Javer et al. 2014).
Notably, Hi-C analysis of Caulobacter cells progressing synchronously through the cell cycle showed that chromosomal interaction domains get re-established coincident with or shortly after replication, which may help prevent the two newly synthesized chromosomes from becoming entangled, aiding chromosome segregation (Le et al. 2013).
Although purely physical forces play a major, and perhaps dominant, role in bulk chromosome segregation, NAPs and other chromosome-associated proteins likely contribute as well (Junier et al. 2014). For instance, nucleoid-associated proteins that compact DNA, such as HU and IHF, probably facilitate the segregation of recently duplicated DNA to opposite sides of cells (Hong & McAdams 2011, Swiercz et al. 2013). Indeed, strains deleted of various NAPs often exhibit increased production of anucleate cells, an indicator of defective chromosome segregation (Huisman et al. 1989, Kaidow et al. 1995)
Similarly, supercoiling likely promotes bulk chromosome segregation by compacting DNA, and mutations in gyrase, topo IV, and topo I can each lead to defects in chromosome segregation (reviewed in Vos et al. 2011). Topo IV, which resolves the precatenanes that can form between sister chromosomes, may be particularly critical. When Topo IV activity is disrupted, cells can complete chromosome replication, but sisters often remain colocalized (Wang et al. 2008).
Another key player in chromosome segregation is the SMC/ScpA/ScpB complex, or the related MukB/MukE/MukF complex found in E. coli and other γ-proteobacteria (Fig. 5b-c) (Britton et al. 1998, Danilova et al. 2007, Jensen & Shapiro 1999, Niki et al. 1991). In E. coli, the absence of MukB prevents the formation of the usual left-ori-right chromosome organization pattern and leads to an increase in anucleate cell formation, indicating that MukB/E/F may promote proper chromosome segregation. MukBEF complexes cluster around the origin region, although the mechanism of recruitment is unknown (Danilova et al. 2007) (Fig. 5c). It also remains unclear precisely how MukBEF contributes to chromosome segregation and whether it primarily affects the origin, or whether it also contributes to bulk chromosome segregation. Additionally, a major challenge is to determine whether MukBEF promotes chromosome segregation indirectly by condensing DNA, which may make other mechanisms of segregation operate more efficiently, or whether the MukBEF complex plays a more active role in directly partitioning sister chromosomes. Recent studies have demonstrated that MukBEF directly stimulates Topo IV, implying that MukBEF contributes to the disentangling of sister chromosomes (Hayama & Marians 2010, Li et al. 2010, Nicolas et al. 2014).
In B. subtilis, SMC proteins are also thought to promote chromosome segregation, as cells lacking SMC exhibit a range of chromosome partitioning defects (Britton et al. 1998, Gruber & Errington 2009, Sullivan et al. 2009). In particular, rapid depletion of SMC has revealed a requirement of SMC for origin segregation (Gruber et al. 2014, Wang et al. 2014a,b). And, as in E. coli, SMC is recruited to the origin-proximal regions of the chromosome, via a direct interaction with ParB. However, in contrast to E. coli MukB, SMC does not appear to function by promoting Topo IV activity (Wang et al. 2014b). Instead, SMC may act primarily to condense chromosomal DNA, helping sister chromosomes from becoming entangled and increasing the overall efficiency of chromosome segregation. Notably, ChIP studies in B. subtilis showed that SMC proteins are also found at regions of high transcription. Thus, in addition to origin condensation, SMC may also aid in bulk chromosome condensation, ensuring fast and efficient segregation (Gruber & Errington 2009).
Terminus segregation
While much of chromosome segregation is accomplished by partitioning the origins and by the ensuing bulk segregation of DNA, the final segregation of chromosome termini, the ter regions, requires dedicated machinery, in part because replication of circular chromosomes can result in catenated or dimeric chromosomes if sister chromosomes recombine (Adams et al. 1992, Peter et al. 1998, Steiner & Kuempel 1998). One major component of the ter segregation apparatus is the DNA translocase FtsK, which localizes with cell division proteins to mid-cell (Bigot et al. 2007, Lesterlin et al. 2004). There, FtsK binds and may stimulate Topo IV to decatenate chromosomes (Espeli et al. 2003). Additionally, FtsK can pump chromosomal DNA to opposite sides of the cell (Lesterlin et al. 2008). This pumping also brings together, near FtsK, the ter-proximal dif loci from sister chromosomes. FtsK directly activates the tyrosine recombinase XerCD, which can resolve dimeric chromosomes by catalyzing site-specific recombination between two dif loci (Fig. 5d) (Grainge et al. 2007, Steiner et al. 1999).
Bringing the dif loci together requires that FtsK-dependent pumping be directional. In E. coli, FtsK recognizes short motifs, called KOPS (FtsK Orienting Polar Sequences) that are overrepresented in the chromosome and heavily biased in their orientation toward dif (Bigot et al. 2005, Löwe et al. 2008, Sivanathan et al. 2006). Although FtsK translocase activity is not essential in E. coli, presumably because dimeric chromosomes are produced in only ∼15% of cells per replication cycle, ftsK is essential for viability in the absence of MukBEF. This latter finding underscores the idea that FtsK contributes to bulk chromosome segregation, in addition to specifically promoting the decatentation and resolution of sister chromosomes. Moreover, FtsK is present in bacteria with linear chromosomes where decatenation and dimer resolution are not essential for terminus segregation (Chaconas & Kobryn 2010, Flärdh & Buttner 2009); in these organisms, FtsK probably functions mainly to pump DNA from sister chromosomes to opposite sides of the division plane. The FtsK homolog SpoIIIE in B. subtilis also pumps DNA, localizing to the septum formed during sporulation between a mother cell and forespore compartment. As discussed above, the polarly-localized protein RacA helps anchor one origin inside the forespore, with SpoIIIE then pumping most of the rest of the chromosome into that compartment (Fig. 5e) (Ben-Yehuda et al. 2003, Wu & Errington 1994, 1998).
In most bacteria, cytokinesis is actively delayed until sister chromosomes are fully segregated to opposite sides of the cell, preventing the guillotining of DNA. In E. coli the mechanism responsible, called nucleoid occlusion, involves a protein called SlmA that binds to specific DNA sites that are enriched in the terminus-proximal region of the chromosome. SlmA also binds to and blocks FtsZ polymerization (Bernhardt & de Boer 2005). Hence, SlmA blocks cell division until the terminus-proximal regions of the chromosome have been segregated away from mid-cell. In B. subtilis a similar mechanism occurs, involving the unrelated protein Noc, which binds to DNA sequences across the chromosome. Noc does not specifically target FtsZ, or other cell division protein, and instead appears to form large nucleoprotein complexes that physically occlude the division apparatus (Adams et al. 2015, Wu & Errington 2004, Wu et al. 2009).
Chromosome segregation in the absence of replication
Although chromosome segregation is usually concomitant with, and linked to, DNA replication, cells may sometimes need to segregate regions of their chromosomes independent of replication. For example, DNA damage can require major movements of chromosomal DNA if homologous chromosomes must pair to promote recombination-based repair. Recent work in E. coli showed that DNA near the site of a double-strand break can move, pair with its homologous partner, and then be resegregated to its approximate, original position (Lesterlin et al. 2014). This movement appears to involve large RecA filaments, but the nature of these filaments and how they drive homolog pairing, and whether they also participate in locus resegregation, is unknown as yet. Nevertheless, this initial work suggests that bacteria have mechanisms to move and resegregate portions of their chromosomes; it will be critical to determine whether the mechanisms responsible overlap with or are different from those used to drive the segregation that occurs concomitantly with DNA replication.
Future Perspectives.
There are many outstanding questions and challenges in understanding the principles and mechanisms of chromosome organization and segregation in bacteria. New, powerful tools have been developed, including Hi-C and super-resolution fluorescence microscopy, which are enabling investigations in unprecedented ways and at many different spatial scales. Studies of model organisms continue to provide new insights, with work on other species helping to reveal the general, conserved properties of bacterial chromosomes and the idiosynchracies of specific bacteria. Future work will undoubtedly continue to provide important new insights into the fundamental organization and functioning of bacterial chromosomes. Because of the central importance of chromosomes, this work promises to impact our understanding of nearly every physiological function of bacteria. Some of the immediate goals and questions for future studies are:
How does chromosome organization influence gene expression and vice versa?
How does chromosome organization, including its domain structure, influence DNA-based transactions such as DNA replication, DNA repair, and recombination?
Many NAPs are individually dispensible, but display synthetic effects when deleted in combination; how do NAPs work together to organize the chromosome, support chromosome segregation, and regulate transcription?
How do the biochemical and biophysical properties of SMC and NAPs ultimately enable the cellular-level phenomena of chromosome compaction and segregation?
How do chromosomes successfully segregate in species that do not have the ParAB-parS system?
What drives bulk chromosome segregation, and what are the relative contributions of purely physical forces and protein-based systems?
Acknowledgments
We thank X. Wang and D. Grainger for comments on the manuscript, and acknowledge H. Shin and B. Oh for providing the SMC-ScpA-ScpB structural model. This work has supported by NIH grant R01GM082899 (M.T.L), a Gordon and Betty Moore Foundation postdoctoral fellow of the Life Sciences Research Foundation (T.B.K.L), and a Human Frontiers Science Program Postdoctoral Fellowship (A.B.). M.T.L. is an Early Career Scientist of the Howard Hughes Medical Institute.
Literature Cited
- Adams D, Shekhtman E, Zechiedrich E, Schmid M, Cozzarelli N. The role of topoisomerase-IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71(2):277–88. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- Adams DW, Wu LJ, Errington J. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J. 2015 doi: 10.15252/embj.201490177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auner H, Buckle M, Deufel A, Kutateladze T, Lazarus L, et al. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J Mol Biol. 2003;331(2):331–44. doi: 10.1016/s0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- Austin SJ, Mural RJ, Chattoraj DK, Abeles AL. Trans- and cis-acting elements for the replication of P1 miniplasmids. J Mol Biol. 1985;183(2):195–202. doi: 10.1016/0022-2836(85)90212-8. [DOI] [PubMed] [Google Scholar]
- Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5(8):613–26. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181(20):6361–70. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85(1):21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121(6):899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. Cross-talk between topoisomerase I and HU in Escherichia coli. J Mol Biol. 1996;256(2):292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Fujita M, Liu XS, Gorbatyuk B, Skoko D, et al. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol Cell. 2005;17(6):773–82. doi: 10.1016/j.molcel.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299(5606):532–36. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- Berlatzky IA, Rouvinski A, Ben-Yehuda S. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci U A. 2008;105(37):14136–40. doi: 10.1073/pnas.0804982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell. 2005;18(5):555–64. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, et al. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24(21):3770–80. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64(6):1434–41. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134(6):945–55. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12(9):1254–59. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butan C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, et al. Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J Bacteriol. 2011;193(6):1341–50. doi: 10.1128/JB.01061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G, Kobryn K. Structure, function, and evolution of linear replicons in Borrelia. Annu Rev Microbiol. 2010;64(1):185–202. doi: 10.1146/annurev.micro.112408.134037. [DOI] [PubMed] [Google Scholar]
- Chen S, Calvo JM. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J Mol Biol. 2002;318(4):1031–42. doi: 10.1016/S0022-2836(02)00187-0. [DOI] [PubMed] [Google Scholar]
- Claret L, Rouviere-Yaniv J. Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J Mol Biol. 1997;273(1):93–104. doi: 10.1006/jmbi.1997.1310. [DOI] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJL. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444(7117):387–90. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28(18):3504–10. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65(6):1485–92. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries R. DNA condensation in bacteria: Interplay between macromolecular crowding and nucleoid proteins. Biochimie. 2010;92(12):1715–21. doi: 10.1016/j.biochi.2010.06.024. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8(3):185–95. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- Di Ventura B, Knecht B, Andreas H, Godinez WJ, Fritsche M, et al. Chromosome segregation by the Escherichia coli Min system. Mol Syst Biol. 2013;9:686. doi: 10.1038/msb.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaigne P, Tonthat NK, Espéli O, Whitfill T, Boccard F, Schumacher MA. Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome. Mol Cell. 2012;48(4):560–71. doi: 10.1016/j.molcel.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P. Streptomyces: Molecular Biology and Biotechnology. Horizon Scientific Press; 2011. [Google Scholar]
- Easter J, Gober JW. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell. 2002;10(2):427–34. doi: 10.1016/s1097-2765(02)00594-4. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134(6):956–68. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Ringgaard S, Møller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol Microbiol. 2006;61(6):1428–42. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- Espeli O, Levine C, Hassing H, Marians KJ. Temporal regulation of topoisomerase IV activity in E. coli. Mol Cell. 2003;11(1):189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol. 2008;68(6):1418–27. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- Fekete RA, Chattoraj DK. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol. 2005;55(1):175–83. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Keren K, Theriot JA. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol Microbiol. 2006;60(5):1164–78. doi: 10.1111/j.1365-2958.2006.05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JK, Bourniquel A, Witz G, Weiner B, Prentiss M, Kleckner N. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153(4):882–95. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7(1):36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20(23):3269–82. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Minsky A. Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis and D. radiodurans. J Struct Biol. 2006;156(2):311–19. doi: 10.1016/j.jsb.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Sack R, Englander J, Shimoni E, Eisenstein M, et al. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J Bacteriol. 2004;186(11):3525–30. doi: 10.1128/JB.186.11.3525-3530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141(6):927–42. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Graham TGW, Wang X, Song D, Etson CM, van Oijen AM, et al. ParB spreading requires DNA bridging. Genes Dev. 2014;28(11):1228–38. doi: 10.1101/gad.242206.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainge I, Bregu M, Vazquez M, Sivanathan V, Ip SC, Sherratt DJ. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26(19):4228–38. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Goldberg MD, Busby SJW. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34(16):4642–52. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJW. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci U S A. 2005;102(49):17693–98. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137(4):685–96. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, Errington J. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr Biol. 2014;24(3):293–98. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Adhya S. Spiral structure of Escherichia coli HUαβ provides foundation for DNA supercoiling. Proc Natl Acad Sci U S A. 2007;104(11):4309–14. doi: 10.1073/pnas.0611686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Sajid A, Sharma K, Ghosh S, Arora G, et al. HupB, a nucleoid-associated protein of Mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J Bacteriol. 2014;196(14):2646–57. doi: 10.1128/JB.01625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF. Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol. 2012;86(6):1318–33. doi: 10.1111/mmi.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CD, Cozzarelli NR. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57(6):1636–52. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- Harms A, Treuner-Lange A, Schumacher D, Sogaard-Andersen L. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 2013;9(9):e1003802. doi: 10.1371/journal.pgen.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107(44):18826–31. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, et al. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52(4):569–84. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J Bacteriol. 1996;178(10):2825–35. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, McAdams HH. Compaction and transport properties of newly replicated Caulobacter crescentus DNA. Mol Microbiol. 2011;82(6):1349–58. doi: 10.1111/j.1365-2958.2011.07899.x. [DOI] [PubMed] [Google Scholar]
- Huisman O, Faelen M, Girard D, Jaffé A, Toussaint A, Rouvière-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol. 1989;171(7):3704–12. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LC, Vecchiarelli AG, Han YW, Mizuuchi M, Harada Y, et al. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013;32(9):1238–49. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NW, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176(17):5320–29. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol. 1963;28:329–48. [Google Scholar]
- Jain IH, Vijayan V, O'Shea EK. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc Natl Acad Sci U A. 2012;109(34):13638–43. doi: 10.1073/pnas.1211144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, Żydek P, Kois A, Zakrzewska-Czerwińska J, Chater KF. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol Microbiol. 2007;65(3):625–41. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
- Javer A, Kuwada NJ, Long Z, Benza VG, Dorfman KD, et al. Persistent super-diffusive motion of Escherichia coli chromosomal loci. Nat Commun. 2014;5:3854. doi: 10.1038/ncomms4854. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U A. 1999;96(19):10661–66. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Wang SC, Shapiro L. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 2001;20(17):4952–63. doi: 10.1093/emboj/20.17.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, et al. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci. 2011;108(7):2765–70. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi MC, Magnan D, Montminy TP, Lies M, Stepankiw N, Bates D. Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 2013;9(8):e1003673. doi: 10.1371/journal.pgen.1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier I, Boccard F, Espéli O. Polymer modeling of the E. coli genome reveals the involvement of locus positioning and macrodomain structuring for the control of chromosome conformation and segregation. Nucleic Acids Res. 2014;42(3):1461–73. doi: 10.1093/nar/gkt1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci U A. 2006;103(33):12388–93. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Microbiol. 2010;8(8):600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee ASN, Prieto AI, Ibberson D, Schmidt S, et al. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39(6):2073–91. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidow A, Wachi M, Nakamura J, Magae J, Nagai K. Anucleate cell production by Escherichia coli Δhns mutant lacking a histone-like protein, H-NS. J Bacteriol. 1995;177(12):3589–92. doi: 10.1128/jb.177.12.3589-3592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Edgar R, Adhya S. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc Natl Acad Sci U S A. 2005;102(45):16397–402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavenoff R, Ryder OA. Electron microscopy of membrane-associated folded chromosomes of Escherichia coli. Chromosoma. 1976;55(1):13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Fisher JK, Stouf M, White MA, Bates D, Witz G. The bacterial nucleoid: nature, dynamics and sister segregation. Curr Opin Microbiol. 2014;22:127–37. doi: 10.1016/j.mib.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux G, Jacobs-Wagner C. Spatiotemporal control of PopZ localization through cell cycle–coupled multimerization. J Cell Biol. 2013;201(6):827–41. doi: 10.1083/jcb.201303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282(5393):1516–19. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28(15):2272–82. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol Microbiol. 2005;55(4):978–85. doi: 10.1111/j.1365-2958.2004.04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Löwe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer--a conserved biological switch. EMBO J. 2005;24(2):270–82. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506(7487):249–53. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Barre FX, Cornet F. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol Microbiol. 2004;54(5):1151–60. doi: 10.1111/j.1365-2958.2004.04356.x. [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F. Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet. 2008;4(12):e1000288. doi: 10.1371/journal.pgen.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342(6159):731–34. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TB, Laub MT. New approaches to understanding the spatial organization of bacterial genomes. Curr Opin Microbiol. 2014;22C:15–21. doi: 10.1016/j.mib.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 2000;19(4):710–18. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby EA, Roggiani M, Goulian M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc Natl Acad Sci U S A. 2012;109(19):7445–50. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92(5):675–85. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol. 2007;189(23):8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Stewart NK, Berger AJ, Vos S, Schoeffler AJ, et al. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc Natl Acad Sci. 2010;107(44):18832–37. doi: 10.1073/pnas.1008678107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell. 2008;31(4):498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2(8):e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. Histone-like protein HU and bacterial DNA topology: suppression of an HU deficiency by gyrase mutations. J Mol Biol. 1996;256(1):66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- Mera PE, Kalogeraki VS, Shapiro L. Replication initiator DnaA binds at the Caulobacter centromere and enables chromosome segregation. Proc Natl Acad Sci U S A. 2014;111(45):16100–105. doi: 10.1073/pnas.1418989111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, et al. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135(3):475–85. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Easter J, Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42(3):741–55. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, DiMario PJ, Grove A. Mycobacterium smegmatis histone-like protein Hlp is nucleoid associated. FEMS Microbiol Lett. 2009;291(2):232–40. doi: 10.1111/j.1574-6968.2008.01458.x. [DOI] [PubMed] [Google Scholar]
- Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006;25(19):4586–95. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135(1):74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Murray H, Ferreira H, Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol. 2006;61(5):1352–61. doi: 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313(5784):236–38. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev MMBR. 2000;64(3):548–72. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Upton AL, Uphoff S, Henry O, Badrinarayanan A, Sherratt D. The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. mBio. 2014;5(1):e01001–13. doi: 10.1128/mBio.01001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006a;61(2):383–93. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006b;62(2):331–38. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- Niki H, Jaffé A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 Kda protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10(1):183–93. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14(2):212–23. [PMC free article] [PubMed] [Google Scholar]
- Nolivos S, Sherratt D. The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol Rev. 2014;38(3):380–92. doi: 10.1111/1574-6976.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, et al. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29(12):1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94(6):819–27. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18(14):1766–79. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee ASN, Luscombe NM. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012;40(8):3524–37. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Gahlmann A, Bowman GR, Perez AM, van Diezmann ARS, et al. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci. 2014;111(19):E2046–55. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12(8):791–98. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106(32):13541–45. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133(1):90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87(7):1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci U S A. 2009;106(46):19369–74. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow C, Kellenberger E. The bacterial nucleoid revisited. Microbiol Rev. 1994;58(2):211–32. doi: 10.1128/mr.58.2.211-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno P, Larsson J, Bucca G, Laing E, Smith CP, Flärdh K. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J Bacteriol. 2009;191(21):6489–6500. doi: 10.1128/JB.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Romero MA, Busby SJW, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of SeqA protein across the chromosome of Escherichia coli K-12. mBio. 2010;1(1):e00012–10. doi: 10.1128/mBio.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Austin S. Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci U S A. 2000;97(4):1671–76. doi: 10.1073/pnas.030528397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Travers A, Kutateladze T, Muskhelishvili G. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol Microbiol. 1999;34(5):953–64. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29(18):3068–81. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol. 2011;79(4):1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Shapiro L. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol Microbiol. 2011;82(6):1359–74. doi: 10.1111/j.1365-2958.2011.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebelut CW, Guberman JM, van Teeffelen S, Yakhnina AA, Gitai Z. Caulobacter chromosome segregation is an ordered multistep process. Proc Natl Acad Sci U S A. 2010;107(32):14194–98. doi: 10.1073/pnas.1005274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR, Pettijohn DE. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981;78(1):224–28. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014;28(3):214–19. doi: 10.1101/gad.234336.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanathan V, Allen MD, de Bekker C, Baker R, Arciszewska LK, et al. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol. 2006;13(11):965–72. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Grossman AD. The Transcriptional regulator rok binds A+T-Rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6(11):e1001207. doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W, Liu G, Donachie WD, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31(2):579–83. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998;180(23):6269–75. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RA, Deng S, Higgins NP. Measuring chromosome dynamics on different time scales using resolvases with varying half-lives. Mol Microbiol. 2005;56(4):1049–61. doi: 10.1111/j.1365-2958.2005.04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Cascio D, Johnson RC. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24(8):814–26. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137(4):697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiercz JP, Nanji T, Gloyd M, Guarné A, Elliot MA. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 2013;41(7):4171–84. doi: 10.1093/nar/gkt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU–DNA cocrystal structures. EMBO J. 2003;22(14):3749–60. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse S, Mascarenhas J, Kösters B, Hasilik A, Graumann PL. Genetic interaction of the SMC complex with topoisomerase IV in Bacillus subtilis. Microbiology. 2005;151(11):3729–37. doi: 10.1099/mic.0.28234-0. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126(1):147–62. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Thiel A, Valens M, Vallet-Gely I, Espéli O, Boccard F. Long-range chromosome organization in E. coli: a site-specific system isolates the Ter macrodomain. PLoS Genet. 2012;8(4):e1002672. doi: 10.1371/journal.pgen.1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro E, Hong SH, McAdams HH, Shapiro L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci. 2008;105(40):15435–40. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Huang CH, Tessmer I, Erie DA, Chen CW. Linear Streptomyces plasmids form superhelical circles through interactions between their terminal proteins. Nucleic Acids Res. 2011;39(6):2165–74. doi: 10.1093/nar/gkq1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44(2):252–64. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23(21):4330–41. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013;9(5):e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Han YW, Tan X, Mizuuchi M, Ghirlando R, et al. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78(1):78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Mizuuchi K, Funnell BE. Surfing biological surfaces: exploiting the nucleoid for partition and transport in bacteria. Mol Microbiol. 2012;86(3):513–23. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Neuman KC, Mizuuchi K. A propagating ATPase gradient drives transport of surface-confined cellular cargo. Proc Natl Acad Sci. 2014;111(13):4880–85. doi: 10.1073/pnas.1401025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U A. 2004;101(25):9257–62. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12(12):827–41. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333(6048):1445–49. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20(13):1727–31. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci U S A. 2014a;111(35):12877–82. doi: 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22(17):2426–33. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Rudner DZ. Spatial organization of bacterial chromosomes. Curr Opin Microbiol. 2014;22:66–72. doi: 10.1016/j.mib.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sherratt DJ. Independent segregation of the two arms of the Escherichia coli ori region requires neither RNA synthesis nor MreB dynamics. J Bacteriol. 2010;192(23):6143–53. doi: 10.1128/JB.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014b;24(3):287–92. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci U A. 2012;109(19):7338–43. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T, Nordström K, Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol Microbiol. 1999;34(1):157–68. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci U S A. 2010;107(11):4991–95. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400(6739):83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- Wright MA, Kharchenko P, Church GM, Segrè D. Chromosomal periodicity of evolutionarily conserved gene pairs. Proc Natl Acad Sci U S A. 2007;104(25):10559–64. doi: 10.1073/pnas.0610776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264(5158):572–75. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27(4):777–86. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol Microbiol. 2003;49(6):1463–75. doi: 10.1046/j.1365-2958.2003.03643.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117(7):915–25. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28(13):1940–52. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Bruckner R, Ringgaard S, Möll A, Cameron DE, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26(20):2348–60. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Niki H. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 2004;23(1):221–33. doi: 10.1038/sj.emboj.7600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MC, Losick R. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J Bacteriol. 2001;183(17):5180–86. doi: 10.1128/JB.183.17.5180-5186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren B, Nielsen HJ, Jun S, Austin S. The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 2014;28(1):71–84. doi: 10.1101/gad.231050.113. [DOI] [PMC free article] [PubMed] [Google Scholar]