Abstract
Chemical probes are powerful reagents with increasing impacts on biomedical research. However, probes of poor quality or that are used incorrectly generate misleading results. To help address these shortcomings, we will create a community-driven wiki resource to improve quality and convey current best practice.
About a decade ago, academia substantially increased its efforts in chemical biology and drug discovery. These efforts arose in part because of the availability of large numbers of uncharacterized potential drug targets emerging from genome sequencing efforts, from the development and commoditization of new screening technologies and from the possibility of inventing new medicines. Some of these efforts, perhaps in appreciation of the complexity and capriciousness of drug discovery, set out a more measured objective: to generate small-molecule tools (chemical probes) to help elucidate the roles of the targeted proteins in healthy and diseased cells and tissues.
Over the past decade, we have learned some important lessons from these forays into chemical biology. First, chemical biology has had a major impact on our understanding of human biology and the treatment of human disease. New chemical biology technologies, such as the cellular thermal shift assay for assessing direct target engagement in cells1 and click chemistry as a means for bioorthogonal functionalization2, are increasingly used in the broader scientific community. High-quality chemical probes have served both as powerful research tools and as seeds to spur the development of new medicines (Table 1).
Table 1. Examples of high-impact chemical probes.
| Probe | Target | Mode of action |
|---|---|---|
| (+)-JQ1, I-BET, PFI-1 (ref. 4) | BET family bromodomains | Inhibitor |
| Rapamycin | mTOR | Allosteric inhibitor |
| GW683965 (ref. 12) | LXRα and LXRβ | Agonist |
| PF-04457845 (ref. 34) | Fatty acid amide hydrolase | Irreversible inhibitor |
| GNF-5 (ref. 35) | Bcr-Abl | Allosteric inhibitor |
| Cyclopamine | Smoothened | Orthosteric inhibitor |
Second, chemical reagents, akin to any other protein-targeted reagent, are only useful if they are potent, have known selectivity and have a proven mechanism of action. During the past decade, we have gained a greater appreciation that probes of this quality are difficult to produce and require substantial resources, commitment and skills. We learned that many of the chemical probes in use today had initially been characterized inadequately and have since been proven to be nonselective or associated with poor characteristics such as the presence of reactive functionality that can interfere with common assay features3 (Table 2). The continued use of these probes poses a major problem: tens of thousands of publications each year use them to generate research of suspect conclusions, at great cost to the taxpayer and other funders, to scientific careers and to the reliability of the scientific literature.
Table 2. Examples of widely used low-quality probes.
| Compound | Putative target | Problems |
|---|---|---|
| Flavones | Many, varied | Often promiscuous and can be pan-assay interfering (PAINS) compounds |
| Epigallocatechin-3-gallate | DYRK1A | Promiscuous PAINS compound |
| LiCl | GSK3β | Typically used at high (mM) concentrations; known to inhibit other targets36 |
| WY14643 (ref. 37) | PPARα | Significant activity difference in human versus murine orthologs of target |
| Valproicacid38 | HDAC | Used at concentration regimes (mM) where nonspecific mechanisms are likely |
| Resveratrol | Sirtuin | Assay artifact39 |
Third, attempts by experts to disseminate accurate and reliable information to the research community regarding both well-characterized and poorly characterized chemical probes do not seem to be having sufficient impact. Despite a large number of outstanding reviews on the aspirational properties of high-quality probes4–6, excellent papers describing frequently occurring artifacts in chemical screening and chemical biology3,7 and countless ‘case-by-case’ papers describing the serious deficiencies of specific chemical probes, nonselective and/or poorly characterized compounds continue to be widely used. Thus, the evidence suggests that the literature is an ineffective vehicle to provide guidance to the community about the quality of new chemical probes or to reduce the use of low-quality chemical probes. We argue that a complementary approach is needed.
In this Commentary, we will first, for clarity, provide a working definition of a ‘chemical probe’ and then, for perspective, highlight some selected examples of high-quality chemical probes (those that are currently believed to be) and probes of lesser value. We will then describe our plans to create a web-based resource annotated by the chemical biology community comprising the most appropriate chemical probe (or probes) for a given protein target. This site, which we have named the Chemical Probes Portal, will be available to scientists, reviewers and editors to aid in their experiments and deliberations.
What is a chemical probe?
A chemical probe is simply a reagent—a selective small-molecule modulator of a protein's function that allows the user to ask mechanistic and phenotypic questions about its molecular target in biochemical, cell-based or animal studies. Chemical probes have proven to be very impactful not only because they are complementary to genetic approaches, such as CRISPR and RNAi8, but also because they have unique advantages. They can rapidly and reversibly inhibit a protein or a protein domain in cells or animals, be used in almost any cell type and reveal temporal features of target inhibition. When coupled with RNAi, they can distinguish between effects due to scaffolding and effects due to inhibition of catalytic or protein-interaction activity. In this way, chemical probes can be quite effective at invalidating drug targets9. Multiple chemical probes can also be used in synthetic lethal screens to investigate the connectivity between distinct pathways. Finally, and importantly, the results obtained with chemical probes are more relevant for translational studies as they are more likely to mimic the pharmacology realized when a therapeutic small-molecule drug is used.
Only high-quality chemical probes generate meaningful biological data. Quality is a difficult parameter to quantify, but excellent guides to the properties required in useful chemical probes have been put forth by the expert community5,6. As a working example, within the Structural Genomics Consortium (http://www.thesgc.org/), a large collaboration of academic and industry medicinal chemists and chemical biologists, probes for epigenetics targets are required minimally to have in vitro potency at the target protein of <100 nM, possess >30-fold selectivity relative to other sequence-related proteins of the same target family, be profiled against an ‘industry standard’ selection of pharmacologically relevant off-targets (for example, http://www.eurofins.com/pharma-services/pharma-discovery-services/services/in-vitro-pharmacology.aspx) and against large protein families of relevance to drug discovery and, finally, have demonstrated on-target effects in cells at <1 μM. The consortium has strongly encouraged and now requires that the chemical probe be accompanied by an inactive close analog of the compound to serve as a negative control. Chemical probes for other protein targets or for use in animal models would require different criteria as appropriate, but all should go through similar profiling cascades.
Providing one chemical probe for a given protein is also insufficient. Each protein should be targeted by another equally well-characterized ‘orthogonal’ chemical probe having a completely different chemical structure which would reduce the probability of having common off-targets.
Historically, demonstrating on-target activity in cells, and especially in animal models, has proven very challenging, particularly for those proteins with no known cellular function. Although this task still remains a challenge, chemical biologists can now tackle the problem with any of a suite of genetic10, chemical proteomic and biophysical assays11 and ideally using a combination of orthogonal methods. The use of molecular genetics to map or engineer drug resistance is also a powerful way to link the target, the chemical probe and the bioactivity.
In summary, for any given protein, the ideal scenario would be to have two structurally distinct chemical probes, each with singular activity and exquisite selectivity, as well as two inactive derivatives. However, this aim is often neither realistic nor always necessary. Indeed, for each chemical probe, it is only reasonable to expect that the most comprehensive analysis possible is made available when the probe is generated, that the counter-screening data are made available and that probes are subjected to ongoing and openly shared characterization as new technologies and screening panels emerge. In some instances, these follow-on studies might highlight serious deficiencies that reduce or eliminate the utility of the probe in certain settings or experimental systems, or set the stage for medicinal chemistry efforts to produce more potent or selective second-generation probes. Of course, first-generation probes with known off-target activities might still be useful provided that the off-target activities are judged to be irrelevant to the experiment at hand.
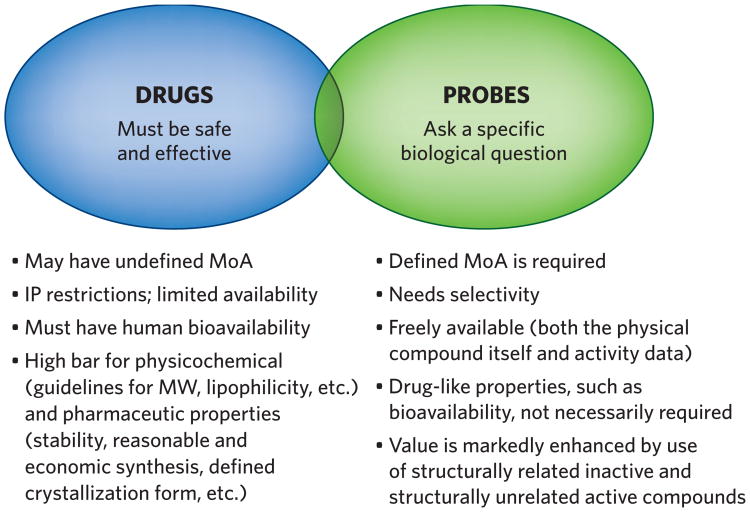
It is also important to understand that small-molecule drugs and chemical probes can be very different in their characteristics and their purposes (Fig. 1). For example, a drug need not have a selective activity profile, and indeed many medicines manifest their clinical effects through polypharmacology. In turn, chemical probes do not need to meet the same requirements as a successful medicine, such as good pharmacodynamics and oral bioavailability, but they must exhibit high potency, selectivity and on-target action to be useful probes of biological questions.
Figure 1.
Different purposes and requirements for chemical probes and drugs. IP, intellectual property; MoA, mechanism of action; MW, molecular weight.
High quality means high impact
Chemical probes have had a major impact in enabling and accelerating discoveries along the path to pioneer medicines (Table 1). They have helped to improve our understanding of targets and pathways and have created opportunities for proprietary drug discovery efforts to an extent that would not have been possible otherwise (Fig. 2a).
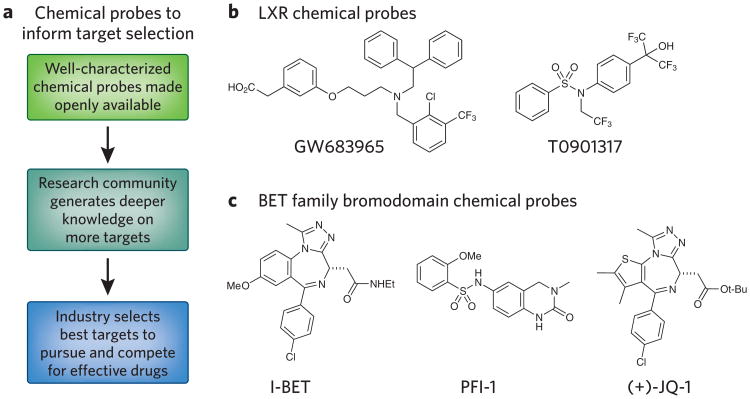
Figure 2.
Chemical probes are valuable research tools. (a) Open access to quality chemical probes can inform initial target selection. (b,c) Examples of chemical probes for the LXRs (b) and BET family bromodomains (c).
For example, the release of chemical probes for several orphan nuclear receptors led to a large increase in drug discovery efforts in this target family. One example is the liver X receptor (LXR), which was the target of the agonist GW683965 (Table 1 and Fig. 2b). This compound was optimized by iterative medicinal chemistry both for cell potency (submicromolar half-maximum effective concentration) and oral bioavailability in mice. In conjunction with the structurally distinct LXR agonist T0901317, these compounds made an excellent toolset to elucidate LXR biology and to uncover its potential as a therapeutic target12. On the basis of these data, the research community produced data suggesting the LXRs as potential therapeutic targets in inflammation, atherosclerosis and Alzheimer's disease, and several LXR agonists have been progressed for evaluation in humans13.
The BET family bromodomain probes (+)-JQ1, I-BET and PFI-1 constitute another example of the impact made by quality chemical probes (Table 1 and Fig. 2c)4. The availability of these compounds, accompanied by structurally related inactive negative control compounds, enabled the research community to interrogate BET family function in diverse areas such as oncology, inflammation, virology and male contraception as well as identify several opportunities for drug discovery14. As a specific consequence of openly publishing this work and making the tools available for independent validation, several established pharmaceutical companies and start-up companies have initiated discovery and clinical programs targeting BET proteins only a few years after the initial publication of the chemical probes15.
More generally, evidence presented elsewhere16 demonstrates that the availability of a high-quality chemical probe (or probes) for a target greatly stimulates research activity on that protein. Indeed, bibliometric evidence suggests that chemical probes comprise the most impactful of all classes of biomedical reagents, as judged by citations and usage16. Unfortunately, the impact of chemical probes also extends to those probes of lesser quality.
Caveat emptor
In the previous section, we highlighted some examples of well-characterized chemical probes and their impact on scientific understanding and translational medicine. Unfortunately, most chemical inhibitors are not characterized appropriately or have outdated characterization. As a result, they may have major off-target properties, and their use can contribute to misleading or incorrect conclusions.
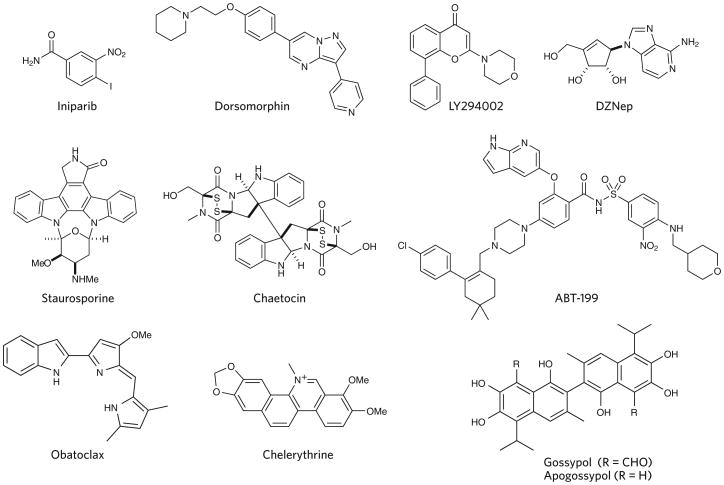
Another set of compounds in widespread use (Table 2) can deregulate biological systems nonspecifically, for example, by affecting the redox state, by forming covalent or irreversible adducts with large numbers of proteins, or by forming aggregates3,7; these compounds are frequent hits in phenotypic screens. To the experienced chemist, these classes of compounds are well known as molecules either to avoid if possible or to be treated very cautiously, yet they are used profligately in the literature. A lamentable example is BSI-201 (iniparib; Fig. 3), which was developed and advertised as a PARP inhibitor, used in thousands of publications and progressed to phase 3 clinical trials, where it failed, and was only later shown to modify cysteine-containing proteins nonspecifically17.
Figure 3.
Structures for selected compounds.
In the following three sections, we highlight a few of the many egregious examples, selecting from different areas of cell biology.
Cellular signaling
The discoveries that protein kinases could be inhibited weakly by isoquinolinesulfonamides18 and potently by the bacterial natural product staurosporine at nanomolar concentrations19 were true breakthroughs, revealing that protein kinases were ‘druggable’ in cellular systems and paving the way to the discovery of approximately 30 approved drugs that target the protein kinase domain20. Staurosporine was initially described as an inhibitor of protein kinase C (PKC), but over the following decades, as the molecule was characterized more fully and ever-larger kinase screening panels became available, it became clear that it was not selective for PKC and was instead a pan-kinase inhibitor. At some time in this period, the molecule should have been discarded in favor of more selective inhibitors that were being discovered. Unfortunately, this did not happen.
The misuse of many other early kinase inhibitors continues to plague the literature. For instance, LY294002 was originally described in 1994 as a selective inhibitor of PI3 kinase21 and remains advertised as such by nearly all vendors. Yet by 2005, it was already clear that the compound inhibited many other proteins at the concentrations used to inhibit PI3 kinase22. In the meantime, a large number of more selective and more well-characterized PI3 kinase inhibitors have become available. The availability of these new inhibitors certainly obviated the need for LY294002 as a chemical probe, and it should be discarded as a selective research tool. Yet a search of Google Scholar in 2014–2015 alone for ‘LY294002 and PI3 kinase’ returned ∼1,100 documents.
Dorsomorphin is a chemical probe that was first published as a nanomolar inhibitor of TGF-β signaling and is advertised and used as such23. It was also published as a nanomolar inhibitor of AMPK signaling and is advertised and used as such24. In 2014–2015, a search of Google Scholar retrieved ∼300 documents using dorsomorphin as a probe of either TGF-β receptor kinases or AMPK. Which of these activities is responsible for the observed biological effects? Perhaps neither. There are at least ten other kinases that are more potently inhibited by dorsomorphin than either AMPK or TGF-β receptor kinases25.
A brief review of the protein kinase literature reveals that these are not isolated cases; many nonselective and insufficiently characterized inhibitors continue to be used as tools to connect specific kinases to biological effects.
Epigenetics
In 1986, DZNep was published as a picomolar inhibitor of S-adenosyl homocysteine (SAH) hydrolase26, a key enzyme involved in the biosynthetic pathway of S-adenosyl methionine, the cofactor of nearly all cellular methyltransferases. In 2007, when used at a concentration a million times higher than that required to inhibit SAH hydrolase in vitro, DZNep was reported to reduce methylation of histones, including H3K27, the target of the EZH2 methyltransferase, ostensibly by downregulating expression of the EZH2 methyltransferase27. Subsequent publications using DZNep have erroneously implied and interpreted data as though DZNep is a catalytic inhibitor of EZH2. A Google Scholar search for ‘DZNep and EZH2’ publications in the past year returned ∼250 documents. A search for EZH2 together with the higher-quality inhibitors EPZ005687, EPZ-6438, GSK343 or GSK126 returned ∼400 documents over the same period.
The natural product chaetocin contains a pair of disulfide bonds, a substructure that can confound assays through nonspecific redox behavior, covalent modification or both28. Chaetocin was reported in 2005 to be a selective inhibitor of the Drosophila histone methyltransferase SU(VAR)3-9 (ref. 28). In the intervening years, the community realized that this compound indeed had activity on many other proteins, and in 2013 chaetocin was shown to form covalent adducts with numerous proteins, most likely explaining its promiscuity29,30 and confirming its lack of utility as a valuable chemical probe. Nevertheless, a Google Scholar search for ‘chaetocin and histone’ returned ∼100 documents since 2014.
Apoptosis
The relevance of apoptosis as a target for therapy is being tested through the use of ABT-199, also known as GDC-0199 (venetoclax, a nanomolar inhibitor of the BH3–Bcl-2 interaction that is in phase 3 clinical trials in a variety of cancers31). ABT-263 and ABT-737, earlier inhibitors of Bcl-2 family proteins that satisfy all of the criteria of high-quality probes, are triumphs of chemical biology and modern drug discovery. They are rationally designed potent and selective protein-protein interaction inhibitors whose mechanisms of action have conclusively been shown to be due to inhibition of the relevant target in cells.
Other reported bioactive inhibitors of the BH3–Bcl-2 interaction, including obatoclax, chelerythrine, EM20-25, gossypol and apogossypol, are inadequate as probes. Indeed, since 2009, we have known that none of these agents exert their biological effects through Bcl-2 alone and perhaps not through Bcl-2 at all32, and many of these inhibitors contain structural motifs that would raise concern with any experienced medicinal chemist3. Nevertheless, a Google Scholar search for any of these compounds and Bcl in 2014 or 2015 returned about as many documents as did a search for ‘ABT-737 and Bcl’.
The sins of the past
On the basis of these examples, our collective experience over the past decade or so highlights four factors that contribute to the continued use of nonselective chemical probes.
First, it is difficult for the research community to keep abreast of recent developments and to remain well informed about the most appropriate probe for a given target. Chemical probe experts, including the authors here, are routinely queried as to whether there is a high-quality probe available for a given target or for advice on which probe to use. In the absence of expert advice, and with no other recourse available, the selection of a probe compound seems to be guided by precedent and availability rather than appropriateness or quality.
Second, some of the high-quality probes are not commercially available and are beyond the reach of scientists who may not have access to synthetic chemistry expertise. On other rare occasions, commercially available probes are of insufficient purity, stability or quality; sometimes what is in the bottle is not what is printed on the label33.
Third, even if high-quality probes are available, they are sometimes used at concentrations at which they become nonselective and render the biological insights derived from these experiments uninterpretable.
Fourth, dissemination of these insights and provision of guidelines via the peer-reviewed literature has clearly proven inadequate to improve the situation. As shown above, recent papers (and grant applications) making mechanistic conclusions based on the use of nonselective probes continue to be published by the thousands despite clear and convincing publications that point out their flaws. The financial implications are especially important to note as these studies are often supported by public funds.
Toward a rosier future
“Discontent is the first necessity of progress.”
—Thomas Edison
The well-documented use of suboptimal probes and the misuse of high-quality probes suggest that alternative mechanisms to disseminate information are necessary. We believe that an expert community-driven wiki-like site is one possible solution and one that we will implement (http://www.chemicalprobes.org/). In this resource, which we call the Chemical Probes Portal, we plan to crowdsource medicinal chemistry and pharmacology expertise to answer the most common questions we receive: Is there a probe for my target protein? Which ones should I use? How should I use this probe properly? Is this probe suitable for use in animal models?
In this resource we will suggest the best available chemical probe (or probes) for a protein target and ensure that each probe is accompanied by the most current available information on activity and selectivity, including information about the best available compounds to use as controls. We also expect to provide experimental guidance on how to best use the probes. For those proteins that have suboptimal but still useful probe compounds available, we will describe both their benefits and limitations, along with specific guidance on their use. Ideally, the scientific community will reciprocate by adding its feedback and by placing any new data into any of the outstanding chemical biology databases (PubChem, ChEMBL and so on).
The authors will oversee the generation and maintenance of this resource to begin with, but ultimately its success depends on community input (‘wikification’) and use. This resource will need to be supported and used by academic and industrial researchers, publishers, funders and investors, groups that share a common interest in increasing the reliability of the published literature and the robustness of target validation. As with many other wiki resources, the chemical probe validation information from the community would be curated by specialists with expertise about the probes in question.
The Chemical Probes Portal will also help address the challenges faced by peer reviewers in evaluating grants or manuscripts that describe or make use of small-molecule tools. However, our resource may be insufficient or incomplete and cannot be used to judge new chemical probes, and thus we also recommend that reviewers and editors provide a checklist of areas to address in the preparation and review of manuscripts where chemical probes are generated and used to drive key biological conclusions (Box 1). Additionally, because the communities that generate chemical probes and those that use them are often disconnected, we strongly believe that no paper where a chemical probe is reported or used should be reviewed without including someone who is deeply familiar with proper usage of chemical probes, especially when a tool is used in animal models where pharmacokinetics and metabolism will have dominant roles. We also encourage vendors to provide selectivity and metabolic stability profiles and realistic guidelines indicating relevant concentrations for cell culture or animal studies. Where available, vendors are also encouraged to offer the inactive analog of the probe for use as a negative control.
Box 1. Checklist for chemical probe–based experiments.
As authors, editors and referees evaluate studies containing experiments using chemical probes, they should ask themselves the following questions:
Is the potency and selectivity of the probe suitable for drawing the conclusions of the experiment?
Is the probe used at an appropriate concentration relative to its XC50 (the concentration at which half-maximal activity change is observed, encompassing both IC50 and EC50) values at the primary protein target and any known off-target proteins?
Is evidence presented that the chemical probe is engaging its target in cells?
Are the appropriate control compounds used? Specifically, does the study include a structurally related inactive compound for the same target? Does it include parallel data with a structurally unrelated chemical probe?
Is the source and purity of the compound documented?
Is the chemical structure of the probe compound (including stereoisomerism, if applicable) reported?
If planning in vivo experiments, does the probe have appropriate pharmacokinetics and pharmacology to be utilized in animal models?
Chemical probes can be powerful research tools in studies of protein function. The resource that we outline here will increase the proper use of the best available probe compounds and will reduce the use of inadequate probes. If successful, it will help to increase understanding of fundamental biology and identify new therapeutic opportunities for the discovery of medicines.
Footnotes
Competing financial interests: The authors declare competing financial interests: details accompany the online version of the paper.
References
- 1.Martinez Molina D, et al. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 2.Thirumurugan P, Matosiuk D, Jozwiak K. Chem Rev. 2013;113:4905–4979. doi: 10.1021/cr200409f. [DOI] [PubMed] [Google Scholar]
- 3.Baell J, Walters MA. Nature. 2014;513:481–483. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 4.Bunnage ME, Chekler EL, Jones LH. Nat Chem Biol. 2013;9:195–199. doi: 10.1038/nchembio.1197. [DOI] [PubMed] [Google Scholar]
- 5.Frye SV. Nat Chem Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 6.Workman P, Collins I. Chem Biol. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassano MF, Doak AK, Roth BL, Shoichet BK. J Med Chem. 2013;56:2406–2414. doi: 10.1021/jm301749y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss WA, Taylor SS, Shokat KM. Nat Chem Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sweis RF. ACS Med Chem Lett. 2015;6:618–621. doi: 10.1021/acsmedchemlett.5b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark K, et al. Proc Natl Acad Sci USA. 2012;109:16986–16991. doi: 10.1073/pnas.1215450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon GM, Niphakis MJ, Cravatt BF. Nat Chem Biol. 2013;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins JL, et al. J Med Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 13.Hong C, Tontonoz P. Nat Rev Drug Discov. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- 14.Filippakopoulos P, Knapp S. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 15.Garnier JM, Sharp PP, Burns CJ. Expert Opin Ther Pat. 2014;24:185–199. doi: 10.1517/13543776.2014.859244. [DOI] [PubMed] [Google Scholar]
- 16.Edwards AM, et al. Nature. 2011;470:163–165. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, et al. Clin Cancer Res. 2012;18:510–523. doi: 10.1158/1078-0432.CCR-11-1973. [DOI] [PubMed] [Google Scholar]
- 18.Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 19.Tamaoki T, et al. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 20.Knapp S, Sundstrom M. Curr Opin Pharmacol. 2014;17:58–63. doi: 10.1016/j.coph.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 21.Vlahos CJ, Matter WF, Hui KY, Brown RF. J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 22.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Cancer Res. 2010;70:2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu PB, et al. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou G, et al. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogt J, Traynor R, Sapkota GP. Cell Signal. 2011;23:1831–1842. doi: 10.1016/j.cellsig.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Glazer RI, et al. Biochem Biophys Res Commun. 1986;135:688–694. doi: 10.1016/0006-291x(86)90048-3. [DOI] [PubMed] [Google Scholar]
- 27.Tan J, et al. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 29.Cherblanc FL, Chapman KL, Brown R, Fuchter MJ. Nat Chem Biol. 2013;9:136–137. doi: 10.1038/nchembio.1187. [DOI] [PubMed] [Google Scholar]
- 30.Cherblanc FL, et al. J Med Chem. 2013;56:8616–8625. doi: 10.1021/jm401063r. [DOI] [PubMed] [Google Scholar]
- 31.Leverson JD, et al. Sci Transl Med. 2015;7:279ra240. doi: 10.1126/scitranslmed.aaa4642. [DOI] [PubMed] [Google Scholar]
- 32.Vogler M, et al. Cell Death Differ. 2009;16:1030–1039. doi: 10.1038/cdd.2009.48. [DOI] [PubMed] [Google Scholar]
- 33.Zeiger E, Prokopetz A, Walters DB. Accountability in Research. 1993;3:45–46. [Google Scholar]
- 34.Ahn K, et al. J Pharmacol Exp Ther. 2011;338:114–124. doi: 10.1124/jpet.111.180257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, et al. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davies SP, Reddy H, Caivano M, Cohen P. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinelli A, et al. J Med Chem. 2005;48:5509–5519. doi: 10.1021/jm0502844. [DOI] [PubMed] [Google Scholar]
- 38.Göttlicher M, et al. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pacholec M, et al. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]