SUMMARY
The carbon-phosphorus (C-P) lyase complex is essential for the metabolism of unactivated phosphonates to phosphate in bacteria. Using single-particle cryo-electron microscopy, we determined two structures of the C-P lyase core complex PhnG2H2I2J2, with or without PhnK, respectively. PhnG2H2I2J2 is a two-fold symmetric hetero-octamer. Its two PhnJ subunits provide two identical binding sites for PhnK. Only one PhnK binds to PhnG2H2I2J2 due to steric hindrance. PhnK is homologous to the nucleotide-binding domain (NBD) of ATP-Binding Cassette (ABC) transporters. The α-helices 3 and 4 of PhnK bind to the α-helix 6 and a loop (residues 227–230) of PhnJ, in a different mode from the binding of NBDs to their trans-membrane partners. Moreover, binding of PhnK exposes the active site residue, Gly32 of PhnJ, located near the interface between PhnJ and PhnH. This structural information provides a basis for further deciphering the reaction mechanism of the C-P lyase.
INTRODUCTION
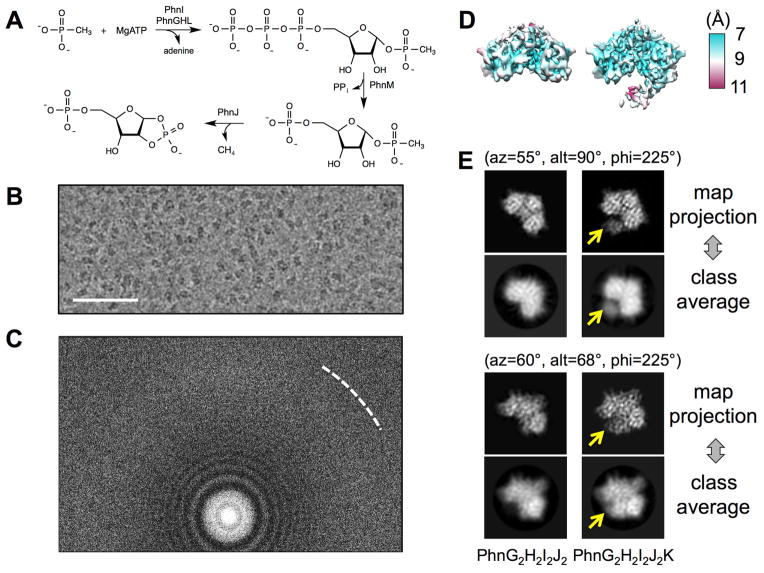
Phosphorus is essential for life. Currently, this element is most abundant as phosphoric acid and phosphate esters (Wackett et al., 1987). However, certain organisms can acquire needed phosphorus under conditions of low phosphate directly from organophosphonate compounds, a class of compounds that contain a carbon-phosphorus (C-P) bond that must be enzymatically cleaved to phosphate (McGrath et al., 2013). Phosphonates have been proposed as a major source of phosphorus on the prebiotic earth and large quantities of phosphonates are discharged annually into the environment as agricultural herbicides and industrial detergents (Ternan et al., 1998). In bacteria such as Escherichia coli, the C-P bond in unactivated organophosphonates can be enzymatically cleaved to form phosphate and an alkane by the multi-subunit C-P lyase complex (Chen et al., 1990). In E. coli, the C-P lyase complex is encoded by the 14 genes (phnCDEFGHIJKLMNOP) contained within the phn operon (Chen et al., 1990; Metcalf and Wanner, 1993a, b). Previous genetic and biochemical studies have demonstrated that seven of the genes in this operon (phnGHIJKLM) are critical for the expression of proteins that are required for the enzymatic cleavage of the C-P bond during the transformation of phosphonates to phosphate (Metcalf and Wanner, 1993b). The individual subunits of the E. coli C-P lyase complex that are essential for the enzymatic cleavage of the C-P bond in methylphosphonate have been reconstituted and characterized in vitro (Kamat et al., 2011; Kamat et al., 2013). The series of enzymatic steps catalyzed by the C-P lyase complex is illustrated in Figure 1A. The initial reaction of the C-P lyase pathway is catalyzed by a nucleotide phosphorylase, PhnI, in the presence of PhnG, PhnH and PhnL in which ATP and methylphosphonate are converted to adenine and α-D-ribose-1-methylphosphonate-5-triphosphate (RPnTP). The RPnTP is hydrolyzed by the phosphohydrolase PhnM to produce pyrophosphate and 5-phosphoribosyl-1-phosphonate (PRPn). PRPn serves as a substrate for the cleavage of the C-P bond by PhnJ through a SAM-dependent radical-based reaction, transforming PRPn to the production of 5-phosphoribosyl-1, 2-cyclic phosphate (PRcP) and methane (or corresponding alkane).
Figure 1. Cryo-EM analysis reveals co-existence of PhnG2H2I2J2 and PhnG2H2I2J2K.
(A) The series of the enzymatic steps of C-P lyase. (B) A region of a representative micrograph showing the particles of PhnG2H2I2J2 and PhnG2H2I2J2K with a defocus of - 1.2 μm. The scale bar denotes 500Å. (C) Power spectrum of the same micrograph in (B). The white dashed curve shows the Nyquist at 3Å. (D) 3D density maps of PhnG2H2I2J2 (left) and PhnG2H2I2J2K (right). The maps were colored based on the local resolutions. (E) Projections of the two maps compared to the reference-free class averages in two representative orientations. The Euler angles were based on the EMAN2 convention. Yellow arrows indicate the location of PhnK. See also Figure S1.
Fragments of the C-P lyase complex containing PhnG2I2, PhnG2H2I2J2 and PhnG2H2I2J2K can be expressed and purified in high yield (Jochimsen et al., 2011). We have used mass spectrometry and H/D exchange methods to construct a low-resolution interaction map of the PhnG2H2I2J2K complex that illustrates how the individual subunits associate with one another to form larger multi-subunit complexes (Ren et al., 2015). Interestingly, the demonstrated stoichiometry indicates that one copy of PhnK binds to the dimeric core complex (PhnG2H2I2J2). The X-ray crystal structure of the core complex PhnG2H2I2J2 has been recently reported (Seweryn et al., 2015). It revealed an intertwined network of subunits PhnG, PhnH, PhnI and PhnJ with self-homologies. However, how PhnK binds to the core complex remains unclear.
From sequence analysis, PhnK is homologous to the nucleotide-binding domain (NBD) of ATP-Binding Cassette (ABC) transporters. PhnK has all the NBD motifs: Walker A, Walker B, ABC signature, A-loop, D-loop, Q-loop and switch H-loop. The role of PhnK in the C-P lyase pathway is unclear: PhnK is apparently not required for any of the essential reactions of the C-P lyase pathway in vitro (Kamat et al., 2011). However, the E. coli phnK strain is phosphonate growth-deficient (Metcalf and Wanner, 1993b). PhnK was previously mapped in a groove close to the two-fold symmetry axis of the core complex PhnG2H2I2J2 based on a low-resolution density map from negative stain electron microscopy (negative stain EM) (Seweryn et al., 2015). The ABC signature was suggested to bind to the core complex based on the assumption that conserved regions comprise the interaction surface. However, this model for the binding of PhnK needs to be validated. Moreover, the following questions still remain unanswered. (1) Why is there only one copy of PhnK bound to a dimeric PhnG2H2I2J2 core complex? (2) How does PhnK bind to the core complex? (3) What is the effect on the core complex after PhnK binds? A higher resolution structure of PhnG2H2I2J2K is clearly needed. Unfortunately, attempts to crystalize the PhnG2H2I2J2K complex have thus far failed.
Recent advances in single-particle cryo-electron microscopy (cryo-EM) have enabled the structure determination of large macromolecular complexes, usually with a molecular weight of more than 400 kDa, to atomic resolutions (Bartesaghi et al., 2015; Campbell et al., 2015; Fischer et al., 2015; Grant and Grigorieff, 2015; Jiang et al., 2015; Yu et al., 2015). This breakthrough is mostly attributed to the development of the instrumentation needed to acquire better images (Bai et al., 2013; Bammes et al., 2012; Fischer et al., 2015; Li et al., 2013), and the software to more reliably process and validate the results (Frank et al., 1981; Hohn et al., 2007; Lyumkis et al., 2013; Scheres, 2012; Tang et al., 2007), especially the use of statistical approach for processing images of particles with conformational and compositional heterogeneity (Lyumkis et al., 2013; Scheres, 2012). Such “resolution revolution” (Kuhlbrandt, 2014) in cryo-EM is expanding to the structure determination of small (~200 kDa in size), nonsymmetrical macromolecular complexes, with a few examples being determined to near-atomic or subnanometer resolutions (Kim et al., 2015; Liang et al., 2015; Lu et al., 2014). In this study, we used single-particle cryo-EM to determine two respective structures of the PhnG2H2I2J2K (248 kDa) and PhnG2H2I2J2 (220 kDa) complexes at 7.8Å resolution from a sample containing both of them. The interaction between the monomeric PhnK and the dimeric PhnG2H2I2J2 was revealed.
Results
Co-existence of PhnG2H2I2J2 and PhnG2H2I2J2K
The genes expressing PhnG, PhnH, PhnI, PhnJ and PhnK were cloned into a pET-28b plasmid with a polyhistidine tag on the N-terminus of PhnG as previously described (Jochimsen et al., 2011; Ren et al., 2015). The expressed proteins were isolated using a nickel column and then further purified by passage through a gel filtration column. The purified complexes were vitrified and imaged (Experimental Procedures). The collected movie stacks, from an electron-counting direct detection camera (Li et al., 2013), showed clear particles and strong power spectra after aligning the frames within each movie (Grant and Grigorieff, 2015) (Figures 1B and 1C). A total of 492,889 particles were semi-automatically picked, and subsequently classified with reference-free two-dimensional (2D) classification and unsupervised three-dimensional (3D) classification in Relion (Scheres, 2012) (Figure S1). Two populations of particles were identified and reconstructed into two density maps, which correspond to the PhnG2H2I2J2 (core complex) and PhnG2H2I2J2K (core complex bound with PhnK), respectively. These two maps were similar except for an extra lobe of density in one of them (Figure 1D). Differences in the extra density were also observed in the reference-free 2D class averages calculated from all particles (Figure 1E). This extra density was later confirmed as the PhnK subunit of the PhnG2H2I2J2K complex. Further refinements against the separated particles yielded the cryo-EM density maps of PhnG2H2I2J2 and PhnG2H2I2J2K, both at 7.8Å resolution, based on the gold-standard Fourier shell correlation (FSC, Figure S1) (Scheres and Chen, 2012). Local resolution was estimated, showing most regions of the maps were at a resolution range between 7–8Å (Figure 1D). At this resolution, the protein secondary structures were clearly visible.
The existence of a complex missing PhnK, in the purified sample expressed from a plasmid containing the phnGHIJK genes, is consistent with previous native gel electrophoresis and mass spectrometry experiments (Jochimsen et al., 2011), suggesting the loose binding of PhnK to the core complex PhnG2H2I2J2. The fact that PhnG2H2I2J2 and PhnG2H2I2J2K co-exist in the same sample apparently hindered previous trials for the crystallization of PhnG2H2I2J2K (Seweryn et al., 2015).
Architecture of PhnG2H2I2J2 with or without PhnK bound
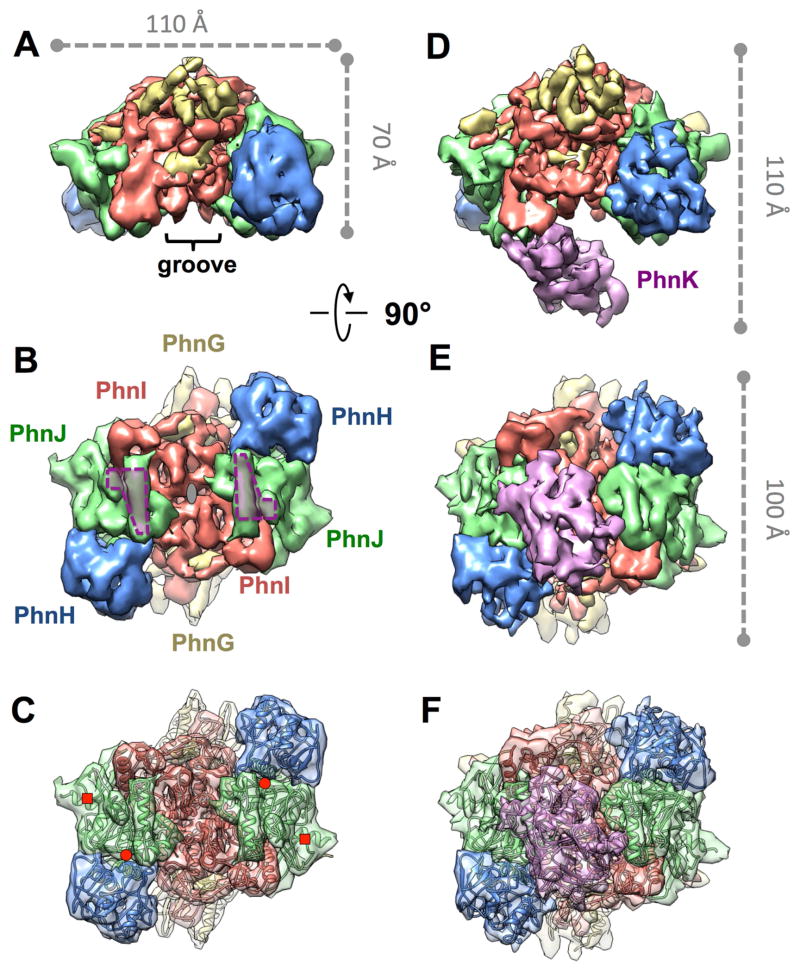
The 3D density maps of the PhnG2H2I2J2 and PhnG2H2I2J2K complexes measure 110Å x 70Å x 100Å and 110Å x 110Å x 100Å in dimension, respectively (Figure 2). The common part of these two complexes is the PhnG2H2I2J2 core. The core retains the same architecture after PhnK binding. In both density maps, the two copies of PhnG, PhnH, PhnI and PhnJ are arranged in a head-to-tail fashion in a sequence of H-J-I-G as labeled in Figure 2B. While the cryo-EM maps were independently determined and not biased by the recently determined crystal structure of PhnG2H2I2J2 (Experimental Procedures), the crystal structure fits well into the cryo-EM density map of the core complex PhnG2H2I2J2 with matching secondary structures (Figure 2C).
Figure 2. Architecture of PhnG2H2I2J2 without or with PhnK bound.
(A, B) Front and bottom view of PhnG2H2I2J2. In (A), the groove on the core complex PhnG2H2I2J2 is labeled. In (B), the grey ellipse denotes the two-fold symmetry axis, and the two purple polygons denote the two identical binding sites for PhnK. (C) The crystal structure of PhnG2H2I2J2 fit into the cryo-EM map of PhnG2H2I2J2 in the same orientation as (B). The red circles and squares denote the positions of Gly32 and Cys272 of PhnJ, respectively. (D, E) Front and bottom view of PhnG2H2I2J2K, in the same orientations as (A) and (B), respectively. (F) The crystal structure of PhnG2H2I2J2 and homology model of PhnK fit into the cryo-EM map of PhnG2H2I2J2K in the same orientation as (E). See also Figure S1.
Two identical binding sites for PhnK on PhnG2H2I2J2
The binding site for PhnK is located on the α-helix 6 (residue 147–158) and a nearby loop (residue 227–230) of PhnJ. We term this loop “chock-loop” as it may stabilize the binding of PhnK like a chock. Two PhnJ subunits in the core complex provide two identical binding sites for PhnK (purple polygons in Figure 2B). These two binding sites are approximately 50Å away from each other. Noticeably, between these two binding sites lies a negatively charged groove (Figure S1), which was previously proposed as the PhnK-binding region based on a low-resolution density map of PhnG2H2I2J2K from negative stain EM and protein sequence conservation (Seweryn et al., 2015). However, the entire groove is exposed to solvent and not in contact with PhnK in our cryo-EM map of PhnG2H2I2J2K.
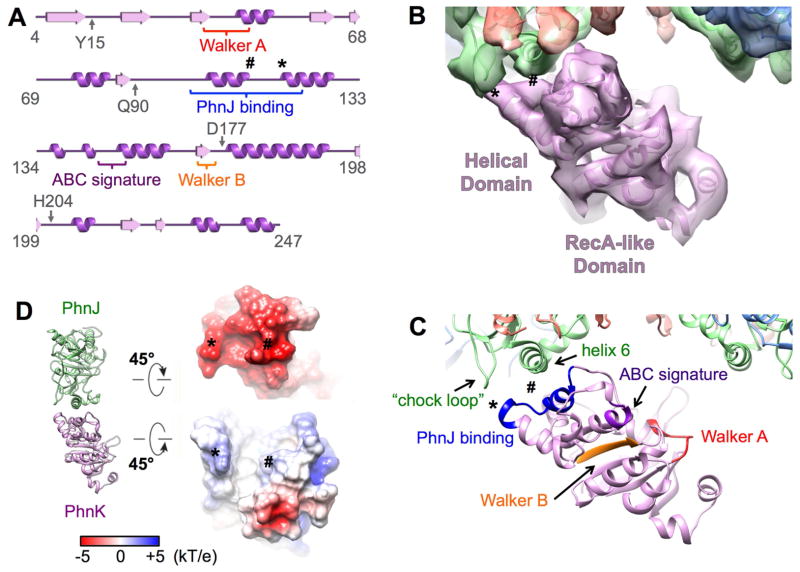
One copy of PhnK, through its helical domain, binds to PhnG2H2I2J2
Even though the core complex provides two identical binding sites for PhnK, only one copy of PhnK was found to bind (Figures 2D, 2E, 2F and 3). PhnK is homologous to the conserved NBD of proteins in the ABC transporter family. Like NBDs, PhnK consists of a helical domain and a RecA-like domain. It has all of the sequence motifs (Walker A, Walker B, ABC signature, A-loop, D-loop, Q-loop and switch H-loop) of the NBD (Figure 3A) (Ambudkar et al., 2006; Davidson et al., 2008). A sequence alignment of PhnK with other structurally characterized NBDs is presented in Figure S2. NBDs are known to interact with a transmembrane domain or protein (TMD) through its helical domain (Schmitt et al., 2003). The L-loop of the TMD inserts into a cavity around the helical domain of the NBD (Figure S3), which forms the major interaction between NBD and TMD (Locher et al., 2002). In contrast, PhnK, an NBD-like protein, interacts with the cytoplasmic protein complex PhnG2H2I2J2. This interaction is via a helix-turn-helix motif (residues 100–120 containing the α-helices 3 and 4) within the helical domain of PhnK to bind to the α-helix 6 and chock-loop of PhnJ (Figures 3A and 3C). When PhnK binds to PhnJ, the cavity of PhnK is completely exposed to solvent. The chock-loop of PhnJ may instead provide additional support to block the movement of PhnK (Figure S3). The binding interface between PhnK and PhnJ shows complementary electrostatic surface potentials. At the interface, PhnK exhibits a net positive charge while PhnJ exhibits a negative charge (Figure 3D).
Figure 3. PhnK is an NBD-like protein and interacts with PhnJ.
(A) Secondary structure of PhnK with the NBD motifs labeled. The symbol * and # label the sites for the interactions between PhnK and PhnJ. (B) One PhnK binds to the core complex with the models fit in the map. (C) The same view as (B) with the NBD motifs color labeled. (D) Electrostatic surface potentials at the interface of PhnJ and PhnK. See also Figures S2 and S3.
Attempts to dock a second copy of PhnK, in the same conformation as the first one, to the empty binding site in the PhnG2H2I2J2K, resulted in considerable collision between their RecA-like domains, which prevents the simultaneous binding of two PhnK subunits.
Binding of PhnK exposes the active site residue Gly32
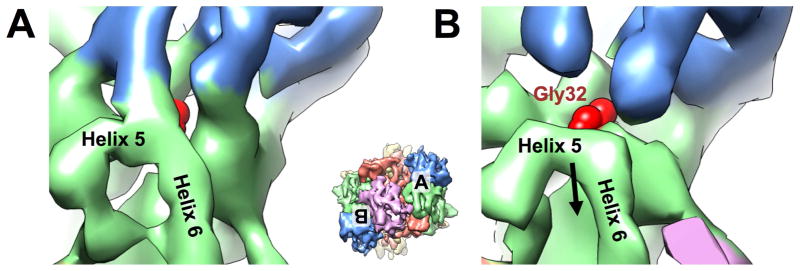
The universally conserved Gly32 of PhnJ was proposed to generate a stable glycyl radical enzyme that supports multiple turnovers without further SAM consumption (Kamat et al., 2013). In the core complex, the two Gly32, each from one subunit of PhnJ, are symmetrically located on the two sides of the complex. Both of these glycine residues are buried in PhnJ near the interface between PhnJ and PhnH. In the map of PhnG2H2I2J2K, the density burying Gly32 of PhnJ shows striking difference between the two copies of PhnJ (Figure 4). On the side where there is no PhnK bound (apo side), the density burying Gly32 is stronger than the corresponding density on the side where there is a PhnK bound (bound side), indicating the interaction between PhnJ and PhnH in the apo side is tighter than in the bound side. On the apo side, the residue Gly32 is hidden, as in our cryo-EM density map of PhnG2H2I2J2 (Figure 4A). On the bound side, the α-helices 5 and 6 of PhnJ move away from PhnH, disrupting the molecular interaction between PhnJ and PhnH to expose the active site residue Gly32 (Figure 4B). Further comparison with the crystal structure of PhnG2H2I2J2 was shown in Figure S4. On the apo side, the density of α-helices 5 and 6 of PhnJ matches well with the crystal structure, while on the bound side, the density of α-helices 5 and 6 of PhnJ is shifted relative to the crystal structure. This shift indicates there is a structural rearrangement in this region to reveal the underneath active site.
Figure 4. Binding of PhnK causes Gly32 of PhnJ more exposed.
(A) On the side without PhnK bound (apo side), the Helix 5 of PhnJ connects PhnH. (B) On the side with PhnK bound (bound side), the densities for H5 and H6 of PhnJ moves away from PhnH to expose the Gly32 of PhnJ. The density map of PhnG2H2I2J2K is rendered at 4.5 s above mean and colored coded as in Figure 2. The red sphere model denotes Gly32 of PhnJ. Inset shows the map of PhnG2H2I2J2K in the same orientation as Figure 2E with the locations of (A) and (B) labeled. See also Figure S4.
The glycyl radical formed at Gly32 was proposed to abstract the hydrogen of Cys272 to form a cysteine radical during PhnJ catalysis (Kamat et al., 2013). In contrast, Seweryn et al. made the startling discovery that Gly32 of PhnJ is more than 30 Å away from Cys272 in the crystal structure of PhnG2H2I2J2 complex (Figure 2C; Seweryn et al., 2015). Here, in our cryo-EM structure of PhnG2H2I2J2 with PhnK bound, the overall structure of PhnJ remains similar to the crystal structure with Gly32 still ~30Å away from Cys272. If the proposed reaction mechanism for C-P lyase, which involves the direct interaction between Cys272 and Gly32, is correct, then substantial structural rearrangements must occur transiently and/or after the binding of substrates or other protein subunits to this complex.
Discussion
We have demonstrated the unique binding mode of the NBD-like PhnK to the C-P lyase core complex PhnG2H2I2J2. Even though there exist two identical binding sites for PhnK on the core complex, only one PhnK can bind at a time, leading to a more exposed active site residue Gly32 of PhnJ on the PhnK bound side. Such rearrangement around the active site may facilitate the delivery of the substrate or release of the reaction product.
PhnK has all the structural motifs to bind the ATP molecule. It might help recruit the ATP as the substrate for the C-P lyase reaction. While the two-fold symmetrical C-P lyase core complex PhnG2H2I2J2 potentially provides two equivalent sets of active sites for two series of the catalytic reactions, the loose and monomeric binding of PhnK suggest that it may alternate between the two binding sites on the core complex to deliver ATPs and facilitate the two series of reactions in a “staggered time shift”. Further biochemical and structural studies are needed to test these hypotheses.
The structural information provided here will help a better understanding of the enzymatic reaction mechanism for phosphonates utilization, especially the function of the NBD-like PhnK. Besides, our results clearly demonstrate the power of cryo-EM for the structural study of small (~200 kDa) asymmetrical multi-subunit protein complexes with compositional heterogeneity.
Experimental Procedures
Protein Purification
The plasmids for the expression of PhnG2H2I2J2K from the C-P lyase complex of E. coli were cloned, transformed and expressed as previously described (Ren et al., 2015). The cells were resuspended (4:1, v/w) in binding buffer (50 mM HEPES, pH 8.5, 150 mM NaCl, 20 mM imidazole and 2 mM tris (2-carboxyethyl) phosphine) containing 0.10 mg/mL PMSF and 0.1% protease cocktail for polyhistidine-tagged protein purification. The cells were lysed by sonication and the supernatant solution was separated from the insoluble debris by centrifugation at 10,000 x g for 20 minutes at 4°C. The pellet was discarded and the supernatant solution was applied to a 5-mL HisTrap (GE Healthcare) column, which was pre-equilibrated with 10 column volumes of binding buffer. The proteins were eluted with a gradient of elution buffer (0.5 M imidazole in binding buffer). The eluted fractions were pooled and applied to a High Load 26/60 Superdex 200 prep grade gel filtration column (GE Healthcare), which was previously equilibrated with running buffer (binding buffer without imidazole). The fractions were pooled and analyzed by SDS-PAGE.
Electron Microscopy
PhnG2H2I2J2K was diluted to 0.1 mg/mL with the dilution buffer (50 mM HEPES, pH 8.5, 150 mM NaCl, 2 mM TCEP). 3 μL of the sample was applied to a C-Flat 1.2/1.3 holey carbon grid at 16°C with 100% relative humidity and vitrified using a Vitrobot Mark III (FEI company, Netherlands). The thin-ice areas that were expected to show clearly visible and mono-dispersed particles were imaged under an FEI Tecnai F20 cryo electron microscope with a field emission gun (FEI company, Netherlands) operated at 200 kV. Data were recorded on a Gatan K2 Summit electron-counting direct detection camera (Gatan, Pleasanton CA) in the electron counting mode (Li et al., 2013). A nominal magnification of 25,000X was used, yielding a pixel size of 1.5 Å. The beam intensity was adjusted to a dose rate of ~10 electrons per pixel per second on the camera. A 25-frame or 50-frame movie stack was recorded for each picture, with 0.2 second per frame for a total exposure time of 5 seconds or 10 seconds, respectively.
Image Processing
967 collected image stacks were iteratively aligned and filtered based on the electron dose using Unblur (Grant and Grigorieff, 2015). 694 sum images were selected according to visibility of particles and power spectra. For particle picking, the sum images were first low-pass filtered to 15 Å to visualize particles unambiguously. Then the “Erase” tool of e2boxer.py in EMAN2 (Tang et al., 2007) was used to erase bad areas that have junk or have no particles. Finally the “Swarm” tool of e2boxer.py was used to semi-automatically pick 492,889 particles using a box size of 120 pixels, during which the threshold for picking was interactively changed so that the false-positive picking was minimized. The coordinates were imported into Relion (Scheres, 2012) to extract the particles from the original sum images with a downscaling factor of 1, 2 or 4. Reference-free 2D classification was performed in Relion to remove bad particles that cannot average well with each other, which generated a dataset of 294,203 particles. We selected 3,956 particles with good contrast to generate an initial model using the PRIME routine in the SIMPLE package (Elmlund et al., 2013), which was low-pass filtered to 60 Å and used for the first round of unsupervised 3D classification in Relion. The 3D classification and refinement were performed hierarchically (Figure S1). Briefly, in the first two rounds of 3D classification, particles were separated into three classes based on its quality and the presence or absence of PhnK. 50,061 good particles from one class with PhnK were selected and further refined with C1 symmetry to produce the density map of PhnG2H2I2J2K. 86,391 particles without PhnK were subjected to an additional round of 3D classification to separate into three classes, which yielded 23,861 good particles in one class. These 23,861 particles were further refined with C2 symmetry to produce the density map of PhnG2H2I2J2. The particle images were downscaled by 4 times in the first two rounds of the 3D classification and downscaled by 2 times in the later 3D classification and refinement. Refinement against the particles without downscaling, did not improve the final resolution of the maps. Blocres of the Bsoft package (Heymann and Belnap, 2007) was used to estimate the local resolution of the cryo-EM maps. Our resolution is limited to 7.8Å, which may be due to the flexible nature of the specimen.
Molecular Modeling and Map Segmentation
Fitting of models into the density map is done in UCSF Chimera (Pettersen et al., 2004). The correct handedness of the density maps of PhnG2H2I2J2 and PhnG2H2I2J2K were initially determined by docking the crystal structure of PhnH (PDB id: 2FSU) (Adams et al., 2008) into both the original maps and the maps with the flipped handedness. The crystal structure of PhnH fit into the flipped maps with all the secondary structures matching, confirming that the original maps should have their handedness flipped. Homology model of PhnK was built using SWISS Model (Guex et al., 2009) based on a NBD (PDB id: 4U00) (Devi et al., 2015). The location of PhnK was determined by the difference map between PhnG2H2I2J2K and PhnG2H2I2J2. Moreover, the homology model of PhnK fit the difference map with all of the secondary structures in place, confirming the correct location of PhnK in PhnG2H2I2J2K. The segmentation of PhnG2H2I2J2 core was done using Segger (Pintilie et al., 2010) in UCSF Chimera based on the crystal structure of PhnG2H2I2J2 (PDB id: 4XB6). The electrostatic surface of the models was calculated using the APBS module (Baker et al., 2001) in UCSF Chimera.
Supplementary Material
Highlights.
Cryo-EM structures of a small, asymmetric C-P lyase complex at 7.8Å resolution.
Only one PhnK binds to the dimeric core complex due to steric hindrance.
The NBD-like PhnK binds to a cytoplasmic protein, distinct from NBD-TMD interaction.
Binding of PhnK exposes the active site residue, Gly32 of PhnJ.
Acknowledgments
We acknowledge the Texas A&M Supercomputing Facility for providing computational resources. We thank Mengqiu Jiang for assistance in data collection. J.Z. would like to acknowledge the Microscopy and Imaging Center at Texas A&M University for providing instrumentation for EM data collection. This work was supported by NIH grant GM103917 to F.M.R. and startup funding from the Department of Biochemistry and Biophysics and the Center for Phage Technology at Texas A&M University to J.Z. K.Y. is partly supported by the Welch foundation grants A-1863 to J.Z.
Footnotes
Accession Numbers
The cryo-EM maps have been deposited in the EM Databank with accession codes EMD-6410 (PhnG2H2I2J2K) and EMD-6411 (PhnG2H2I2J2).
Author Contributions.
Z.R. purified the sample. K.Y. solved the structures. K.Y., Z.R., F.M.R. and J.Z. designed the experiments, interpreted the results and wrote the paper. F.M.R. and J.Z. are the principal investigators.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MA, Luo Y, Hove-Jensen B, He SM, van Staalduinen LM, Zechel DL, Jia Z. Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli. J Bacteriol. 2008;190:1072–1083. doi: 10.1128/JB.01274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambudkar SV, Kim IW, Xia D, Sauna ZE. The A-loop, a novel conserved aromatic acid subdomain upstream of the Walker A motif in ABC transporters, is critical for ATP binding. FEBS Lett. 2006;580:1049–1055. doi: 10.1016/j.febslet.2005.12.051. [DOI] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J Struct Biol. 2012;177:589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JL, Subramaniam S. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MG, Veesler D, Cheng A, Potter CS, Carragher B. 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. eLife. 2015;4:e06380. doi: 10.7554/eLife.06380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Ye QZ, Zhu ZM, Wanner BL, Walsh CT. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli. B J Biol Chem. 1990;265:4461–4471. [PubMed] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi SK, Chichili VP, Jeyakanthan J, Velmurugan D, Sivaraman J. Structural basis for the hydrolysis of ATP by a nucleotide binding subunit of an amino acid ABC transporter from Thermus thermophilus. J Struct Biol. 2015;190:367–372. doi: 10.1016/j.jsb.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Elmlund H, Elmlund D, Bengio S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21:1299–1306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- Frank J, Shimkin B, Dowse H. Spider - a Modular Software System for Electron Image-Processing. Ultramicroscopy. 1981;6:343–357. [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Jiang J, Pentelute BL, Collier RJ, Zhou ZH. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature. 2015;521:545–549. doi: 10.1038/nature14247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochimsen B, Lolle S, McSorley FR, Nabi M, Stougaard J, Zechel DL, Hove-Jensen B. Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway. Proc Natl Acad Sci USA. 2011;108:11393–11398. doi: 10.1073/pnas.1104922108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat SS, Williams HJ, Dangott LJ, Chakrabarti M, Raushel FM. The catalytic mechanism for aerobic formation of methane by bacteria. Nature. 2013;497:132–136. doi: 10.1038/nature12061. [DOI] [PubMed] [Google Scholar]
- Kamat SS, Williams HJ, Raushel FM. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature. 2011;480:570–573. doi: 10.1038/nature10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu S, Tomasiak TM, Mergel C, Winter MB, Stiller SB, Robles-Colmanares Y, Stroud RM, Tampe R, Craik CS, et al. Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature. 2015;517:396–400. doi: 10.1038/nature13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, et al. Three-dimensional structure of human gamma-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JW, Chin JP, Quinn JP. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat Rev Microbiol. 2013;11:412–419. doi: 10.1038/nrmicro3011. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Wanner BL. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene. 1993a;129:27–32. doi: 10.1016/0378-1119(93)90692-v. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Wanner BL. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA' elements. J Bacteriol. 1993b;175:3430–3442. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Ranganathan S, Zinnel NF, Russell WK, Russell DH, Raushel FM. Subunit Interactions within the Carbon-Phosphorus Lyase Complex from Escherichia coli. Biochemistry. 2015;54:3400–3411. doi: 10.1021/acs.biochem.5b00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt L, Benabdelhak H, Blight MA, Holland IB, Stubbs MT. Crystal structure of the nucleotide-binding domain of the ABC-transporter haemolysin B: identification of a variable region within ABC helical domains. J Mol Biol. 2003;330:333–342. doi: 10.1016/s0022-2836(03)00592-8. [DOI] [PubMed] [Google Scholar]
- Seweryn P, Van LB, Kjeldgaard M, Russo CJ, Passmore LA, Hove-Jensen B, Jochimsen B, Brodersen DE. Structural insights into the bacterial carbon-phosphorus lyase machinery. Nature. 2015;525:68–72. doi: 10.1038/nature14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Ternan N, Mc Grath J, Mc Mullan G, Quinn J. Review: Organophosphonates: occurrence, synthesis and biodegradation by microorganisms. World J Microb Biot. 1998;14:635–647. [Google Scholar]
- Wackett LP, Shames SL, Venditti CP, Walsh CT. Bacterial carbon-phosphorus lyase: products, rates, and regulation of phosphonic and phosphinic acid metabolism. J Bacteriol. 1987;169:710–717. doi: 10.1128/jb.169.2.710-717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jiang J, Sun J, Zhou ZH. A putative ATPase mediates RNA transcription and capping in a dsRNA virus. eLife. 2015;4:e07901. doi: 10.7554/eLife.07901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.