Abstract
MHC haplotypes of humans and the African great ape species have one copy of the MHC-A, -B, and -C genes. In contrast, MHC haplotypes of orangutans, the Asian great ape species, exhibit variation in the number of gene copies. An in-depth analysis of the MHC class I gene repertoire in the two orangutan species, Pongo abelii and Pongo pygmaeus, is presented here. This analysis involved Sanger and next-generation sequencing methodologies, revealing diverse and complicated transcription profiles for orangutan MHC-A, -B, and -C. Thirty-five previously unreported MHC class I alleles are described. The data demonstrate that each orangutan MHC haplotype has one copy of the MHC-A gene, and that the MHC-B region has been subject to duplication, giving rise to at least three MHC-B genes. The MHC-B*03 and -B*08 lineages of alleles each account for a separate MHC-B gene. All MHC-B*08 allotypes have the C1-epitope motif recognized by KIR. At least one other MHC-B gene is present, pointing to MHC-B alleles that are not B*03 or B*08. The MHC-C gene is present only on some haplotypes, and each MHC-C allotype has the C1-epitope. The transcription profiles demonstrate that MHC-A alleles are highly transcribed, whereas MHC-C alleles, when present, are transcribed at very low levels. The MHC-B alleles are transcribed to a variable extent and over a wide range. For those orangutan MHC class I allotypes that are detected by human monoclonal anti-HLA class I antibodies, the level of cell-surface expression of proteins correlates with the level of transcription of the allele.
Introduction
The major histocompatibility complex (MHC) emerged with the evolution of jawed vertebrates approximately 400 million years ago (1). The cell-surface proteins encoded by the MHC class I genes play a key role in the adaptive immune response to infections and cancer, and involve the presentation of peptides to CD8+ cytotoxic T cells. The contractions and expansions of the MHC class I gene family, which took place during primate evolution, suggests that different species seem to have fine-tuned their immune capability in the course of resisting and surviving a spectrum of infections (2). In addition to their role in T-cell immunity, MHC class I molecules are also ligands for killer-cell immunoglobulin-like (KIR) receptors. The KIR are principally expressed on natural killer (NK) cells, a subset of lymphocytes involved in innate immunity (3). In humans, HLA-C molecules are the dominant ligands for KIR, and there are two types, which carry either the C1 or the C2 epitope, and engage different KIR subsets (4). In addition to fighting infection, NK cells play a critical role in reproduction. Here the interaction of maternal KIR on uterine NK cells, with fetal HLA-C expressed by extravillous trophoblast (EVT), is essential for embryo implantation and formation of the placenta (5). This raises the possibility that during primate evolution adaptations in the reproduction process may also have influenced the gene content variation and polymorphism of the MHC class I gene family.
Humans and orangutans shared a common ancestor approximately 12-16 million years ago (6), and the orangutan, an inhabitant of Asia, is the only great-ape species living outside of Africa. Two species are officially recognized (7): Pongo abelii (Poab) and Pongo pygmaeus (Popy), inhabiting the islands of Sumatra or Borneo, respectively. Orangutans possess genes that are homologous to HLA-A, -B, and -C genes (8). In the two species, however, there is evidence for differences in the copy number of MHC class I genes. All human haplotypes carry single copies of MHC class I genes, whereas some orangutan haplotypes seem to have at least two MHC-B genes, while others lack MHC-C (9, 10). For gibbons, the small apes that are also inhabitants of Asia, no homolog of HLA-C has been identified (8), and the corresponding genomic region appears to be absent from the MHC (11). In reconstructing the evolution of the MHC-C and its coevolution with KIR, these comparisons point to the orangutan being a pivotal species.
The accessibility of next-generation sequencing (NGS) technologies opened new avenues for the study of gene content in species with gene copy number variation (12). For instance, in rhesus macaque, a species known to have extensive variation in the gene copy number (13, 14), 454 pyrosequencing is a viable tool for studying the MHC class I repertoire (15). Moreover, this technique can be used to analyze the level of transcription of different alleles present in an individual more reliably than was done in the past by conventional sequencing techniques (15-17). In addition to the Sanger method, we have applied NGS technology to characterize MHC polymorphism, and to examine the level of transcription of MHC class I genes in a panel of unrelated orangutans. The cell-surface expression of MHC class I allotypes on orangutan B-cell lines was assessed using a panel of monoclonal antibodies. The data revealed a differential level of transcription/expression for the orangutan MHC allotypes, and shed light on the evolution of the orangutan MHC class I family, as well as the plasticity of this region in primates.
Materials and Methods
Cell lines
EBV-transformed B-cell lines from six unrelated orangutan individuals were studied: three cell lines from individuals of the Pongo abelii species (PPY1, Jinjing, and Guchi) and three cell lines from individuals of the Pongo pygmaeus species (Jago, Katja, and Elmar). The human EBV-transformed B-cell line JY was used as a reference and control.
Sequencing of full-length orangutan MHC class I transcripts using the Sanger method
Conventional Sanger sequencing was performed to define MHC-A, -B, and -C gene variation in the six orangutans using four different primer sets: two generic primer sets amplifying the full length cDNA sequence of all three classical genes (exons 1 to 8 for the MHC-A [1098 bp] and -C [1101 bp] genes, and exons 1 to 7 for the MHC-B [1089 bp] gene), and two gene-specific primer sets (Fig. S1). RNA was extracted from the cultured B-cell lines with the RNeasy Mini kit (Qiagen), in according with the manufacturer's recommendation, and served as a template for the cDNA synthesis using the RevertAid First Strand cDNA synthesis kit (Thermo Scientific). The cDNA was used as starting material in the PCR for the amplification of MHC-A, -B, and -C. The PCR mixes (50 μl total) contained 2 μl cDNA, 1 uM of each primer, 0.25 mM of each dNTP, 2.5 mM MgCl2, 5 μl 10× PCR buffer, and 0.5 units platinum Taq polymerase (Invitrogen). The PCR conditions for primer set A were: 95°C for 30 s, 30 cycles of 94°C for 10 s, 55°C for 60 s, 72°C for 90 s, and a final extension at 72°C for 5 min. For B and C: 95°C for 60 s, 30 cycles of 95°C for 30 s, 63°C for 30 s, 72°C for 90 s, and a final extension at 72°C for 5 min. For D: 95°C for 30 s, 30 cycles of 94°C for 10 s, 60°C for 60 s, 72°C for 90 s, and a final extension at 72°C for 5 min. PCR amplification was verified using gel electrophoresis, and PCR products were purified using the GeneJET gel extraction kit (Thermo Scientific). Subsequently, the PCR products were cloned and sequenced as previously described (18). The data were analyzed using the programs Lasergene Seqman Pro version 11.2.1 (Dnastar, Inc Madison, USA) and Macvector version 12.7.5 (MacVector, Inc Cambridge, UK). An allele was defined unambiguously if it was confirmed in at least two independent PCR reactions. The sequences were deposited in the European Molecular Biology Laboratory (EMBL) database (www.ebi.ac.uk), and given accession-numbers HE801276 to HE801302, HG970960 to HG970962. All sequences were officially named and deposited in the IPD-MHC NHP database (19).
Sequencing of orangutan MHC class I using the 454-Roche platform
RNA and subsequently cDNA were synthesized as described above, and used as starting material for the PCR amplification of MHC class I genes with generic primers, generating a product of 564 bp covering part of exon 2, all of exon 3, and part of exon 4. The PCR mixture (20 μl total) contained 2 μl cDNA, 0.25 μM of each primer (primer set E, Fig. S1), 0.2 mM of each dNTP, 0.6 μl DMSO, 4 μl 5× Phusion buffer HF, and 0.1 μl Phusion Hot Start High Fidelity DNA Polymerase (Thermo Scientific). The cycling parameters consisted of an initial denaturation step of 1 min at 98 °C, followed by 10 cycles at 98 °C 15 s, 68 °C 10 s, 66 °C 10 s, 64 °C 30 s, and 72 °C 1 min, with a final extension of 40 s at 72 °C. The PCR reaction was followed by an exonuclease step (25 μl total) containing the PCR mixture (20 μl), 0.5 μl Exonuclease I and 1 μl 5× Phusion buffer HF. The mixture was incubated for 30 min at 37 °C, followed by 15 min at 80 °C. Subsequently a second PCR was performed on the mixture containing the 454-Roche multiplex identifier (MID)-encoded primers. This PCR (30 μl total) contained the exonuclease step mixture (25 μl), 0.066 mM of each dNTP, 0.17 μM of each primer (454-Primer-A and 454-Primer-B; see Figure S1 for specific primers used to differentiate between each animal), 1 μl 5× Phusion buffer HF, and 0.1 μl Phusion Hot Start High Fidelity DNA Polymerase. Cycling parameters for the PCR consisted of an initial denaturation step of 1 min at 98 °C, followed by 20 cycles at 98 °C 10 s, 68 °C 20 s, and 72 °C 30 s, with a final extension of 40 s at 72 °C. PCR amplification was verified using gel electrophoresis, and PCR products were purified as described above. DNA concentrations were measured with the Quant-it kit (Invitrogen) on the Qubit 2.0 Fluorometer (Invitrogen). The emulsion PCR and 454 sequencing run were performed following the manufacturer's recommendation (Sequencing Method Manual, GS Junior Titanium Series, May 2010, Roche). The web-based Galaxy platform was used to manage and analyze the pyrosequencing data (usegalaxy.org). A stringent (1%/1%) and a less stringent (0.1%/0.1%) filtering method were compared. The implementation of both methods was based on the expectation that the correct sequences (non-PCR and non-sequencing artifacts) were read more frequently within a collection of in-duplicate sequenced samples. PCR errors were reduced by performing the experiment in duplicate, and a sequence was qualified as being correct if it appeared in both duplicates, and/or a sequence appeared to be identical to a sequence identified with the Sanger method. Correction for homopolymer errors was achieved by collapsing them all to one base. Sequencing in both directions, forward and reverse, was performed to minimize systematic errors (e.g. reading a wrong base after a homopolymer of Gs). When counting the number of sequences, and taking into account the above conditions, the correct sequences were distinguished from the incorrect ones. In addition, sequence errors were controlled by obtaining the same sequences in several individuals, by identifying the sequences obtained with the Sanger method, and/or by confirming sequences present in the IPD-MHC NHP database. In the filtering methods, the first percentage is the cut-off point above which a sequence is present (where repetitions of nucleotides within a sequence are concatenated). The second percentage is the cut-off point above which the original sequence is present within the specific pool of concatenated sequences. Macvector version 12.7.5 (MacVector, Inc Cambridge, UK) was used to compare the reads/sequences retrieved after stringent and less stringent filtering with the orangutan MHC class I alleles using the Sanger method in order to identify new sequences and confirm those that had been previously detected.
In the forward primer (Popy454cl1-ex2-Forward) at position 30 a wobble (Y) representing the nucleotide bases C or T was introduced, as the orangutan MHC class I alleles display polymorphism at this position. C- and T-positive alleles could display different transcription levels (Fig. S2), suggesting that the introduction of this wobble did not influence the PCR amplification. Newly detected sequences (exons 2 to 3) using the 454-Roche platform were deposited in the EMBL database (www.ebi.ac.uk) and given accession numbers HG970954 to HG970959, and LN885087. These sequences were also officially named and deposited in the IPD-MHC NHP database (19).
Genomic sequencing of partial MHC class I genes to identify their presence or absence
For genomic DNA, the PCR (50 μl total) contained 100 nanograms of DNA, 0.5 μM of each relevant primer (primer set F, Fig. S1), 0.2 mM of each dNTP, 1.5 mM MgCl2, 5 μl 10× PCR buffer, and 1 unit platinum Taq polymerase (Invitrogen). The cycling parameters consisted of an initial denaturation step of 1 min at 94 °C, followed by 30 cycles at 94 °C 30 s, 64 °C 30 s, 72 °C 50 s, with a final extension of 10 min at 72 °C. The PCR products (349 bp in size, covering part of exon 2, complete intron 2, and part of exon 3) were purified as described above, and cloned and sequenced as described previously (18). The data were analyzed using the programs Lasergene Seqman Pro version 11.2.1 and MacVector version 12.7.5. Three independent PCR reactions were performed for each animal to confirm the presence or absence of the different orangutan MHC class I alleles.
Analysis of cell-surface expression of MHC class I allotypes
Cell-surface expression of MHC class I allotypes on the orangutan EBV-transformed B-cell lines was investigated by flow cytometry using a panel of human HLA class I specific monoclonal antibodies (mAb). The amino acid sequences of the orangutan MHC class I allotypes were examined for the presence of epitopes previously defined from the patterns of reactivity of anti-HLA mAb with HLA class I. Monoclonal antibodies that reacted with these putative trans-species epitopes were selected for analysis. The HLA specificities of the mAbs had been previously determined by complement-dependent cytotoxicity assays against large panels of HLA-typed human PBMC, and confirmed in binding assays using beads coated with single HLA allotypes (20-22). For flow cytometry, the amount of mAb that gives saturation or near saturation was determined (Fig. S2). One hundred thousand orangutan or human cells were placed in wells of a 96 well plate or in tubes. Prior to adding the mAb, excess culture media was removed by two washes with FACS-buffer (PBS + 0.5% BSA made filter sterile). An amount of 25 μl mAb-containing hybridoma supernatant for WK1D12 (12.9 μg/ml), SN607D8 (16.5 μg/ml) and SN66E3 (20.1 μg/ml), 100 μl for VDK8F7 (142 μg/ml) and WK4C11 (24.2 μg/ml), and 200 ul for HDG11G12 (4.8 μg/ml) was added, and incubated on ice in the dark for 1 hour. After one wash with FACS-buffer cells incubated with mAb in FACS-tubes were transferred to a 96 well plate. Following two additional washes with FACS buffer, cells were stained with either F(ab′)2 goat anti human IgG/PE (Jackson ImmunoResearch) or with F(ab′)2 rabbit anti human IgM/FITC (Dako), depending on the isotype of the mAb being tested, for 30 min in the dark on ice. Cells were washed twice and then fixed with 2% paraformaldehyde. Conjugate-only controls were incorporated in the test, and the total MHC class I expression of the EBV-transformed B-cell lines was assessed with the fluorochrome-labeled antibody HLA-ABC/RPE (clone W6/32, Dako). FACS analyses were performed on the LSR II (BD Bioscience), and data analyses were performed with Flowjo software Version 9.7.2 (Flowjo © Tree Star, Inc).
Calculations and statistics
Neighbor-Joining (NJ) trees were constructed with the MEGA program version 4.0.2 (23), while pairwise distances were calculated using the maximum composite likelihood method, and bootstrap values were calculated based on 1000 replicates.
The percentage of transcription of each MHC class I allele was calculated by dividing the number of reads per allele/amplicon/individual by the total number of reads analyzed per amplicon/individual times 100%.
The relative expression of an MHC allotype is defined as:
Where gMFI denotes geometric mean fluorescence intensity, and mAb, conjugate-only control and W6/32 refer to the relevant incubations. The mean of expression in three independent determinations with standard deviation was plotted with Prism 6 Version 6.0d. The threshold value, above which all values are considered positive relative expression, is defined as the mean of expression determined for the orangutans for SN607D8 in the case of IgG and SN66E3 in the case of IgM ± 3SD. Data relating to SN607D8 and SN66E3 antibodies were taken as controls because their target epitopes are not present on orangutan MHC class I molecules.
Results
MHC class I gene variation in orangutans
B-cell lines derived from six unrelated orangutans were used as a source of cDNA. From these samples, full-length mature MHC class I transcripts were independently amplified, cloned, and sequenced using the Sanger method. Intensive sequencing of the samples, using four different primer sets to retrieve as many different transcripts as possible, resulted in the identification of 30 diverse full-length MHC class I alleles, derived from various genes (Fig. 1). Nonetheless, results of the analysis also suggested that certain alleles, in particular MHC-C, are poorly transcribed. Therefore, we wished to investigate the transcription/expression profile of the orangutan MHC class I genes in more detail.
Figure 1. MHC class I alleles present in six unrelated orangutans.
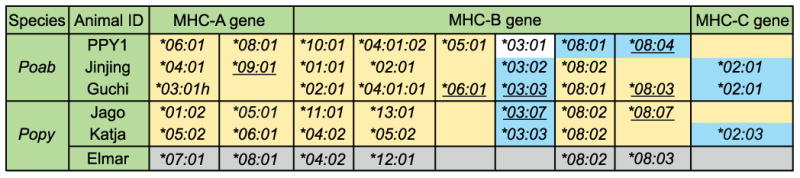
Poab and Popy signify Pongo abelii and Pongo pygmaeus, respectively. All alleles, except those underlined, comprise full-length sequences detected using the Sanger method. The allele names on a yellow background are the alleles detected by pyrosequencing with stringent filtering. The alleles on a blue background are the alleles detected by pyrosequencing with less stringent filtering. Underlined are alleles represented by exon 2 and exon 3 sequences that were confirmed using the Sanger method. For orangutan Elmar, only Sanger sequencing was performed, and the data are presented on a grey background. On a white background is the allele detected only by Sanger sequencing. The “h” indicates that the individual is likely homozygous for this particular allele. The sequences of Popy-A*05:01 and -C*02:03 were published previously (9, 19).
Sequences identified by means of the Sanger method were the model for the design of generic MHC-A, -B, and -C primers targeting conserved regions in exons 2 and 4. Complementary DNA from five of the cell lines was available and subjected to two independent rounds of amplification. Owing to the incorporation of different MIDs, the two different amplicons could be distinguished (Fig. S1). Pyrosequencing of the ten amplicons gave a total number of 50,496 reads, with an average of 10,099 reads per individual (range = 8,863 – 11,983) (Fig. S2).
A stringent filtering of the reads was first performed, and ∼200 reads per individual were analyzed. This approach allowed the identification of 25 different MHC class I alleles (Fig. 1, yellow background). Although most of these alleles had been identified previously by the Sanger method, four of the sequences (Fig.1, underlined alleles on a yellow background) were newly identified, and using the Sanger sequencing method we confirmed their exon 2 and exon 3 sequences. However, certain full-length MHC class I alleles that had been identified using the Sanger method were not obtained upon stringent filtering: notably, all MHC-B*03, some -B*08, and all MHC-C alleles (Fig.1, non-underlined alleles on a blue background).
Hence, a less stringent filtering was applied to the reads, to search for as yet unidentified MHC class I sequences. This approach resulted in the analysis of ∼2800 reads per individual, and all but one of the MHC class I alleles identified using the Sanger method were obtained with this analysis. In addition, three previously unreported alleles, Poab-B*03:03 and -B*08:04 and Popy-B*03:07, were detected (Fig. 1, underlined alleles on a blue background), and for these alleles we also confirmed their exon 2 and exon 3 sequences by means of the Sanger method. Neither stringent nor less stringent filtering yielded sequences representing the Poab-B*03:01 allele identified in orangutan PPY1 using the Sanger method.
An average of 10% of the reads, both in the stringent and less stringent filtering, could not unambiguously be assigned to an allele (Fig. S2). An independent analysis of genomic DNA derived from the cell lines demonstrated that all the MHC class I alleles detected by cDNA had corresponding genes, and indicated that the genomes of PPY1 and Jago did not contain an MHC-C gene.
The Sanger and pyrosequencing methods resulted in the detection of a total number of 35 alleles that had not been previously reported to the IPD-MHC NHP database: 20 for Pongo abelii and 15 for Pongo pygmaeus. Phylogenetic analysis – combining the 35 sequences with previously reported orangutan MHC class I sequences (8-10, 24-26) – showed that for the MHC-A, -B, and -C genes, lineages are shared between the orangutan species. Moreover, unreported lineages in both species could be defined for the MHC-A and -B genes (Fig. 2), illustrating that these genes are highly polymorphic in orangutans. In sharp contrast, the MHC-C gene shows a low level of polymorphism, since of the 35 identified alleles, 10 are MHC-A alleles, 24 are MHC-B alleles, and only 1 is an MHC-C allele. In every orangutan, one or two MHC-A alleles were identified, consistent with the presence of one A gene on an MHC haplotype (Fig. 1). For MHC-B, 4-6 alleles per individual were identified, which indicates the presence of 2-3 different B genes in the orangutan MHC. However, at this stage we cannot exclude the possibility that MHC haplotypes in orangutans show more complex variations with regard to gene copy number variation. In four subjects, PPY1, Guchi, Jago, and Elmar, two different alleles of the MHC-B*08 lineage were detected (Fig. 1), indicating that in these subjects both haplotypes possess an MHC-B*08 allele, and the likelihood that this lineage is encoded by a separate gene. Jinjing and Katja appear to be homozygous for the B*08 allele. Again, as the studied orangutans are unrelated (27), and pedigree analysis could not be performed, it is difficult to make any firm statement regarding the haplotype distribution of the other MHC-B genes/alleles. In the case of MHC-C, three of the orangutans have one allele, and for the others no MHC-C allele was identified. This transcriptional analysis is consistent with previous genomic and cDNA studies, demonstrating that the C gene is absent from ∼50% of orangutan MHC haplotypes (8, 9). Moreover, both forms of haplotype – those possessing and those lacking an MHC-C gene – are present in the Sumatran (Poab) and Bornean (Popy) species of orangutan (Fig. 1).
Figure 2. Phylogenetic comparison of Pongo abelii (Poab) and Pongo pygmaeus (Popy) MHC class I full-length cDNA sequences.
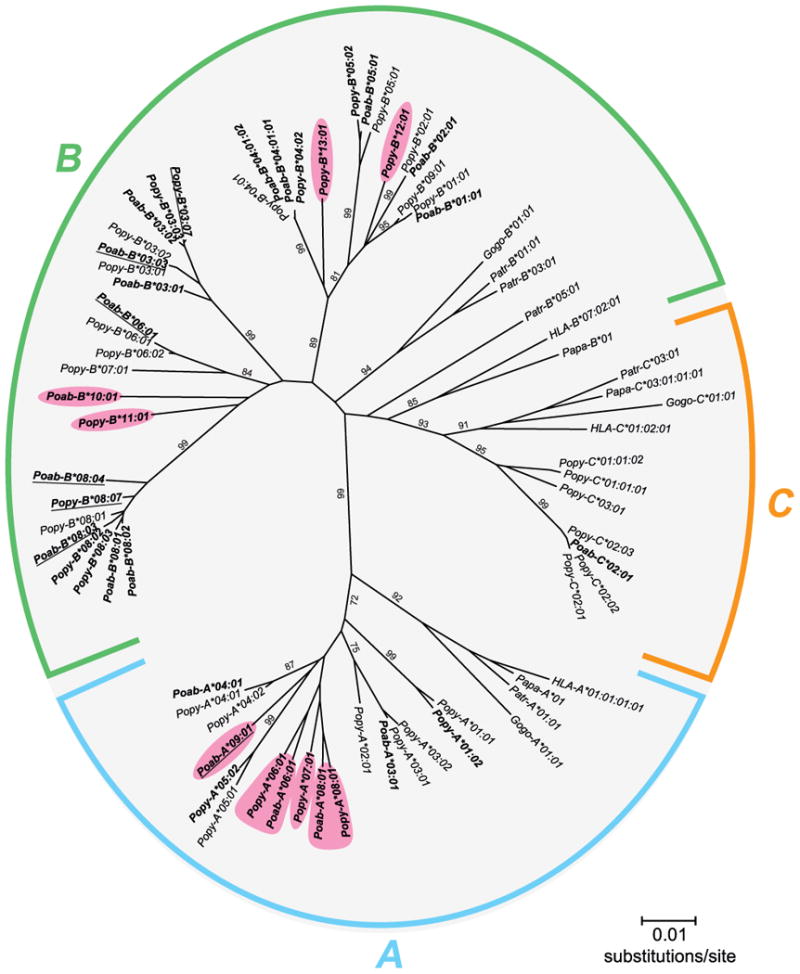
Particular human (HLA), bonobo (Papa), chimpanzee (Patr), gorilla (Gogo) and previously published orangutan MHC class I sequences were included (19). Alleles newly identified in this study are indicated with boldface. The pink boxes indicate new alleles clustering into previously unreported lineages. Relevant bootstrap values (>70) are indicated. The alleles with underlined names were assigned on the basis of only exon 2 and 3 sequences. This, however, did not affect the results of the phylogenetic comparison.
Orangutan MHC class I genes display differential levels of transcription
From the distribution of reads from each amplicon, the percentage transcription was calculated for the various MHC class I alleles detected in each orangutan B-cell line (Fig. S2). The analysis was independently performed for the reads obtained using stringent and less stringent filtering, and for each allele there was concordance between the four assessments. Despite variation in the total number of reads obtained, the relative representation of the different MHC class I transcripts was remarkably similar. The less stringent filtering gathered 14-fold more reads and a better representation of the MHC class I variability present in the orangutans, and was therefore used to plot histograms (Fig. 3).
Figure 3. Comparison of the transcription of MHC class I alleles detected in five orangutan cell lines.
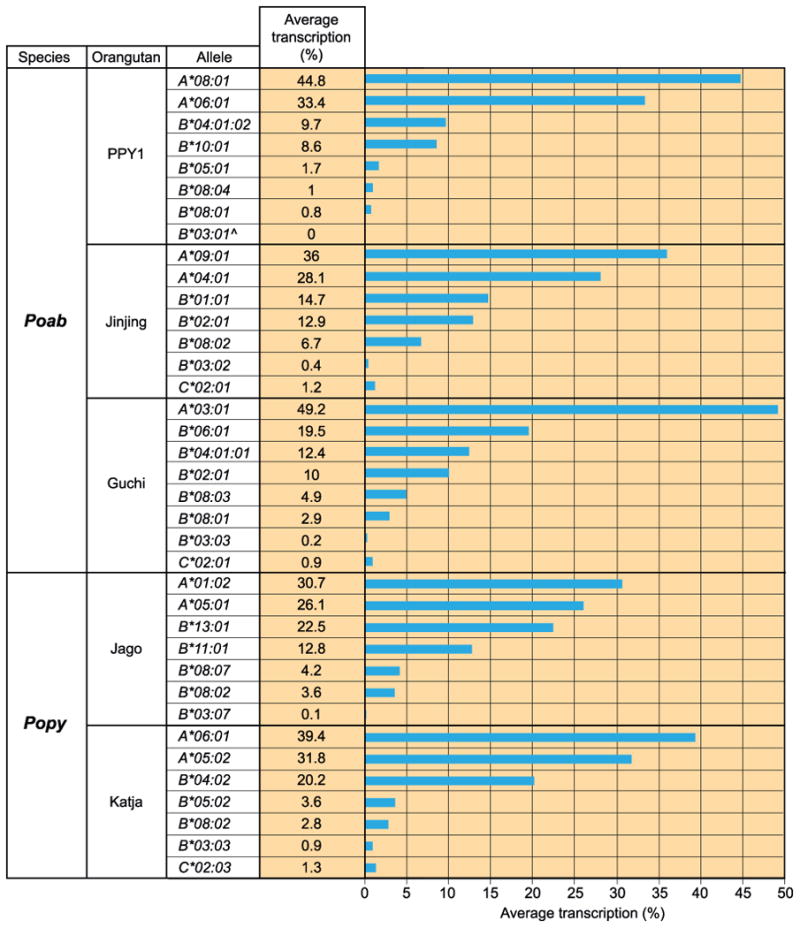
For each of the five orangutan B cell lines (PPY1, Jinjing, Guchi, Jago, and Katja) the relative transcription of each MHC class I allele is given as a percentage of the total. The values are the averages of two determinations (Fig. S2). The analysis was performed on the data obtained with the less stringent filtering. ˆNo reads were obtained for this allele in the pyrosequencing; as a result, the transcription percentage is equal to zero.
The extent to which the MHC class I genes are transcribed varies over a wide and continuous range from 0 to ∼50% of the total MHC class I transcription. The analysis illustrated that the MHC-A gene is transcribed highly (26.1-49.2%), MHC-B shows differential transcription ranging from 0 to 22.5%, and MHC-C, if present, is only weakly transcribed (0.9-1.3%) (Fig. 3).
The individual MHC-A alleles in the four heterozygous animals are transcribed at similarly high levels. In the orangutans heterozygous for the MHC-B*08 gene, the individual alleles also make a similar transcriptional contribution, although the level of transcription is much lower than for the A alleles. Taking all five orangutans into consideration, the relative contribution of the MHC-B*08 genes is 1.8-7.8% compared to 49.2-78.2% for MHC-A. From all alleles identified, the MHC-B*03 alleles are transcribed at a very low level (0-0.9%). This result was obtained for all five orangutans examined, and is consistent with MHC-B*03 being an independent gene. Because the haplotype distribution of the other B alleles cannot be determined, it is challenging to describe their relative contribution to the total transcription level. An upper limit can be estimated, however, by summing the two MHC-B alleles with the highest transcription in each cell line. These sums give values from 18.3-35.3%. The above analysis demonstrates how the orangutan MHC-B genes exhibit more complex transcription profiles than MHC-A and -C genes.
Previously, for the three alleles, Popy-B*03:02, -B*06:01, and -B*07:01, the second nucleotide on the start codon was shown to be deleted, and translation initiation for these alleles is most likely provided by the ATG present on the fourth codon, which lies downstream (9). Equivalents of Popy-B*03:02 and -B*06:01 are present in our panel. All our orangutans transcribe a B*03-lineage allele, and most likely use the ATG present on the fourth codon. Transcription profiles for the MHC-B*03 alleles show that they are less well transcribed than other MHC-B alleles. In contrast, Poab-B*06:01, present in Guchi, is also likely to use the ATG present on the fourth codon, and shows the highest transcription of all MHC-B alleles present in Guchi. This suggests that differential start codon usage does not explain why particular orangutan MHC-B alleles are less well transcribed.
Certain human anti-HLA class I mAbs recognize orangutan MHC class I molecules
The amino acid sequences of the α1 and α2 domains of orangutan MHC class I allotypes were examined for the presence of motifs that are known to be epitopes of a panel of human anti-HLA class I mAbs (Fig. S3). Particular motifs are present in the orangutan MHC class I allotypes (Fig. S3), and to monitor the cell surface expression of specific orangutan MHC class I molecules, 20 mAbs were selected and analyzed for their binding to the orangutan cell lines by flow cytometry. Six of the antibodies tested – three IgG (WK1D12, HDG11G12, and SN607D8) and three IgM (VDK8F7, WK4C11, and SN66E3) – gave informative reactions. The human JY cell line, expressing HLA-A*02:01, -B*07:02, and -C*07:02, was used as a control. Consistent with previous data, high amounts of HLA-A*02:01 and -B*07:02 allotypes were detected on the JY cell line with mAbs SN607D8/SN66E3 and WK1D12, respectively, and a low but distinct amount of the HLA-C*07:02 gene product was detected with WK4C11 (Figs. 4 and S4) (28). The results for JY with mAbs SN607D8 and SN66E3, respectively, an IgG and an IgM antibody binding to HLA-A*02:01, suggested that the IgG gave a ∼ 4 times stronger reaction.
Figure 4. Expression of MHC class I allotypes by five orangutan cell lines defined by six human monoclonal antibodies.
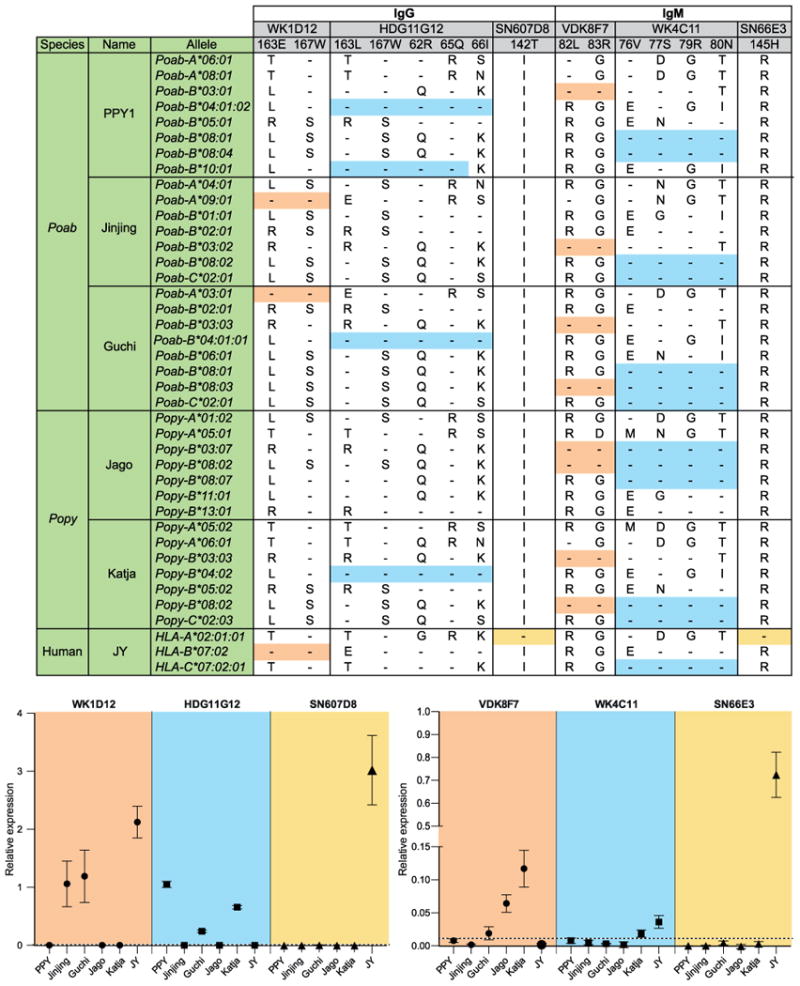
The top panel presents the six human mAbs, and gives the key residues in the human MHC class I sequences that determine the six epitopes and the distribution of those residues in orangutan MHC class I sequences. Identity to the amino acids in the target epitope is indicated by dashes, whereas amino acid replacements are given using the conventional one-letter code. Colored boxes (orange, blue, and yellow) indicate the MHC class I allotypes that have identical or nearly identical amino acids at the epitope-defining positions. In the bottom panel, the relative expression of a MHC class I allotype, as defined by flow cytometry of mAb bound by the five orangutan cell lines, is plotted as the mean with the standard deviation of three experiments. The dotted line represents the threshold value, the point above which all values indicate positive relative expression. The human cell line JY is taken as a control. Poab and Popy signify Pongo abelii and Pongo pygmaeus, respectively.
Differential cell-surface expression of orangutan MHC class I molecules
The specificity and diversity of the binding reactions of the six mAbs demonstrated the cell surface expression of certain orangutan MHC class I allotypes (Figs. 4 and S4). Antibody WK1D12 (IgG) binds to the cell lines of Jinjing and Guchi, but not to the other orangutan cell lines (Figs. 4 and S4). WK1D12 recognizes an epitope containing glutamate 163 and tryptophan 167, which is present in Poab-A*03:01 and -A*09:01 (Fig. 4). Antibody HDG11G12 (IgG) binds to the PPY1, Guchi, and Katja cell lines (Figs. 4 and S4), consistent with leucine 163 and tryptophan 167 in combination with arginine 62, glutamine 65, and isoleucine 66. This epitope characterizes the MHC-B*04 lineage. Four of the five residues of this epitope are present in Poab-B*10:01 (Fig. 4). This allele is transcribed by PPY1 as well, and might also account for HDG11G12 binding to PPY1.
Binding of antibody SN607D8 (IgG) to the highly expressed HLA-A*02:01 on the JY cell line gave an average relative expression of 3.02 (Fig. 4). In comparison, the binding of WK1D12 (average relative expression ranging from 1.06-1.19) indicates that Poab-A*03:01 and Poab-A*09:01 have high cell-surface expression indexes. HDG11G12 (average relative expression ranging from 0.24-1.05) reflects an intermediate to high cell-surface expression index for the orangutan MHC-B*04 allotypes. The seemingly higher expression by PPY1 cells can be explained by the binding of HDG11G12 to Poab-B*04:01:02 and Poab-B*10:01 (Fig. 4).
Antibody VDK8F7 (IgM) recognizes the combination of leucine 82 and arginine 83. This motif is present in orangutan MHC-B*03 and two -B*08 allotypes (Fig. 4). Antibody VDK8F7 binds to Guchi, Jago and Katja cell lines (Figs. 4 and S4), which transcribe MHC-B*03 and -B*08 allotypes having the indicated motif. VDK8F7 does not bind to PPY1 and Jinjing, which transcribe only MHC-B*03 allotypes with the indicated motif. Therefore VDK8F7 most likely demonstrates the expression of MHC-B*08 allotypes in Guchi, Jago and Katja cell lines.
Antibody WK4C11 (IgM) recognizes the C1-epitope, comprising valine 76, serine 77, arginine 79, and asparagine 80. In humans, this epitope is present on particular HLA-C and some HLA-B allotypes. In orangutans, the epitope exists on MHC-B*08 allotypes present in all five orangutans studied as well as on MHC-C allotypes present in Jinjing, Guchi, and Katja (Fig. 4). WK4C11 binds to Katja but not to the other orangutan cell lines (Figs. 4 and S4). Because WK4C11 does not bind to Jago cells, which have the same MHC-B*08:02 allotype as Katja but lack MHC-C, the target for the binding of WK4C11 to Katja is probably the C1 epitope of Popy-C*02:03. The specificity of WK4C11 for C1 was defined by its binding to human HLA class I determinants. The mAb does not recognize the C1-epitopes carried by orangutan MHC-B*08 allotypes. As mAb VDK8F7 demonstrates that orangutan B*08 allotypes are expressed on the cell surface, lack of binding of WK4C11 to B*08 allotypes may be explained by the presence of additional polymorphisms neighboring the C1 epitope in orangutans (Fig. S3).
Binding of antibody SN66E3 (IgM) to the highly expressed HLA-A*02:01 on JY cells gave an average relative expression of 0.72 (Fig. 4). The average relative expression indexes detected with VDK8F7 and WK4C11 ranges from 0.02-0.12 and 0.02, respectively (Fig. 4), and is less then 3-17% of that seen for SN66E3 on JY cells. Thus the cell-surface expression of Popy-C*02:03 is low and for MHC-B*08 ranges from low to intermediate.
The cell-surface expression data and the transcriptional data correlate with each other. Allotypes with low to intermediate cell-surface expression (average relative expression ranging from 0.02-0.12) display a low transcription level (ranging from 0-6.7%). In contrast, allotypes with an intermediate to high cell-surface expression (average relative expression ranging from 0.24-1.19) have intermediate to high transcription levels (ranging from 13.4-34.2%).
Discussion
The MHC class I transcription repertoire was determined for six orangutan B-cell lines using Sanger and NGS methodologies. Thirty-five novel alleles were defined, 10 for MHC-A, 24 for MHC-B, and one for MHC-C. Pyrosequencing allowed us to define transcription levels of the MHC class I alleles in five cell lines. This revealed distinct profiles: a high transcription level for the MHC-A gene, differential transcription levels for the multiple MHC-B genes, and weak transcription of the MHC-C gene, which is only present in three of the cell lines. In addition, the cell-surface expression of orangutan MHC class I proteins was assessed. The six mAbs that gave informative data revealed an intermediate to high expression of the orangutan MHC-A*03, -A*09, -B*04, and -B*10 allotypes. For the MHC-B*08 allotypes a low to intermediate cell-surface expression was observed, and in only one of the three orangutans positive for MHC-C was cell-surface expression detected for this allotype, and it was low.
As in the human MHC, the data corroborate the presence of one active copy of an MHC-A gene per haplotype in orangutans. Orangutans may have haplotypes with at least 2 or 3 apparently functional MHC-B genes, as each individual transcribes between 4 to 6 different MHC-B alleles. However, due to the lack of pedigree information, the haplotype distribution of the MHC-B genes could not be firmly established, and we cannot exclude the possibility that more complex profiles exist. Our data do imply that both the MHC-B*03 and -B*08 alleles are encoded by an independent gene. Thus, in comparison to humans, chimpanzees, and gorillas, the orangutan shows a distinctive feature in that its MHC-B region has been subject to moderate expansion. Expansion of the MHC-B region is also observed in rhesus macaques, an Old World monkey species, and their individual genes are transcribed to different degrees (13, 15, 17). The present data show similar findings in orangutans, where the MHC-B gene and gene products were found transcribed and expressed at levels ranging from low to high. After the expansion of the orangutan MHC-B region, the transcription/expression level of some of its genes/gene-products may subsequently have been reduced. The mechanisms that control such events have yet to be explored.
Evolutionary analyses illustrate that gene duplication can result in the emergence and deletion of genes, or in their subsequent degeneration into pseudogenes (2, 29). Such phenomena may have determined the fate of the MHC-C gene in orangutans. Orangutan haplotypes can lack a MHC-C gene, resulting in some individuals lacking MHC-C. When present, the MHC-C gene is characterised by low levels of polymorphism at the population level and a low level of transcription, and the gene product may not reach the cell surface.
In humans, HLA-C epitopes are well-known ligands for lineage III KIR members. These KIRs specifically recognize the C1 or C2 epitope encoded by HLA-C as well as by HLA-B*46 and -B*73 gene products (30). In orangutans, the lineage III KIRs are expanded (31). We demonstrate that in orangutans the C1 epitope is present on MHC-C and MHC-B*08 allotypes. All orangutans we examined have at least one MHC-B*08 variant, and the VDK8F7 mAb showed that MHC-B*08 allotypes are present on the cell surface (Fig. 4). Thus MHC-B*08 allotypes are potential ligands for KIRs on NK cells.
Various HLA-B and some HLA-A alleles encode the Bw4 motif (77N, 78L, 79R, 80I, 81A, 82L, 83R), a ligand for lineage II KIRs. This KIR lineage is also present in orangutans (24). In orangutans, all MHC-B*03 and some -B*08 allotypes carry a Bw4-like motif (Fig. S3). With VDK8F7, the cell surface expression of MHC-B*08 allotypes was demonstrated, but it still needs to be determined whether the Bw4-like motif in orangutans indeed is a ligand for orangutan lineage II KIRs.
HLA-C is the only classical MHC class I gene expressed on the surface of human EVT cells. During placentation, interaction between HLA-C and the KIR receptors present on uterine NK cells is vital (32). Deep trophoblast invasion as observed in humans is found for chimpanzees and gorillas, and this may be attributed to the co-evolution of MHC-C and their cognate receptors (33, 34). In Old World monkeys and gibbons, both of which lack an equivalent of the HLA-C gene, trophoblast invasion is much shallower. For orangutans, the extent of the trophoblast invasion is not known (34). Although it is virtually impossible to obtain EVT cells/tissue, it would be of interest to study the MHC class I transcription levels on this type of cell in the orangutan. Nonetheless, in the case of MHC-C, one should bear in mind that a substantial proportion of orangutans (∼25%) completely lack the gene.
In summary, we have shown that the different MHC class I genes in orangutans differ in transcription profiles, as is reflected by the levels of cell-surface expression. Although certain orangutans lack an MHC-C gene, all studied orangutans in this study do transcribe an allele of the C1 epitope-encoding MHC-B*08 gene, which may function as a ligand for KIR receptors. Therefore, the duplicated form of this MHC-B gene may compensate for the absence or very poor expression of the -C gene in orangutans.
Supplementary Material
Acknowledgments
The authors thank Donna Devine for editing the manuscript, Henk van Westbroek for preparing the figures, and Dr. E.J. Remarque for assistance with the statistical analyses. We are also deeply grateful to Henk Niphuis for providing the Katja B-cell line.
Footnotes
This study was supported by the Biomedical Primate Research Centre. Lisbeth A. Guethlein and Peter Parham were supported by NIH grant AI31168.
Abbreviations: dNTP deoxynucleotide
EVT extravillous trophoblast
KIR killer-cell immunoglobulin-like receptor
NGS next-generation sequencing
Poab Pongo abelli
Popy Pongo pygmaeus
μM micromolar
mM millimolar
References
- 1.Kasahara M, Suzuki T, Pasquier LD. On the origins of the adaptive immune system: novel insights from invertebrates and cold-blooded vertebrates. Trends Immunol. 2004;25:105–111. doi: 10.1016/j.it.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Piontkivska H, Nei M. Birth-and-death evolution in primate MHC class I genes: divergence time estimates. Mol Biol Evol. 2003;20:601–609. doi: 10.1093/molbev/msg064. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. Natural killer cells: from no receptors to too many. Immunity. 1997;6:371–378. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- 4.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 5.Parham P, Norman PJ, Abi-Rached L, Hilton HG, Guethlein LA. Review: Immunogenetics of human placentation. Placenta. 2012;33(Suppl):S71–80. doi: 10.1016/j.placenta.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A, Ponting CP, Marques-Bonet T, Alkan C, Chen L, Cheng Z, Kidd JM, Eichler EE, White S, Searle S, Vilella AJ, Chen Y, Flicek P, Ma J, Raney B, Suh B, Burhans R, Herrero J, Haussler D, Faria R, Fernando O, Darre F, Farre D, Gazave E, Oliva M, Navarro A, Roberto R, Capozzi O, Archidiacono N, Della Valle G, Purgato S, Rocchi M, Konkel MK, Walker JA, Ullmer B, Batzer MA, Smit AF, Hubley R, Casola C, Schrider DR, Hahn MW, Quesada V, Puente XS, Ordonez GR, Lopez-Otin C, Vinar T, Brejova B, Ratan A, Harris RS, Miller W, Kosiol C, Lawson HA, Taliwal V, Martins AL, Siepel A, Roychoudhury A, Ma X, Degenhardt J, Bustamante CD, Gutenkunst RN, Mailund T, Dutheil JY, Hobolth A, Schierup MH, Ryder OA, Yoshinaga Y, de Jong PJ, Weinstock GM, Rogers J, Mardis ER, Gibbs RA, Wilson RK. Comparative and demographic analysis of orang-utan genomes. Nature. 2011;469:529–533. doi: 10.1038/nature09687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson DE, Reeder DM, editors. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd. Johns Hopkins University Press; Baltimore, MD: 2005. pp. 111–184. [Google Scholar]
- 8.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 9.Adams EJ, Thomson G, Parham P. Evidence for an HLA-C-like locus in the orangutan Pongo pygmaeus. Immunogenetics. 1999;49:865–871. doi: 10.1007/s002510050566. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZW, McAdam SN, Hughes AL, Dogon AL, Letvin NL, Watkins DI. Molecular cloning of orangutan and gibbon MHC class I cDNA. The HLA-A and -B loci diverged over 30 million years ago. J Immunol. 1992;148:2547–2554. [PubMed] [Google Scholar]
- 11.Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, Parham P, Walter L. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babik W. Methods for MHC genotyping in non-model vertebrates. Mol Ecol Resour. 2010;10:237–251. doi: 10.1111/j.1755-0998.2009.02788.x. [DOI] [PubMed] [Google Scholar]
- 13.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulski JK, Anzai T, Shiina T, Inoko H. Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol. 2004;21:2079–2091. doi: 10.1093/molbev/msh216. [DOI] [PubMed] [Google Scholar]
- 15.Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr, Wright C, Harkins T, O'Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene JM, Wiseman RW, Lank SM, Bimber BN, Karl JA, Burwitz BJ, Lhost JJ, Hawkins OE, Kunstman KJ, Broman KW, Wolinsky SM, Hildebrand WH, O'Connor DH. Differential MHC class I expression in distinct leukocyte subsets. BMC Immunol. 2011;12:39. doi: 10.1186/1471-2172-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A. 2005;102:1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Groot NG, Heijmans CM, de Groot N, Doxiadis GG, Otting N, Bontrop RE. The chimpanzee Mhc-DRB region revisited: gene content, polymorphism, pseudogenes, and transcripts. Mol Immunol. 2009;47:381–389. doi: 10.1016/j.molimm.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Groot NG, Otting N, Robinson J, Blancher A, Lafont BA, Marsh SG, O'Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE. Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, Claas FH, Walker JJ, Redman CW, Morgan L, Tower C, Regan L, Moore GE, Carrington M, Moffett A. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J Clin Invest. 2010;120:4102–4110. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrari M, Mostecki J, Mulder A, Claas F, Balazs I, Duquesnoy RJ. Human monoclonal antibody reactivity with human leukocyte antigen class I epitopes defined by pairs of mismatched eplets and self-eplets. Transplantation. 2010;90:1468–1472. doi: 10.1097/TP.0b013e3182007b74. [DOI] [PubMed] [Google Scholar]
- 22.Mulder A, Kardol MJ, Arn JS, Eijsink C, Franke ME, Schreuder GM, Haasnoot GW, Doxiadis II, Sachs DH, Smith DM, Claas FH. Human monoclonal HLA antibodies reveal interspecies crossreactive swine MHC class I epitopes relevant for xenotransplantation. Mol Immunol. 2010;47:809–815. doi: 10.1016/j.molimm.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 24.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 25.Lawlor DA, Warren E, Ward FE, Parham P. Comparison of class I MHC alleles in humans and apes. Immunol Rev. 1990;113:147–185. doi: 10.1111/j.1600-065x.1990.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Laso J, Gomez-Casado E, Arnaiz-Villena A. Description of seven new non-human primate MHC-B alleles. Tissue Antigens. 2006;67:85–88. doi: 10.1111/j.1399-0039.2005.00496.x. [DOI] [PubMed] [Google Scholar]
- 27.Perkins L. International Studbook of the Orangutan (Pongo pygmaeus, Pongo abelli) Lincoln Park Zoo; 2001 North Clark Street, Chicago, IL 60614 USA: 2002. [Google Scholar]
- 28.Johnson DR. Differential expression of human major histocompatibility class I loci: HLA-A, -B, and -C. Hum Immunol. 2000;61:389–396. doi: 10.1016/s0198-8859(99)00186-x. [DOI] [PubMed] [Google Scholar]
- 29.Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci U S A. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 32.Parham P, Guethlein LA. Pregnancy immunogenetics: NK cell education in the womb? J Clin Invest. 2010;120:3801–3804. doi: 10.1172/JCI44559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter AM. Comparative studies of placentation and immunology in non-human primates suggest a scenario for the evolution of deep trophoblast invasion and an explanation for human pregnancy disorders. Reproduction. 2011;141:391–396. doi: 10.1530/REP-10-0530. [DOI] [PubMed] [Google Scholar]
- 34.Carter AM, Enders AC, Pijnenborg R. The role of invasive trophoblast in implantation and placentation of primates. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140070. doi: 10.1098/rstb.2014.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


