Abstract
Repair of double-strand breaks is essential for the maintenance of genome integrity and cell survival. In eukaryotes, double-strand-break repair by homologous recombination requires the Rad52 group of proteins. Human Rad52 protein (HsRad52)-mediated annealing of complementary strands has been studied in detail, but little has been reported on the recombinase activities of HsRad52. For this study, we purified HsRad52 from Escherichia coli. DNase I protection experiments indicated that HsRad52 binds preferentially to single-stranded DNA and protects it against digestion by DNase I. HsRad52 catalyzed D-loop formation in superhelical DNA, as well as strand exchange among oligonucleotide substrates. The formation of a stoichiometric complex between HsRad52 and single-stranded DNA was found to be critical for strand exchange activity, and the coating of both the single- and double-stranded oligonucleotides inhibited the exchange reaction.
Double-strand breaks (DSBs) in DNA can be caused by various exogenous agents, including ionizing radiation, chemical mutagens, and carcinogens, as well as by endogenous factors such as the collapse of replication forks and free radicals. If not repaired, DSBs pose a major threat to the integrity of the genome and cell survival (1). In eukaryotes, homologous recombination, a major pathway for DSB repair, accurately repairs DSBs without generating insertions and deletions (2–7). In addition to DSB repair, homologous recombination plays important roles in replication, segregation of chromosomes, and cell differentiation. It now seems that the major function of homologous genetic recombination in bacteria, and a major function in virtually all cells, is the recombinational repair of stalled or collapsed replication forks. This hypothesis is built upon the convergence of many independent lines of research (8–12), and it suggests an essential caretaker role for recombination proteins in maintaining genomic integrity.
In eukaryotes, the mechanisms by which homologous recombination repairs DSBs are not completely understood. The Rad52 epistasis group of genes, including genes for Rad51, Rad52, and Rad54, among others, has been implicated in playing important roles in homologous recombination and DNA repair (13–16). These proteins are highly conserved among all eukaryotes and so are the principal steps of recombination (2, 17–19). In vitro, yeast and human Rad51 proteins, the homologs of Escherichia coli RecA protein, have been shown to mediate central events in recombination: the pairing of homologous DNA molecules and the initiation of strand exchange between the paired partners. In doing so, Rad51 is assisted by several accessory proteins, including Rad52, Rad54, Rad55, and replication protein A (18, 20–23). Recently, two in vivo studies in yeast demonstrated the association of these proteins on a homologous donor sequence during strand invasion and indicated that these proteins, along with Rad51, assemble sequentially at the site of DSBs (21, 23).
Rad52 protein is unique to eukaryotes because it shows no clear homology to known recombination proteins in bacteria. Genetic studies in Saccharomyces cerevisiae have indicated that Rad52 functions in several recombination pathways, including gene conversion, single-strand annealing, and DNA break-induced replication (24, 25). In some of these pathways, it acts along with Rad51 protein, whereas in others, it functions independently of Rad51. The yeast and human Rad52 proteins have been purified and shown to bind preferentially to single-strand DNA (ssDNA), forming a nucleoprotein complex, which promotes annealing of complementary strands (26–30). By using electron microscopic and biochemical techniques, several studies have demonstrated that Rad52 protein self-associates to form ring-shaped oligomers as well as higher-order complexes of these rings (27, 31–34). Recently, human Rad52 protein (HsRad52) was shown to promote homologous pairing between an oligonucleotide and a superhelical plasmid (35) and between two oligonucleotide substrates (36). In addition to these Rad51-independent functions, yeast and human Rad52 proteins also seem to play an early role in promoting the assembly of Rad51 proteins on replication protein A-coated ssDNA (20, 22). In a recent study, HsRad52 protein was shown to interact with the Werner syndrome protein WRN. The biochemical activities of both proteins were stimulated by this interaction (37). An understanding of the enzymology of Rad52 is important for the further understanding of its role in homologous recombination and DNA repair. In this study, we demonstrate that HsRad52 protein promotes strand exchange activity with oligonucleotide substrates. Furthermore, we suggest that the formation of a stoichiometric complex between HsRad52 and ssDNA is critical for the catalysis of strand exchange.
Materials and Methods
DNA Substrates. Three 83-mer oligonucleotides were used in this study: GC19 (19% GC), GC29 (29% GC), and GC37 (37% GC). In the D-loop assay, a G16(-) oligonucleotide (16% GC) was used; the nucleotide sequence of this oligonucleotide has been published in ref. 38. The ssDNAs used to form the presynaptic complex were designated as minus-strands and the respective complementary strands as plus-strands. The sequences of oligonucleotides used in this study are as follows: GC19(-), 5′-AAG TGA ACA TAA AGT AAA TAA GTA TAA CGA TAA TAC AAA ATA AGT AAA TGA ATA AAG ATA GAA AAT AAA GTA AAG GAT ATG AA-3′; GC29(-), 5′-ATG TCC TTT CTT TAA ACT TCT ATC ATT GAT TCT TAC TAG TCT TTA CCT TAC TAT ACT TCT ATC AGT TTA TCG ATT CTT CTG TA-3′; and GC37(-), 5′-TTG ATA AGA GGT CAA TTT TGC GGA TGG CTT AGT GCT TAA TTG CTG AAT CAG GTG CTG TAG CTC AAC TTG TTT TAA ATA TGC AA-3′. These sequences were derived from the published sequences of oligonucleotides G16, G26, and G37 (38) after several modifications. Minus strands were labeled at the 5′ end with [γ-32P]ATP by using the enzyme polynucleotide kinase (New England Biolabs). Radiolabeled minus-strands were annealed with the respective unlabeled plus-strands by thermal annealing as described in ref. 39. The superhelical substrates used in the D-loop assay were prepared by a conventional method in which Triton X-100 was used for the cell extraction, followed by cesium chloride ultracentrifugation (40). All DNA concentrations refer to moles of nucleotide residues, except for duplex DNAs, which are expressed as moles of base pairs.
Purification of HsRad52. To express HsRad52 protein, E. coli BL21 DE3 pLys cells (Invitrogen) carrying the human Rad52 expression vector (a gift from Efim Golub of Yale University, New Haven, CT) were grown in terrific broth medium and induced in logarithmic phase by the addition of 1 mM isopropyl β-d-thiogalactopyranoside. Plasmid pEG161 is derived from the pET15b vector (Invitrogen), in which the human Rad52 gene is inserted between NdeI and BamHI sites. The expressed HsRad52 protein has six histidine residues at its N terminus. Purification of the recombinant protein included chromatographic steps through Q-Sepharose, SP-Sepharose, Ni2+-agarose, and DNA-cellulose column. HsRad52 was purified to homogeneity; one band of 48 kDa was seen by SDS/PAGE analysis of 2 μg of purified preparation (data not shown). The presence of nuclease and helicase activities in the purified HsRad52 preparation was checked by the method described by Gupta et al. (41). The following activities were undetectable (<2% activity after a 1-h incubation): endo- and exonuclease on single- and double-stranded 83-mer oligonucleotides, endonuclease or topoisomerase on φX174 superhelical DNA, and helicase on blunt-end and partially duplex oligonucleotides.
DNA Binding Analysis by Gel Mobility-Shift Assay. A typical reaction mixture consisted of 25 mM Mops (pH 6.8)/1 mM DTT/0.1 mg BSA per ml/0.1 mM MgCl2. For DNA binding, 3 μM 32P-labeled GC19(-) ssDNA or GC19(-/+) double-strand DNA (dsDNA) was incubated with 0.1–2 μM concentrations of HsRad52 protein in the reaction mixture (total volume 10 μl) at 37°C for 10 min to allow for the formation of the DNA–protein complex. The putative complex was covalently cross-linked by the addition of 1 μl of 2% glutaraldehyde followed by an additional 10-min incubation at 37°C. The reaction was terminated by the addition of 2 μl of stop solution (final concentration of 0.5% SDS/25 mM EDTA/0.04% bromophenol blue/0.04% xylene cyanol) and subjected to electrophoresis on a 1% agarose gel. The gel was run at 5 V/cm for 3 h in TAE buffer (40 mM Tris-acetate/1 mM EDTA) and dried, and the reaction products were quantitated by PhosphorImager analysis (Molecular Dynamics).
DNase I Protection. HsRad52 (0.1–2 μM) or RecA (0.1–2 μM) was incubated with 3 μM of 32P-labeled GC19(-) ss- or dsDNA in a reaction mixture (total volume 10 μl) as described in Gel-Shift Assay at 37°C for 10 min. The concentration of MgCl2 was increased to 5 mM before the addition of DNase I (MBI Fermentas, Hanover, MD): 1.5 units for ssDNA and 0.5 unit for dsDNA. The incubation was continued for 3 min, and the reaction was quenched by the addition of SDS to 0.5% and EDTA to 25 mM. Yeast tRNA was added as a carrier at a final concentration of 4 μg/μl before the precipitation of labeled DNA by cold trichloroacetic acid at a final concentration of 20%, and the radioactivity of the acid-soluble supernatant was measured by scintillation counting. This measurement reflects the extent of degradation of DNA molecules that are not protected through protein binding.
Assay for D-Loop Formation. HsRad52 (0.5 μM), RecA (1.2 μM), or HsRad51 (1.2 μM) was preincubated with 3 μM G16(-) ssDNA at 37°C for 10 min in a reaction mixture (total volume 10 μl), as described above, to allow the formation of the nucleoprotein complex. The concentration of MgCl2 then was increased to 1 mM, followed by the addition of a 3.0-kb supercoiled plasmid pEG47 carrying G16 sequences to a final concentration of 75 μM. At different time points, aliquots were deproteinized, and the samples were run on a 1% agarose gel, as described above. The gel was dried onto DEAE chromatography paper (Whatman), and the products were quantitated by using a PhosphorImager. The yield of D-loops was expressed as a percentage of the pEG47 plasmid incorporated into D-loops. For the heterologous control reaction, we used the plasmid pEG61, which is similar to pEG47 but possesses an 83-mer sequence that is heterologous to the G16 sequence in pEG47. In another heterologous control, G16(-) ssDNA was replaced by the GC29(-) ssDNA in the preincubation step.
Strand Exchange Activity. Preincubation of HsRad52 (0.25 μM) or RecA (1.2 μM) with 3 μM GC19(-) was performed as described above. After 10 min, the concentration of MgCl2 was increased to 1 mM, and the reaction was initiated by the addition of 2.5 μM [5′-32P]GC19(-/+) duplex oligonucleotide. Incubation was continued at 37°C and stopped as described above. In the heterologous control, GC19(-) ssDNA was replaced by the GC29(-) ssDNA in the preincubation step. The samples were analyzed on a nondenaturing 12% acrylamide gel run at 10 V/cm for 2 h in TBE buffer (90 mM Tris–borate, pH 8.3/2 mM EDTA). Products were quantified by PhosphorImager analysis. In the studies presented here, the positions at which single- and double-stranded oligonucleotides migrated on an acrylamide gel are different from the positions observed in previous studies (42). This altered pattern is probably caused by different gelling properties of the acrylamide and a different strength of TBE buffer used in these studies.
Results
Binding of HsRad52 to Oligonucleotides. We used 83-mer oligonucleotide substrates to detect DNA–protein complexes made by HsRad52. Two assays were used: a gel mobility-shift assay and an assay based on protection of DNA against digestion by DNase I (DNase I protection assay) (see Materials and Methods). In the gel-shift assay, DNA–protein complexes were covalently cross-linked by the addition of 0.2% glutaraldehyde followed by deproteinization with SDS and EDTA. On addition of HsRad52, the DNA–protein complex shifted to a position at or near the well on an agarose gel (Fig. 1). A shift near the well suggests the formation of large DNA–protein aggregates, which is also evident from refs. 30, 34, 35, and 43. In comparison with ssDNA binding, a relatively higher concentration of HsRad52 protein was needed for maximal binding to dsDNA. For example, maximal binding to ssDNA occurred at a protein concentration of ≈0.25 μM (Fig. 1 A), whereas ≈1 μM HsRad52 was needed to saturate dsDNA (Fig. 1B). This difference suggests that HsRad52 preferentially binds to ssDNA. A similar preference was observed when DNA binding by HsRad52 was tested by using a DNase I protection assay (Fig. 1C). ssDNA was almost completely protected by HsRad52 because only 15–20% of the total radioactivity was found in the acid-soluble supernatant, suggesting that the ssDNA inside the nucleoprotein complex is covered over its entire length by HsRad52. The extent of ssDNA protection was similar to that afforded by E. coli RecA protein. The results obtained from both assays further suggest that HsRad52 binds a significant percentage of dsDNA at the concentration of protein that is sufficient to saturate ssDNA.
Fig. 1.
Analysis of DNA–protein complexes made by HsRad52. (A and B) Gel mobility-shift assay. For this reaction, 3 μM radiolabeled 83-mer oligonucleotide GC19(-) ssDNA (A) or GC19(-/+) dsDNA (B) was incubated with the indicated concentrations of HsRad52 at 37°C for 10 min, followed by crosslinking with 0.2% glutaraldehyde, as described in Materials and Methods. Reaction products were electrophoresed on a 1% agarose gel. Radioactive material at the top of the gel represents higher-order DNA–protein complexes. U, unbound DNA. (C) Graphical representation of DNase I protection of ssDNA and dsDNA bound to HsRad52 and RecA. The reaction was performed as described in Materials and Methods. The amount of DNase I used (0.15 unit/μl for ssDNA and 0.05 unit/μl for dsDNA) was sufficient to digest all unprotected DNAs in 3 min.
In comparison with the gel-shift assay, the DNase I protection assay required a relatively higher stoichiometric concentration of protein for maximal binding, suggesting that perhaps DNA–protein complexes are not stable and dissociate upon the perturbation caused by increasing MgCl2 concentration (needed for optimal DNase I activity) or by the addition of DNase I.
Homologous Pairing Promoted by HsRad52. Next, we tested the ability of the HsRad52–ssDNA complex to catalyze the formation of D-loops and compared it with the similar reactions promoted by E. coli RecA and HsRad51. The D-loop assay detects the ability of a ssDNA to invade duplex DNA to initiate homologous pairing and strand exchange. In the reaction, an 83-mer oligonucleotide was preincubated with HsRad52, RecA, or HsRad51, and then superhelical DNA was added to initiate the reaction. D-loops formed by these proteins all migrated at the same position (Fig. 2A). Unlike the RecA-promoted D-loop (lanes 3–6), which dissociated at later time points during the reaction, the yields of D-loops catalyzed by HsRad52 (lanes 7–10) and HsRad51 (lanes 11–14) showed no tendency to decrease once a maximum was reached at ≈10 min (Fig. 2B). D-loop formation by HsRad52 depended upon homology of the interacting DNA. No reaction was seen when either ssDNA (lane 16) or superhelical DNA (lane 2) was replaced by DNA containing heterologous sequences. Our data suggest that HsRad52-mediated D-loop formation is slower than the reaction promoted by RecA, and it does not exhibit the pseudoreversible pattern observed for the RecA reaction. The D-loop-formation activity by HsRad51 as reported here and in ref. 44 demonstrates the activity in the absence of other accessory proteins. It should be mentioned here that in a recent study Sigurdsson et al. (45) found that at 30°C, a DNA unwinding protein, HsRad54, is required for HsRad51 to promote D-loop activity.
Fig. 2.
D-loop formation by RecA, HsRad52, and HsRad51 proteins. A reaction mixture containing radiolabeled 83-mer ssDNA (3 μM) was incubated with 0.5 μM HsRad52, 1.2 μM RecA, or 1.2 μM HsRad51 for 10 min. The reaction was initiated by adding superhelical DNA and was incubated further for 2, 5, 10, and 20 min. The reaction products were deproteinized and fractionated on a 1% agarose gel (A). Het represents control reactions in which heterologous sequences were used; heterologous superhelical DNA (lane 2) and heterologous ssDNA (lanes 15–18) were substituted for the homologous sequences in the preincubation step. The percent D-loop formed was plotted against time for RecA, HsRad52, and HsRad51 proteins (B).
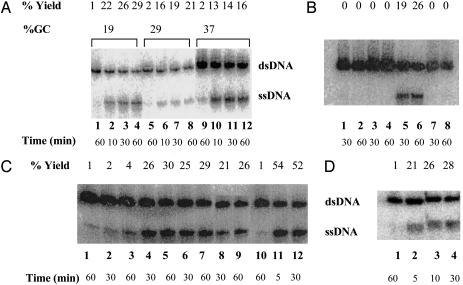
Strand Exchange Promoted by HsRad52. We used three different oligonucleotide substrates with various GC contents to explore strand exchange activity by HsRad52: GC19 (19% GC), GC29 (29% GC), and GC37 (37% GC). In all these reactions, HsRad52 was preincubated with a single-stranded oligonucleotide followed by the addition of radiolabeled homologous duplex. Displacement of radiolabeled ssDNA from the labeled duplex upon strand exchange was monitored. As shown in Fig. 3A, HsRad52 promoted strand exchange with all three substrates. The yield of the strand-exchange reaction at any time point was inversely related to the GC content of the oligonucleotides. This inhibitory effect of high GC content is similar to the published observations on the effect of GC content on strand exchange mediated by several prokaryotic and eukaryotic recombination proteins, including E. coli RecA and RecT and human Rad51 and Dmc1 proteins (41, 46, 47). This observation suggests that perhaps HsRad52 uses the same mechanism for recognition of homology as used by the above-mentioned recombination proteins, a mechanism that involves the preferential switching of A·T base pairs.
Fig. 3.
Strand exchange promoted by HsRad52. For this assay, the presynaptic complex was formed on single-stranded oligonucleotides, and radiolabeled duplex was added to initiate the reaction. Displacement of labeled ssDNA from this duplex as a result of strand exchange was monitored by nondenaturing PAGE. (A) Strand exchange reactions were performed with oligonucleotides GC19, GC29, and GC37. Lanes 1, 5, and 9 represent the reactions in which HsRad52 was omitted. (B) Strand exchange reaction was performed with the GC19 substrate. All lanes contained products from the reaction mixtures with the following omissions or substitutions: lanes 1 and 2, no protein; lanes 3 and 4, the heterologous GC29(-) oligonucleotide substituted for GC19(-); lanes 5 and 6, complete homologous reaction; lanes 7 and 8, no ssDNA. (C) Strand exchange reaction was performed with GC19 oligonucleotides with the following controls: lane 1, no protein; lanes 2 and 3, HsRad52 incubated with Ni2+ agarose beads as indicated in Materials and Methods, and the supernatant was then assayed for strand exchange activity; lanes 4 and 5, HsRad52 incubated with the buffer in which Ni2+ agarose beads were suspended; lanes 6 and 7, HsRad52 heated at 95°C for 10 min before preincubation with ssDNA; lanes 8 and 9, complete homologous reaction with HsRad52; lane 10, complete reaction with E. coli SSB protein; and lanes 11 and 12, complete reaction with RecA protein. (D) Strand exchange reaction with 3′ end-labeled duplex. HsRad52-mediated strand exchange reaction was performed with GC19(-) ssDNA and GC19(-/+) duplex labeled with 32P at its 3′ end. Lane 1, no protein; lanes 2–4, complete homologous reaction with HsRad52 protein.
For the experiments described below, GC19 oligonucleotides were used as the substrates. The strand exchange activity by HsRad52 depended on the homology of DNA substrates because no product was seen when heterologous ssDNA (in this case GC29) was used to form the filament (Fig. 3B, lanes 3 and 4). No reaction was seen when either HsRad52 (lanes 1 and 2) or ssDNA (lanes 7 and 8) was omitted. In another experiment with the GC19 substrate, we compared the strand exchange activity of HsRad52 with that of RecA (Fig. 3C). The RecA-promoted reaction (Fig. 3C, lanes 11 and 12) was both faster and more efficient than the similar reaction catalyzed by HsRad52 (lanes 8 and 9). After a 5-min incubation, the yield for the RecA mediated reaction was 54%, whereas the yield for a HsRad52 promoted reaction was 21% and 26% at 30 and 60 min, respectively. E. coli ssDNA binding protein (SSB), which can bind to ssDNA with high affinity, did not promote strand exchange activity under the similar reaction conditions (lane 10). We failed to detect SSB-mediated strand exchange under numerous different reaction conditions used. These included various concentrations of SSB, ssDNA, dsDNA, and MgCl2, as well as different temperatures and pH (data not shown).
To confirm that the strand exchange reaction was mediated by HsRad52 and was not an artifact due to some other components present in the protein preparation, we specifically eliminated HsRad52 (containing a histidine tag) by incubating it with Ni2+ agarose beads for 1 h, followed by centrifugation. The supernatant obtained was devoid of ≈80% of the protein as judged by protein estimation. In a control experiment, HsRad52 was processed similarly, except that the Ni2+ beads were replaced by an equal volume of buffer, in which the beads were suspended. Supernatants obtained after centrifugation were used to detect strand exchange activity. Elimination of HsRad52 by Ni2+ agarose drastically reduced strand exchange activity (<5% after 60 min) (Fig. 3C, lanes 2 and 3), whereas a control experiment indicated no change in activity (lanes 4 and 5) when compared with the HsRad52 reaction without these treatments (lanes 8 and 9). These results suggest that HsRad52 protein is needed for the strand exchange activity observed in our purified protein preparation. Strikingly, the yield of strand exchange was virtually unaffected when HsRad52 was heated at 95°C for 10 min before the formation of filament (Fig. 3C, lanes 6 and 7). By contrast, the same treatment inhibited strand exchange reactions by E. coli RecA and HsRad51 proteins (data not shown). This finding is consistent with the biophysical studies that examined the thermostability of HsRad52 protein (48).
When purified HsRad52 was checked for the presence of contaminating nuclease and helicase activities, no such activities were detected in our purified preparation (see Materials and Methods). To further exclude the possibility that the strand exchange is an artifact due to contaminating nuclease activity, we performed strand exchange with duplex substrate labeled at its 3′ end of the noncomplementary strand (Fig. 3D). The reaction efficiency was compared with another reaction in which the duplex was labeled at the 5′ end of the noncomplementary strand (Fig. 3C, lanes 8 and 9). The yield was similar when the noncomplementary strand was labeled at the 5′ end or the 3′ end. This finding further suggests that the displacement of ssDNA in our strand exchange reaction is not due to exonucleolytic digestion from either end of the duplex. Moreover, we performed an experiment similar to the one described in the accompanying paper by Bi et al. (49) to exclude the effect of contaminating helicase activity on HsRad52 mediated strand exchange. We failed to detect any evidence of such effect in our experiments (data not shown).
Effect of HsRad52–dsDNA Complex on Strand Exchange Activity. Our observations suggested that HsRad52 binds to both ss- and dsDNA and that the formation of a HsRad52–ssDNA complex leads to pairing and strand exchange reaction. Next, we examined the effect of varying the ratio of HsRad52 to ssDNA on strand exchange activity. For these experiments, 3 μM GC19(-) oligonucleotide was incubated with different concentrations of HsRad52 protein before the addition of the duplex. The optimal strand exchange activity occurred at the ratio of 1 HsRad52 molecule to ≈6–12 nucleotides (Fig. 4A), which is similar to the stoichiometry for optimal binding to ssDNA as deduced from the gel-shift assay (Fig. 1). A further increase in HsRad52 concentration inhibited the strand exchange reaction. At the ratio of 1 HsRad52 molecule to 1.5 nucleotides, the exchange reaction was obliterated. One possible explanation for this inhibition is that the excess of HsRad52 protein (after the formation of the HsRad52–ssDNA complex) binds to and sequesters dsDNA, thus inhibiting both homologous recognition and subsequent strand exchange (50). This explanation is consistent with the observation that HsRad52 binds to dsDNA and protects it against digestion by DNase I. To directly test the effect of binding of dsDNA by HsRad52 on pairing with the HsRad52–ssDNA complex, we incubated dsDNA separately in another tube with various concentrations of HsRad52 and then mixed it with preformed HsRad52–ssDNA complex. The yield of strand exchange product was compared with that found in a reaction in which HsRad52–ssDNA complex was treated with naked dsDNA (50). As shown in Fig. 4B, precoating of dsDNA with HsRad52 strongly inhibited strand exchange activity. When dsDNA (3 μM) was incubated with HsRad52 at the ratios of 12 and 6 nucleotides per HsRad52 molecule, the reaction efficiency was reduced to 57% and 29%, respectively, to that observed with naked dsDNA. The reaction was completely eliminated at the ratio of 3 nucleotides per HsRad52 molecule, the ratio at which HsRad52 completely saturates dsDNA (Fig. 1B). These data suggest that the simultaneous coating of both single- and double-stranded oligonucleotides inhibits the strand exchange reaction by HsRad52. The optimal strand exchange occurs when HsRad52 is bound to ssDNA and the dsDNA is uncoated.
Fig. 4.
Effect of HsRad52–dsDNA complex on strand exchange activity. (A) HsRad52 at 0.25. 0.5, 1.0, 1.5, and 2.0 μM concentrations was incubated with 3 μM of GC19(-) ssDNA at 37°C for 10 min. The radiolabeled duplex then was added to initiate the reaction. The yield of strand exchange promoted by HsRad52 was plotted as a function of protein concentration. (B) HsRad52 at different concentrations (0.25, 0.5, 1.0, 1.5, and 2.0 μM) was incubated with 3μM dsDNA for 10 min at 37°C. This mixture was then added to a preformed Rad52–ssDNA complex (3 μM ssDNA + 0.25 μM HsRad52) and incubated further for 60 min. The reactions were deproteinized, and the products were analyzed by gel electrophoresis. The yields of strand exchange products were normalized with respect to the yield from the reaction in which dsDNA was not coated (18%), which was assigned as 100%. HsRad52 concentration on the x-axis refers to the concentration of HsRad52 protein used for preincubation with dsDNA.
Discussion
Human Rad52 protein was previously shown to catalyze the annealing between two complementary strands and homologous pairing between a single- and a double-stranded DNA molecule. Herein, we report that HsRad52 protein also promotes strand exchange between two homologous DNA molecules, a hallmark of the recombination reaction.
E. coli cells overexpressing HsRad52 protein were used as a source for the purification of HsRad52. HsRad52 formed higher-order complexes with oligonucleotides, which shifted near the well in a gel-shift assay. In earlier studies, the formation of such higher-order complexes by HsRad52 was shown to be critical for the efficient catalysis of strand annealing and end joining (32, 34). Human Rad52 catalyzed the formation of D-loops in superhelical DNA, a standard homologous pairing reaction. Similar to the reaction catalyzed by HsDmc1 (51) and HsRad51 (Fig. 3), the reaction promoted by HsRad52 is slower than that promoted by RecA, has a lower yield, and fails to exhibit the pseudoreversibility characteristic of the RecA reaction. In RecA, the pseudoreversibility has been ascribed to the ATP-dependent “processive unwinding” of superhelical DNA (52). The human proteins may lack a similar ability to unwind DNA. Furthermore, because human proteins hydrolyze ATP much more slowly, this difference may presumably slow the turnover of these proteins from the ssDNA. This slowdown, in turn, may prevent the second round of strand invasion from the displaced strand, which otherwise could dissociate D-loops.
HsRad52 catalyzed a strand exchange between two homologous oligonucleotides. HsRad52-catalyzed strand exchange activity is slower than that of RecA, a property shared by other human recombination proteins, such as Rad51 and Dmc1 (42, 51). For the reasons mentioned below, we believe that the strand exchange activity reported here is not an artifact due to the presence of contaminating nuclease or helicase activities. First, when radiolabeled oligonucleotide substrates were used, no contaminating endonuclease, exonuclease, or helicase activity was detected in our HsRad52 preparation. Second, a similar yield of strand exchange products was observed when the noncomplementary strand (designated minus strand) in the duplex was labeled at either the 5′ or 3′ end. This observation excludes the possibility that the observed strand exchange activity is due to the exonucleolytic digestion from either end of the duplex, which creates a single-stranded region to initiate annealing. Third, the yield of strand exchange product was comparable when the plus strand was labeled in the duplex and HsRad52 was preincubated with the unlabeled plus strand (data not shown). Furthermore, the catalysis of strand exchange by HsRad52 protein, which had been preheated at 95°C, suggests that the observed activity is not due to the presence of contaminating thermolabile nucleases, helicases, or recombinases.
We suggest that the strand exchange reaction by HsRad52 is mediated via a stoichiometric complex of HsRad52 on ssDNA. Optimal strand exchange occurs at the concentration of HsRad52 that is enough to saturate ssDNA. Simultaneous coating of both single- and double-stranded oligonucleotides has a strong tendency to inhibit the strand exchange reaction, presumably by sequestering dsDNA. Kagawa et al. (35) showed that the incubation of HsRad52 with dsDNA before the addition of ssDNA inhibits D-loop formation. When these observations are taken together, it seems that the HsRad52–ssDNA complex provides catalytic surface for promoting pairing and strand-exchange activity. Earlier, we demonstrated that the preferential breathing of A·T base pairs in the duplex DNA plays a critical role in promoting homologous pairing by human Rad51 and Dmc1 and bacterial RecT proteins (38, 41, 46). Further studies are needed to understand the mechanism by which the HsRad52–ssDNA complex recognizes homology in a duplex DNA.
Acknowledgments
We thank Dr. Efim Golub and Dr. Charles Radding (Yale University School of Medicine, New Haven, CT) for providing the human Rad52 expression vector pEG161 and for sharing their results before publication; and Dr. Richard Cunningham and Dr. Richard Zitomer for critically reviewing this manuscript. This research was supported by the University at Albany's College of Arts and Sciences financial start-up package, the University at Albany Faculty Research Award Program, and the Research Foundation's Biological Sciences research support grant.
Abbreviations: DSB, double-strand break; dsDNA, double-stranded DNA; HsRad52, human Rad52 protein; ssDNA, single-stranded DNA.
References
- 1.Pierce, A. J., Stark, J. M., Araujo, F. D., Moynahan, M. E., Berwick, M. & Jasin, M. (2001) Trends Cell Biol. 11, S52-S59. [DOI] [PubMed] [Google Scholar]
- 2.Cromie, G. A., Connelly, J. C. & Leach, D. R. (2001) Mol. Cell 8, 1163-1174. [DOI] [PubMed] [Google Scholar]
- 3.Haber, J. E. (2000) Curr. Opin. Cell Biol. 12, 286-292. [DOI] [PubMed] [Google Scholar]
- 4.Thompson, L. H. & Schild, D. (2001) Mutat. Res. 477, 131-153. [DOI] [PubMed] [Google Scholar]
- 5.Modesti, M. & Kanaar, R. (2001) Genome Biol. 2, Reviews 1014.1-1014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thacker, J. (1999) Biochimie 81, 77-85. [DOI] [PubMed] [Google Scholar]
- 7.van den Bosch, M., Lohman, P. H. & Pastink, A. (2002) Biol. Chem. Hoppe-Seyler 383, 873-892. [DOI] [PubMed] [Google Scholar]
- 8.Cox, M. M. (2001) Annu. Rev. Genet. 35, 53-82. [DOI] [PubMed] [Google Scholar]
- 9.Kowalczykowski, S. C. (2000) Trends Biochem. Sci. 25, 156-165. [DOI] [PubMed] [Google Scholar]
- 10.Flores-Rozas, H. & Kolodner, R. D. (2000) Trends Biochem. Sci. 25, 196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marians, K. J. (2000) Curr. Opin. Genet. Dev. 10, 151-156. [DOI] [PubMed] [Google Scholar]
- 12.Michel, B., Flores, M. J., Viguera, E., Grompone, G., Seigneur, M. & Bidnenko, V. (2001) Proc. Natl. Acad. Sci. USA 98, 8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Game, J. C. (1993) Cancer Biol. 4, 73-83. [PubMed] [Google Scholar]
- 14.Petes, T. D., Malone, R. E. & Symington, L. S. (1991) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, eds. Broach, J. R., Pringle, J. R. & Jones, E. W. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 407-521.
- 15.Friedberg, E. C., Siede, W. & Cooper, A. J. (1991) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics, eds. Broach, J. R., Pringle, J. R. & Jones, E. W. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 147-192.
- 16.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paques, F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sung, P., Trujillo, K. M. & Van Komen, S. (2000) Mutat. Res. 451, 257-275. [DOI] [PubMed] [Google Scholar]
- 19.Bianco, P. R., Tracy, R. B. & Kowalczykowski, S. C. (1998) Front. Biosci. 3, 570-603. [DOI] [PubMed] [Google Scholar]
- 20.West, S. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 435-445. [DOI] [PubMed] [Google Scholar]
- 21.Sugawara, N., Wang, X. & Haber, J. E. (2003) Mol. Cell 12, 209-219. [DOI] [PubMed] [Google Scholar]
- 22.Sung, P., Krejci, L., Van Komen, S. & Sehorn, M. G. (2003) J. Biol. Chem. 278, 42729-42732. [DOI] [PubMed] [Google Scholar]
- 23.Wolner, B., van Komen, S., Sung, P. & Peterson, C. L. (2003) Mol. Cell 12, 221-232. [DOI] [PubMed] [Google Scholar]
- 24.Kraus, E., Leung, W. Y. & Haber, J. E. (2001) Proc. Natl. Acad. Sci. USA 98, 8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haber, J. E. (2000) Trends Genet. 16, 259-264. [DOI] [PubMed] [Google Scholar]
- 26.Mortensen, U. H., Bendixen, C., Sunjevaric, I. & Rothstein, R. (1996) Proc. Natl. Acad. Sci. USA 93, 10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinohara, A., Shinohara, M., Ohta, T., Matsuda, S. & Ogawa, T. (1998) Genes Cells 3, 145-156. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama, T., New, J. H. & Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy, G., Golub, E. I. & Radding, C. M. (1997) Mutat. Res. 377, 53-59. [DOI] [PubMed] [Google Scholar]
- 30.Parsons, C. A., Baumann, P., Van Dyck, E. & West, S. C. (2000) EMBO J. 19, 4175-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stasiak, A. Z., Larquet, E., Stasiak, A., Muller, S., Engel, A., Van Dyck, E., West, S. C. & Egelman, E. H. (2000) Curr. Biol. 10, 337-340. [DOI] [PubMed] [Google Scholar]
- 32.Van Dyck, E., Stasiak, A. Z., Stasiak, A. & West, S. C. (1999) Nature 398, 728-732. [DOI] [PubMed] [Google Scholar]
- 33.Ranatunga, W., Jackson, D., Lloyd, J. A., Forget, A. L., Knight, K. L. & Borgstahl, G. E. (2001) J. Biol. Chem. 276, 15876-15880. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd, J. A., Forget, A. L. & Knight, K. L. (2002) J. Biol. Chem. 277, 46172-46178. [DOI] [PubMed] [Google Scholar]
- 35.Kagawa, W., Kurumizaka, H., Ikawa, S., Yokoyama, S. & Shibata, T. (2001) J. Biol. Chem. 276, 35201-35208. [DOI] [PubMed] [Google Scholar]
- 36.Navadgi, V. M., Dutta, A. & Rao, B. J. (2003) Biochemistry 42, 15237-15251. [DOI] [PubMed] [Google Scholar]
- 37.Baynton, K., Otterlei, M., Bjoras, M., von Kobbe, C., Bohr, V. A. & Seeberg, E. (2003) J. Biol. Chem. 278, 36476-36486. [DOI] [PubMed] [Google Scholar]
- 38.Gupta, R. C., Folta-Stogniew, E., O'Malley, S. O., Takahashi, M. & Radding, C. M. (1999) Mol. Cell 4, 705-714. [DOI] [PubMed] [Google Scholar]
- 39.Rao, B. J., Chiu, S. K. & Radding, C. M. (1993) J. Mol. Biol. 229, 328-343. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 41.Gupta, R. C., Golub, E., Bi, B. & Radding, C. M. (2001) Proc. Natl. Acad. Sci. USA 98, 8433-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta, R., Bazemore, L. R., Golub, E. I. & Radding, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Dyck, E., Stasiak, A. Z., Stasiak, A. & West, S. C. (2001) EMBO Rep. 2, 905-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurumizaka, H., Aihara, H., Kagawa, W., Shibata, T. & Yokoyama, S. (1999) J. Mol. Biol. 291, 537-548. [DOI] [PubMed] [Google Scholar]
- 45.Sigurdsson, S., Van Komen, S., Petukhova, G. & Sung, P. (2002) J. Biol. Chem. 277, 42790-42794. [DOI] [PubMed] [Google Scholar]
- 46.Noirot, P., Gupta, R. C., Radding, C. M. & Kolodner, R. D. (2003) EMBO J. 22, 324-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta, R. C., Folta-Stogniew, E. & Radding, C. M. (1999) J. Biol. Chem. 274, 1248-1256. [DOI] [PubMed] [Google Scholar]
- 48.Ranatunga, W., Jackson, D., Flowers, I. R., II, & Borgstahl, G. E. (2001) Biochemistry 40, 8557-8562. [DOI] [PubMed] [Google Scholar]
- 49.Bi, B., Rybalchenko, N., Golub, E. I. & Radding, C. M. (2004) Proc. Natl. Acad. Sci. USA 101, 9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung, P. & Robberson, D. L. (1995) Cell 82, 453-461. [DOI] [PubMed] [Google Scholar]
- 51.Li, Z., Golub, E. I., Gupta, R. & Radding, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 11221-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwabuchi, M., Shibata, T., Ohtani, T., Natori, M. & Ando, T. (1983) J. Biol. Chem. 258, 12394-12404. [PubMed] [Google Scholar]