Abstract
Previous reports suggested that U11, in contrast to U12 or other small nuclear (sn)RNAs of the U12-type spliceosome, might be either highly divergent or absent in Drosophila melanogaster. Affinity purification of Drosophila U12-containing complexes has led to the identification of the fly U11 snRNA, which contains a potential U12-type 5′ splice-site-interacting sequence, but whose sequence and length differs significantly from vertebrate and plant U11. Analysis of U12-type introns revealed an A-rich region directly downstream of Drosophila, but not human, U12-type 5′ splice sites. This finding, coupled with the presence of a highly divergent U11 snRNA, and the apparent absence of Drosophila homologs of human U11 proteins, suggest that U12-type 5′ splice site recognition might be different in flies. A comparison of U11 snRNAs that we have identified from vertebrates, plants, and insects, suggests that an evolutionarily divergent U11 snRNA may be unique to Drosophila and not characteristic of insects in general.
In higher eukaryotes, pre-mRNA introns are excised by two distinct spliceosomes. The major or U2-type spliceosome, which catalyzes the removal of most introns, is composed of five small nuclear ribonucleoprotein particles (snRNPs) (U1, U2, U4, U5, and U6) and numerous protein factors (1). The minor or U12-type spliceosome recognizes a rare class of introns (<1% of all human introns; ref. 2). It is comprised of a distinct set of low abundance snRNPs, namely U11, U12, and U4atac/U6atac, but shares U5 with the major spliceosome (1). U12-type introns are present in a wide variety of organisms, including plants, vertebrates, and insects (3). Formation of the U12-type prespliceosome involves base pairing of U11 and U12 to the 5′ splice site (SS) and branch site of the pre-mRNA, respectively (4–7). During prespliceosome formation, the U11 and U12 snRNPs bind concomitantly as a preformed 18S di-snRNP (8).
Most U11 and U12 snRNPs in HeLa nuclear extract are present as an 18S di-snRNP; however, 12S U11 and 15S U12 monoparticles are also observed (9). Human 18S U11/U12 snRNPs share several proteins with the U2 snRNP, including all subunits of the heteromeric splicing factor SF3b (10, 11), which interacts with U2-type pre-mRNAs at or near the branch site (12). In contrast, U1-specific proteins (i.e., U1–70K and U1-C) that facilitate formation of the U1 small nuclear (sn)RNA/5′ SS duplex in U2-type spliceosomes, are not present. U11/U12 snRNPs additionally possess seven unique proteins not found in the U2-type spliceosome (11).
The Drosophila melanogaster U1, U2, U4, U5, and U6 snRNAs are encoded by multiple genes, and closely resemble their vertebrate orthologs in size, structure, and sequence (13). In contrast, only one or two genes appear to encode the minor U12 and U6atac or U4atac snRNAs, respectively (13–15). The lengths and sequences of the U6atac and U4atac snRNAs are very similar between Drosophila and other species. Drosophila U12 (dU12), on the other hand, is 88 nt longer than its human homolog and its sequence is only moderately conserved (14). Intriguingly, the fully annotated D. melanogaster genome did not appear to contain genes encoding the U11 snRNA, or U11-associated proteins, suggesting that the U11 snRNP might be either absent or highly divergent in Drosophila (11, 13, 15, 16).
Materials and Methods
Isolation of Drosophila snRNPs. Splicing active nuclear extracts were prepared from D. melanogaster S2 cells (17). UsnRNPs were purified from nuclear extract by anti-m3G affinity chromatography at 150 mM NaCl and were fractionated on 10–30% glycerol gradients (18). RNA was recovered from gradient fractions, analyzed on 10% polyacrylamide/7 M urea gels, and visualized by Northern blotting with 32P-labeled probes against D. melanogaster U11 or U12 snRNA (18). dU12 and dU11 snRNPs were purified by using biotinylated 2′-O-methyl oligonucleotides complementary to nucleotides 11–28 of U12 or nucleotides 27–45 of U11 and streptavidin beads (10).
RNA Sequencing. RNA X was gel-purified, poly(A)-tailed, converted to cDNA by using a 32P-labeled primer (HindIII-T15; 5′-CGCCAAGCTTT15-3′) and a poly C-tail was added by using terminal dideoxytransferase. The cDNA was PCR-amplified by using the primers HindIII-T15 and EcoRI-G15 (5′-CCGAATTCG15-3′), cloned into pBluescript KS (-), and sequenced.
Database Searches. Nucleic acid and protein blast searches were performed by using the default parameters of the following blast servers:
U12-type introns were identified by scoring all annotated Drosophila introns from the euchromatic part of the genome from www.fruitfly.org (Genome release 3.1), by using the published consensus sequences for the 5′ SS, branch site, and 3′ SS (2, 3).
Results
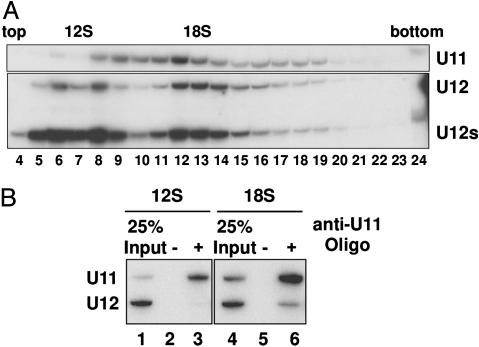
Identification of the dU11 snRNA. Spliceosomal snRNPs were purified from Drosophila S2 cell nuclear extract by anti-m3G affinity chromatography and separated on a glycerol gradient. Northern analyses revealed a full-length dU12 snRNA of 238 nt, as well as two dU12 fragments of 174 nt (U12s) and 64 nt (data not shown) that result from cleavage at position 174 (as evidenced by primer extension analyses) during snRNP isolation (Fig. 1A Lower). Full-length and cleaved snRNAs exhibited the same sedimentation behavior, suggesting that the cleaved fragments are present in intact U12 snRNPs. U12-containing snRNPs peaked in the 12S (fractions 6–8) and 18S (fractions 12–14) regions of the gradient. To determine whether the 18S dU12 particle represents a di-snRNP complex containing U11, 18S snRNPs were affinity-selected with a biotinylated anti-dU12 2′-O-methyl oligonucleotide (oligo) and streptavidin beads. The affinity-selected material was enriched in snRNPs containing U12 snRNA (Fig. 2A, compare lanes 1 and 2) and an RNA denoted X that was subsequently sequenced after amplification by RT-PCR. RNA X is 275 nt in length and shares only 28.3% sequence similarity with human U11. Significantly, it contains a potential U12-type, 5′ SS-interacting region at its 5′ end, as well as an Sm-binding site (Fig. 2C). Based on its sequence, as well as secondary structure and biochemical properties (see below), we conclude that RNA X is the Drosophila U11 (dU11) snRNA. Northern analysis with probes against dU12 and dU11 confirmed that the newly identified dU11 is coselected with dU12 (Fig. 2B, lane 2). Note that in this snRNP preparation most of the U12 snRNA was cleaved and is present as U12s.
Fig. 1.
dU11 and dU12 are present in an 18S di-snRNP. (A) Drosophila UsnRNPs were immunoaffinity purified from S2 nuclear extract and were fractionated on a 10–30% glycerol gradient. SnRNAs were isolated, separated by denaturing PAGE, and identified by Northern blotting with probes against dU11 (Upper) and dU12 (Lower). The peak positions of 12S and 18S dU12 snRNPs are indicated at the top. (B) SnRNAs of affinity-selected 12S (lanes 1–3) and 18S (lanes 4–6) snRNPs, isolated in the presence (lanes 3 and 6) or absence (lanes 2 and 5) of a biotinylated anti-U11 2′-O-methyl oligo, were identified by Northern blotting.
Fig. 2.
Identification of the dU11 snRNA. (A) SnRNAs coselected with U12 snRNPs from the 18S region of the gradient (see Fig. 1 A) in the presence (lane 2) or absence (lane 3) of anti-U12 2′-O-methyl oligo, were separated by denaturing PAGE and were visualized by silver staining. Major snRNAs (Left), and U12 snRNA, coenriched RNA X and the 2′-O-methyl oligo are indicated. (B) Northern analysis of affinity-selected 18S U12 snRNP particles, isolated as described in A by using 32P-labeled probes against dU12 (Lower) or dU11 (Upper) RNA. The apparent lower level of dU12 is likely due to different hybridization efficiencies of the two probes. (C) Genomic sequence encoding the dU11 snRNA (uppercase letters) and flanking sequences. RNA polymerase II-type promoter elements PSE A and B are in bold. The putative U12-type 5′ SS-interacting sequence is underlined, and the Sm-binding site is boxed. (D) Comparison of the PSE A and B promoter elements of the minor U11 and U12 snRNAs. The consensus sequence was obtained from 20 Drosophila U1, U2, U4, and U5 snRNA genes. Nucleotides matching the consensus are underlined.
The dU11 snRNA gene appears to be present as a single copy on chromosome 3L. It possesses an snRNA promoter (Fig. 2D) that contains a highly conserved, RNA polymerase II-type PSE A element and a poorly conserved PSE B element (19, 20). A weakly conserved PSE B element is also found in the dU12 snRNA gene (13) and might thus be a general characteristic of minor snRNA genes in Drosophila. Consistent with it being transcribed by RNA polymerase II, dU11 contains an m3G cap and is associated with Sm proteins, as evidenced by immunoprecipitation (data not shown). Fluorescence in situ hybridization studies further revealed that dU11 is localized in the nucleoplasm of S2 cells, but not in nucleoli (data not shown).
dU11 Sediments as 15S and 18S snRNPs. The distribution of dU11 after glycerol gradient centrifugation of Drosophila snRNPs was determined by Northern blotting (Fig. 1 A Upper). A large portion of dU11 cosedimented with dU12 in the 18S region of the gradient (fractions 12–14), which is consistent with the presence of a U11/U12 di-snRNP. Additionally, approximately 15S (fractions 9–11) and, to a much lesser extent, 12S U11 snRNPs (fractions 6–8), both of which likely represent monoparticles, were observed. To provide additional evidence for a dU11/U12 di-snRNP, we affinity-selected snRNPs from the 12S or 18S region of the gradient with an oligo against dU11 (Fig. 1B). dU11 snRNPs were selected from both regions of the gradient (compare lane 1 with 3, and 4 with 6), whereas dU12 was coselected solely from the 18S region (compare lanes 3 and 6), confirming their presence in an 18S di-snRNP complex.
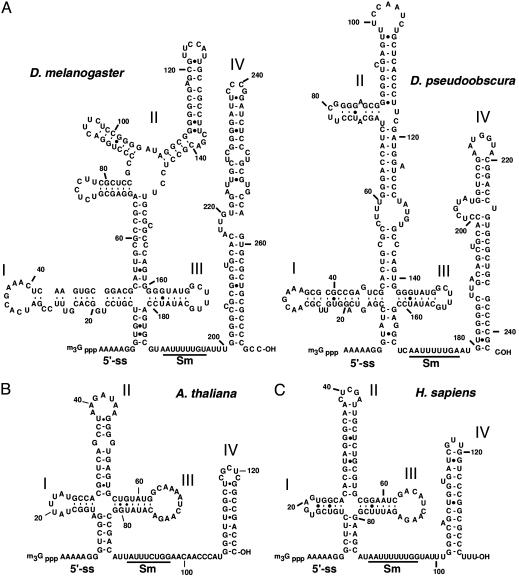
Secondary Structure of the dU11 snRNA. A putative secondary structure of the dU11 snRNA was generated by using the mfold prediction program (21) and by phylogenetic comparison with U11 from Drosophila pseudoobscura (see below). dU11 could be folded into a structure with a four-way junction and a long stem-loop 3′ of the Sm-binding site. Whereas stem-loops III and IV are comparable in length between human and fly, and can be folded in similar ways (Fig. 3), stem loops I and II of dU11 are much longer and theoretically can be folded into several distinct structures (data not shown). To confirm which of these is correct, lead cleavage followed by primer extension analysis was performed on native, phenol-extracted dU11 snRNA (Fig. 5, and Supporting Materials and Methods, which are published as supporting information on the PNAS web site). The experimental results were then compared with each of the theoretical structures, and were most consistent with the secondary structure shown in Fig. 3A.
Fig. 3.
Structure of fly, plant, and vertebrate U11 snRNAs. The proposed secondary structures of D. melanogaster (BK005210) and D. pseudoobscura (BK005209) (A), A. thaliana (BK005207) (B) and Homo sapiens (X58716) (C) U11 snRNAs. Secondary structures were generated by the mfold program. 5′ ss, putative U12-type 5′ SS-interacting sequence; Sm, Sm-binding site (underlined).
Identification of U11 snRNAs in Other Organisms. To date, a fulllength U11 snRNA has only been reported in humans (22). To identify additional U11 snRNAs, we performed blast searches by using both general and specialized databases with the complete or partial sequence of the dU11 or human U11 snRNA. By using the D. melanogaster sequence, we identified an apparent U11 snRNA in D. pseudoobscura. The D. pseudoobscura U11 gene contains correctly positioned RNA polymerase II-type snRNA promoter elements and encodes a 243-nt RNA. Surprisingly, the overall sequence similarity (61%) is not particularly high between the two flies. Database searches with human U11 revealed homologs in Mus musculus, Gallus gallus, Tetraodon nigroviridis, Fugu rubripes, and Danio rerio (Fig. 6, which is published as supporting information on the PNAS web site), but no candidates in invertebrate genomes.
A plant U11 could not be identified by means of blast searches by using vertebrate or fly sequences. However, a closer inspection of the recently published collection of small nonmessenger RNAs in Arabidopsis thaliana (23) led to the identification of the Arabidopsis U11 snRNA (clone Ath-63). The U11 gene contains conserved plant snRNA promoter elements (USE and TATA box), and Northern blotting confirmed the expression of the full-length RNA (130 nt), and a 70-nt fragment (23). By using this sequence, we also identified a putative U11 snRNA ortholog in Oryza sativa, with a predicted length of 121 nt, whose gene also contains the expected snRNA promoter elements (Fig. 6).
Structural Comparison of Drosophila, Plant, and Vertebrate U11 snRNAs. The predicted secondary structures of fly, A. thaliana, and human U11 are shown in Fig. 3 (see Fig. 6 for additional U11 snRNAs). The lengths of the vertebrate and plant U11 snRNAs are very similar, whereas the U11 snRNAs from both Drosophila species are considerably longer (i.e., 275 and 243 vs. 135 nt in human). Although the regions upstream of the Sm site in fly vs. plant/vertebrate U11 differ considerably in sequence and in length, each can be folded into a structure with a four-way junction. Furthermore, functionally important regions like the single-stranded 5′ end that is complementary to the U12-type 5′ SS or the Sm site flanked by a stem loop (IV) are conserved in all species. In contrast, the lengths and sequences of both stem loops I and II are considerably different between fly and plant/vertebrate U11 snRNAs and the sequences of loop I and II are not conserved even within each subgroup (i.e., vertebrates, plants, and flies), suggesting that they do not carry out important functions. In contrast, the length of stem III is highly conserved in all species. The sequence of loop III varies between flies and plants/vertebrates, but it is completely conserved between both Drosophila species and also partially between vertebrates and plants (the latter share the 7-nt sequence AUCAAGA), suggesting it might be a protein-binding site or involved in basepairing interactions.
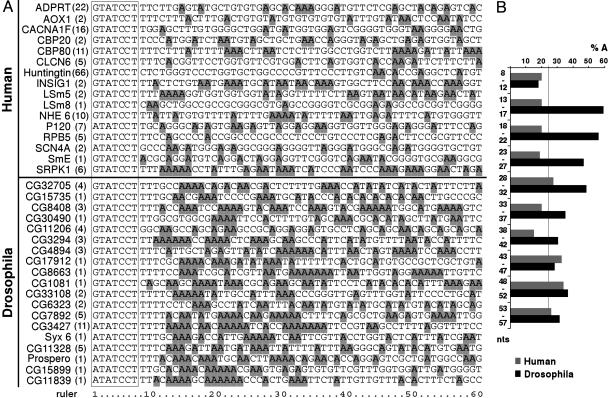
dU12-Type 5′ SSs Are Followed by an A-Rich Region. The identification of an apparently highly divergent U11 snRNP in Drosophila suggests that protein–protein and/or protein–RNA interactions contributing to 5′ SS recognition might be different in flies vs. vertebrates, and that pre-mRNA sequences at or near the 5′ SS may also differ. The 5′ SS consensus sequence ([A/G] TATCCTT) is highly conserved in flies, plants, and vertebrates (2, 3, 24), although only three dU12-type introns (i.e., in the Prospero, Syx6, and cg11839 genes) have been reported so far (14). To analyze the dU12-type 5′ SS in more detail, we identified 16 additional, putative U12-type introns through a computational scan of the Drosophila genome and compared their 5′ SSs with those of human U12-type introns (Fig. 4). Whereas the nucleotide distribution downstream of the 5′ consensus sequence is nearly random in humans, the region downstream of the Drosophila 5′ SS consensus (i.e., intron positions 13–37) is clearly enriched in adenosines. In contrast, an A-rich region was not observed downstream of dU2-type 5′ SSs (data not shown). Thus, the dU12-type 5′ SS appears to contain an additional element that is potentially recognized by RNA and/or proteins.
Fig. 4.
Comparison of human and dU12-type 5′ SSs. (A) A subset of human (Upper) and putative dU12-type (Lower)5′SS sequences is shown. The intron positions in the respective genes are indicated in brackets. The highly conserved consensus sequence [A/G]TATCCT is boxed and adenosines are highlighted. (B) Frequency of adenosines (%A) downstream of human (gray) and dU12-type (black) 5′ SS was averaged for groups of 5 nt (indicated as one bar). Nucleotide positions are relative to the 5′ end of the intron.
Discussion
dU11 snRNA Is Evolutionarily Divergent. We have identified dU11 by isolating U12-containing snRNPs. In contrast to other minor snRNAs, the sequence of dU11 is highly divergent from that of vertebrate and plant U11 snRNAs; only 28.3 and 23% similarity is observed between dU11 and human or A. thaliana U11, respectively. Despite these differences, all U11 snRNAs identified share (i) a single-stranded 5′ end complementary to the U12-type 5′ SS, (ii) a four-way junction, (iii) an Sm site flanked bya3′ stem loop, and (iv) a stem III of similar length. The former suggests that U12-type 5′ SS recognition is mediated, at least in part, by base-pairing between U11 and the pre-mRNA, not only in vertebrates, but also in flies and plants. The length and sequence of stem loops I and II, which by analogy to U1 might bind proteins, are highly variant in all U11 snRNAs identified. The lack of conservation of these stem-loop structures between vertebrates and flies is consistent with the apparent absence of Drosophila homologs of the human U11-associated proteins (Table 1, which is published as supporting information on the PNAS web site). However, the sequences of loops I and II are not even conserved among vertebrate U11 snRNAs, suggesting they generally do not serve as protein-binding sites. In contrast, the sequence of loop III is conserved between both Drosophila species and partially between vertebrates and plants, suggesting a functional role for loop III, for example, in binding a U11-specific protein. It is presently unclear which region of U11, other than the Sm site, is bound by protein. Future structural analyses of U11 snRNPs may answer this question.
Protein Composition of Fly and Human U11 and U12 snRNPs. Drosophila homologs of proteins shown to be U11-associated in humans (i.e., 59K, 48K, 35K, and 25K) were not detected by highly sensitive blast searches (Table 1 and ref. 11), indicating that dU11 snRNPs likely possess significantly different proteins. Consistent with this idea, the dU11 snRNP exhibited a faster sedimentation coefficient (15S) than its human counterpart, which sediments as a 12S particle (9). To clarify this point we have attempted to identify proteins associated with purified dU11 and dU12 snRNPs by using MS, but due to contamination problems these studies have, to date, not been successful. The presence of a highly divergent dU11 snRNP suggests that protein–protein bridging interactions within the U11/U12 di-snRNP and protein interactions contributing to U12-type 5′ SS recognition may be quite different in Drosophila vs. vertebrates.
The dU12 snRNA sedimented as a 12S RNP particle, much slower than the human 15S U12 monoparticle. The 12S dU12 snRNPs appear to contain Sm proteins and one or more U12-specific proteins but, based on their lower S value, likely lack the splicing factor SF3b. The latter is a stable component of human 18S U11/U12 snRNPs and is thought to be present in the human 15S U12 snRNP (11). The absence of a 15S dU12 snRNP suggests that SF3b is stably associated solely in the fly 18S U11/U12 snRNP. Thus, protein–protein and/or protein–RNA contacts stabilizing the association of SF3b with U12 in Drosophila may be different from those in humans. Likely candidates for U12-specific proteins are the 65K, 31K, and 20K proteins, which are found in human U11/U12 snRNPs, but not in 12S U11 monoparticles (Table 1 and ref. 11). Potential Drosophila orthologs of the 65K (gi|21626517) and 20K (gi|24580811) proteins were identified by blast searches and initial MS analyses indicate that the 65K protein is present in both dU11/U12 di-snRNPs and 12S dU12 snRNPs.
A Unique dU12-Type 5′ SS. Drosophila, but not human, U12-type 5′ SSs are followed by an ≈25-nt A-rich, but otherwise nonconserved sequence. U12-type introns in A. thaliana also appear to lack such an A-rich region (24). The latter might serve as a binding site for either a U11-associated protein or a non-snRNP splicing factor that recruits U11 to the 5′ SS. A protein that interacts with U-rich sequences downstream of U2-type 5′ SS and facilitates U1 snRNP binding has been reported in yeast (i.e., the U1-associated protein Nam8p; ref. 25) and higher eukaryotes (TIA-1; ref. 26). In Drosophila, U1 snRNP binding to selected U2-type 5′ SSs is probably facilitated by Rox 8, the homolog of human TIA-1. Additional experiments are needed to clarify whether the A-rich sequence downstream of the dU12-type 5′ SS is indeed recognized by a specific factor that facilitates dU11 snRNP association.
Evolutionary Aspects. Assuming that human and fly U11 share a common ancestor, did this early form of U11 more closely resemble the fly or vertebrate U11? A potential answer can be found if we look at the situation in plants, which diverged from animals >1 billion years ago. In contrast to both Drosophila species, the lengths and secondary structures of both Arabidopsis (130 nt) and rice (121 nt) U11 snRNAs are very similar to those of vertebrates (Table 1). Further, highly conserved homologs of at least one human U11 protein (35K) are found in A. thaliana and other plants. These observations support the idea that flies, and not vertebrates, have diverged dramatically from a very early form of the U11 snRNP. This finding might be due to the highly accelerated divergence times in certain insect lineages. Indeed, the fact that the U11 snRNA from D. melanogaster and D. pseudoobscura show only 61% sequence similarity indicates that these Drosophila species have diverged quite rapidly over the estimated 36–46 million years because they had a common ancestor.
Alternatively, the dU11 may have arisen from a U11 paralog or, less likely, not even be a true homolog of the vertebrate/plant U11 snRNA (i.e., they may have a nonhomologous origin). That is, whereas an ancient form of U11 may still have been present in a common ancestor of insects and plants (see below), it might have been lost in Drosophila and functionally replaced by an analogous RNA (e.g., by nonhomologous gene replacement).
Database searches did not allow the detection of U11 in other insects. However, homologs of the human U11–35K protein are found in the mosquito Anopheles gambiae and the honey bee Apis mellifera (Table 1), suggesting that U11 in these insects may more closely resemble the vertebrate/plant U11. Thus, a highly divergent U11 snRNP might not be characteristic of insects in general, but rather, specific for the genus Drosophila, which diverged from mosquito ≈250 million years ago (27, 28). Remarkably, the Anopheles genome is twice the size of that of Drosophila. A likely explanation for this difference is that D. melanogaster has lost intergenic and intronic sequences during divergence from A. gambiae (27). Thus, the original dU11 gene may indeed have been lost. The complete transcription unit of dU11 is located on chromosome 3L on the strand opposite the 543-nt intron 1 of the CG1079 gene, which encodes a protein of unknown function (Q9VZL8) that is similar (58%) to a predicted protein (ENSANGP00000011276) in A. gambiae. The corresponding intron 1 of the mosquito gene is much shorter (112 nt) and does not contain a U11 gene. This finding suggests that an snRNA gene was inserted into intron 1 of CG1079 by recombination or retroposition, a not uncommon route of gene duplication for small nonmessenger RNAs (29), after Drosophila and mosquito diverged from their common ancestor.
Interestingly, the other snRNA genes of the dU12-type spliceosome are also located within introns of other genes, either in direct orientation (U4atac) or on the opposite strand (U12 and U6atac), whereas the mosquito U12 and human U11, U12, and U6atac snRNA genes are not located in introns (13, 14). Thus, in Drosophila, minor snRNA genes appear to be generally present in introns. However, in contrast to many small nucleolar RNAs, they contain their own promoter elements and thus appear to be independently transcribed.
Evolutionary Divergence of U11 and U12 snRNAs. The significant divergence of the Drosophila vs. vertebrate and plant U11 snRNA contrasts somewhat with the more conserved U12. Although the dU12 snRNA is much longer than U12 in other organisms, there is nearly complete conservation of nucleotides involved in splicing (e.g., branch site- and U6atac-interacting regions) and most of the size difference can be attributed to additional nucleotides inserted between the Sm-binding site and the highly conserved 5′ and 3′ ends (14). The generally higher sequence conservation of U12 could result from several factors. The extended regions of presumably nonfunctional sequence in the dU12 snRNA are significantly shorter than those in dU11. The accumulation of mutations in a large portion of U12 was most likely restricted by the extensive base-pairing interactions of U12 with the U6atac snRNA, which form the catalytic center of the minor spliceosome (30). Additional constraints were probably present due to the fact that the U12 snRNP shares protein components (i.e., SF3b subunits) with the major, highly abundant U2 snRNP (10). Thus, mutations in the dU12 sequence may have been less well tolerated, because they were not easily compensated by the codivergence of SF3b.
Assuming that the dU11 and dU12 snRNAs each share a common ancestor with their human counterparts, why has the dU11 diverged so significantly? Sequences, for example in stem loops I and II of dU11, which account for most of the length difference between vertebrates and flies, are likely not essential for the basic function of the U11 snRNA, i.e., base-pairing with the pre-mRNA. Thus, they might have readily accumulated functionally silent changes that may also have been accompanied by changes in the dU11-associated proteins. Indeed, the sequences of these apparent insertions are not conserved between both Drosophila species. It is also conceivable that U11 proteins play a less essential role in 5′ SS recognition in Drosophila and thus Drosophila may have more readily tolerated significant alterations in U11 proteins and the consequent divergence of the U11 snRNA. The presence of an A-rich region downstream of dU12-type (and not vertebrate/plant) 5′ SSs, which potentially facilitates U11 association, might compensate for the proposed less significant contribution by dU11 proteins.
Supplementary Material
Acknowledgments
We thank G. Heyne, B. Hildebrandt, and P. Kempkes for excellent technical assistance; H. Urlaub for MS analyses of Drosophila proteins; A. Zeman for U11 gene searches in A. thaliana; N. Watkins for assistance with fluorescence in situ hybridization; B. Kastner, E. Makarov, R. Rivera-Pomar, R. Rauhut, and E. Westhof for helpful discussions; and S. Mount and E. Zheng for providing a preliminary list of dU12-type introns. This work was supported by the German Human Genome Project through Bundesministerium für Bildung; Wissenschaft, Forschung und Technologie Grant 01KW9966 (to J.B.); Academy of Finland Project Grant 50527 (to M.J.F.); and Deutsche Forschungsgemeinschaft Grant LU294/12-1 (to R.L.); and Fonds der Chemischen Industrie.
Abbreviations: snRNP, small nuclear ribonucleoprotein particle; snRNA, small nuclear RNA; dU12, Drosophila U12; dU11, Drosophila U11; SS, splice site.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. BK005202–BK005210).
References
- 1.Burge, C. B., Tuschl, T. & Sharp, P. A. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed., pp. 525-560.
- 2.Levine, A. & Durbin, R. (2001) Nucleic Acids Res. 29, 4006-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burge, C. B., Padgett, R. A. & Sharp, P. A. (1998) Mol. Cell 2, 773-785. [DOI] [PubMed] [Google Scholar]
- 4.Hall, S. L. & Padgett, R. A. (1996) Science 271, 1716-1718. [DOI] [PubMed] [Google Scholar]
- 5.Tarn, W. Y. & Steitz, J. A. (1996) Cell 84, 801-811. [DOI] [PubMed] [Google Scholar]
- 6.Yu, Y. T. & Steitz, J. A. (1997) Proc. Natl. Acad. Sci. USA 94, 6030-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolossova, I. & Padgett, R. A. (1997) RNA 3, 227-233. [PMC free article] [PubMed] [Google Scholar]
- 8.Frilander, M. J. & Steitz, J. A. (1999) Genes Dev. 13, 851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wassarman, K. M. & Steitz, J. A. (1992) Mol. Cell. Biol. 12, 1276-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Will, C. L., Schneider, C., Reed, R. & Lührmann, R. (1999) Science 284, 2003-2005. [DOI] [PubMed] [Google Scholar]
- 11.Will, C. L., Schneider, C., Hossbach, M., Urlaub H., Rauhut, R., Elbashir, S., Tuschl, T. & Lührmann, R. (2004) RNA 10, 929-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Will, C. L. & Lührmann, R. (1997) Curr. Opin. Cell Biol. 9, 320-328. [DOI] [PubMed] [Google Scholar]
- 13.Mount, S. M. & Salz, H. K. (2000) J. Cell Biol. 150, 37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otake, L. R., Scamborova, P., Hashimoto, C. & Steitz, J. A. (2002) Mol. Cell 9, 439-446. [DOI] [PubMed] [Google Scholar]
- 15.Misra, S., Crosby, M. A., Mungall, C. J., Matthews, B. B., Campbell, K. S., Hradecky, P., Huang, Y., Kaminker, J. S., Millburn, G. H., Prochnik, S. E., et al. (2002) Genome Biol. 3, 0083.1-0083.22. [Google Scholar]
- 16.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- 17.Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider, C., Will, C. L., Makarova, O. V., Makarov, E. M. & Lührmann, R. (2002) Mol. Cell. Biol. 22, 3219-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo, P. C. & Mount, S. M. (1990) Nucleic Acids Res. 18, 6971-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, R. C., Wang, Y., Hardin, S. B. & Stumph, W. E. (1998) Nucleic Acids Res. 26, 616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker, M. (2003) Nucleic Acids Res. 31, 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montzka, K. A. & Steitz, J. A. (1988) Proc. Natl. Acad. Sci. USA 85, 8885-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marker, C., Zemann, A., Terhorst, T., Kiefmann, M., Kastenmayer, J. P., Green, P., Bachellerie, J. P., Brosius, J. & Hüttenhofer, A. (2002) Curr. Biol. 12, 2002-2013. [DOI] [PubMed] [Google Scholar]
- 24.Zhu, W. & Brendel, V. (2003) Nucleic Acids Res. 31, 4561-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puig, O., Gottschalk, A., Fabrizio, P. & Séraphin, B. (1999) Genes Dev. 13, 569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Förch, P., Puig, O., Kedersha, N., Martinez, C., Granneman, S., Seraphin, B., Anderson, P. & Valcárcel, J. (2000) Mol. Cell 6, 1089-1098. [DOI] [PubMed] [Google Scholar]
- 27.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M., Wides, R., et al. (2002) Science 298, 129-149.12364791 [Google Scholar]
- 28.Zdobnov, E. M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R. R., Christophides, G. K., Thomasova, D., Holt, R. A., Subramanian, G. M., et al. (2002) Science 298, 149-159. [DOI] [PubMed] [Google Scholar]
- 29.Brosius, J. (2003) Genetica (The Hague) 118, 99-116. [PubMed] [Google Scholar]
- 30.Tarn, W. Y. & Steitz, J. A. (1996) Science 273, 1824-1832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.