Abstract
Background
Mutations or single nucleotide polymorphisms (SNPs) within the gene region of microRNAs play an important role for the development of hepatocellular carcinoma (HCC). Extensive studies have tried to investigate the susceptibility role of miR-146a rs2819164 and miR-196a-2 rs11614913. However, these results are still inconsistent and inconclusive. We undertook a meta-analysis containing primarily Asian studies to assess the associations of the two SNPs with HCC risk.
Methods
19 studies including miR-146a (7170 cases and 9443 controls) and 15 studies including miR-196a-2 (6417 cases and 7627 controls) were used for meta-analysis. Odds ratios and 95% CI were calculated to assess the association in five different genetic models.
Results
For the rs2910164 polymorphism of miR-146a, significantly increased risks for HCC were observed when all studies were pooled under two models (CG vs CC: OR = 1.11, 95% CI = 1.02–1.21, P = 0.021; GG + CG vs CC: OR = 1.11, 95% CI = 1.01–1.22, P = 0.035). For the rs11614913 polymorphism of miR-196a-2, significant increased risks for HCC development were observed when all studies were pooled under four models (C vs T: OR = 1.14, 95% CI = 1.06–1.23, P = 0.001; CC vs TT: OR = 1.31, 95% CI = 1.12–1.53, P = 0.001; CC + TC vs TT: OR = 1.16, 95% CI = 1.03–1.31, P = 0.018; CC vs TC + TT: OR = 1.14, 95% CI = 1.00–1.30, P = 0.043).
Conclusion
Our results show that the two common SNPs within the miRNAs were associated with modest increased risk of HCC (OR < 1.6), especially in the Asian population. Larger population-based studies validating these results are needed.
Keywords: HCC, MiRNA
1. Introduction
Hepatocellular carcinoma (HCC) is a global health problem and causes a huge economic burden for both patients and society. It ranks as the sixth most common cancer in the world, with the highest incidence in China (Venook et al., 2010). The major environmental risk factors for HCC include chronic hepatitis B and C infection (Yang & Roberts, 2010). Recent genome-wide association studies have suggested that genetic factors also play an important role in the development of HCC (Jiang et al., 2013, Zhang et al., 2010). However, the specific genetic factors contributing to HCC have still remained largely unknown (Jiang et al., 2013).
MicroRNAs (miRNAs) are a family of noncoding small RNAs with 21- to 25-nucleotides, which primarily target the 3′ untranslated regions of targeting messenger RNAs (mRNAs), thus resulting in gene silencing or abnormal expression (Zhou et al., 2014a). miRNAs have been predicted to regulate 30% of the human genome, including genes in inflammation, proliferation and apoptosis pathways (Lewis et al., 2005). miRNAs' deregulation have been shown to be responsible for initiation and progression of HCC (Yin et al., 2015). A mutation or single nucleotide polymorphism (SNP) within the gene region of miRNAs could alter miRNA expression and thence affect downstream target genes.
The rs2910164 SNP is located within the stem region of an miR-146a precursor and the C to G mutation results in a change from a U:C to a U:G mismatch, while the C to T mutation of the rs11614913 SNP located within the stem region of an miR-196a-2 precursor leads to a change from a G:C to a G:U mismatch (Gong et al., 2012). Functional studies have shown that such mismatches for miR-146a (Jazdzewski et al., 2008) and for miR-196a-2 (Li et al., 2010) could result in reduced expression of both mature miRNAs. Therefore, several studies (Li et al., 2010, Akkiz et al., 2011a, Cong et al., 2014, Kim et al., 2012, Zhang et al., 2013, Xu et al., 2008) have investigated the two SNPs and HCC risk. Thus far, however, results have been inconsistent. For example, for rs2910164 (miR-146a), Xu et al. (2008) found that the GG genotype increased the risk for HCC, while Zhang et al. (2013) did not identify any significant association with HCC. For rs11614913 (miR-196a-2), Li et al. (2010) found that the CC genotype increased the risk for HCC, while Han et al. (2013) failed to replicate the findings. We therefore performed this updated meta-analysis using all available papers published in English and Chinese.
2. Material and methods
2.1. Literature search
We did a comprehensive literature search in PubMed, EMBASE, Cochrane Library, Chinese National Knowledge Infrastructure (CNKI) database and Wanfang database up to April 01, 2015 using the following search terms: (miR-146a OR microRNA-146a OR rs2910164 OR miR-196a-2 OR microRNA-196a-2 OR rs11614913) AND (hepatocellular carcinoma OR liver cancer OR HCC). Searches were done without restricting on sample size, publication date, language or type of report. We also reviewed references within retrieved articles. For published papers without the required data, corresponding authors were contacted by email for more information. The meta-analysis met the requirements of the PRISMA 2009 checklist (Moher et al., 2009).
2.2. Criteria for study selection
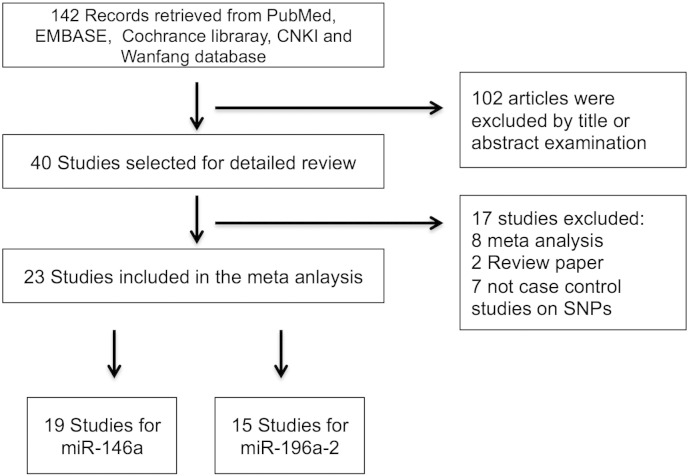
The relevant published papers had to meet all the following criteria in order to be included in the meta-analysis: 1) studies focus on human beings; 2) studies use a case–control design evaluating the association between rs2910164 and/or rs11614913 and HCC development; 3) histopathologically confirmed on HCC diagnosis; and 4) detailed information on the allele and genotype frequencies of rs2910164 and/or rs11614913 required for estimating odds ratio (OR) and 95% confidence interval (CI). The details of included and excluded papers are shown in Fig. 1.
Fig. 1.
Flow chart of the selection of the studies and reasons for exclusion from the meta-analysis.
2.3. Data extraction
Two authors (MC and FL) independently reviewed potentially eligible studies. The following information were extracted from each: first author's name & other publication data, country wherein the study population was recruited, cohort ethnicity, genotyping method, sources of controls, cases and control numbers, and case & control genotype frequencies. The two authors checked the data extraction results and reached consensus on all the data recorded.
2.4. Statistical analysis
The goodness-of-fit test was used to check the Hardy–Weinberg Equilibrium (HWE) in the control samples of each study (P < 0.05 was considered as deviating from HWE). ORs and 95% CI were calculated to assess the association between rs2910164, rs11614913 and HCC risk in five different models: allele, homozygous, heterozygote, dominant and recessive models. The significance of pooled ORs was considered as statistically significant by the Z test with P < 0.05. Cochran's Q-test (PQ) (Handoll, 2006) was used to assess the heterogeneity between studies. The fixed-effect model (Mantel–Haenszel method) (Mantel & Haenszel, 1959) was used when PQ > 0.10; otherwise, a random-effects model (DerSimonian and Laird method) (DerSimonian & Laird, 1986) was applied. We also performed subgroup analyses by ethnicity, sources of controls and HWE status to further explore the heterogeneity between studies. Sensitivity analyses were performed by removing each study stepwise to examine the robustness of the results. Begg's funnel plot and Egger's regression asymmetry test were performed to evaluate publication bias (Egger et al., 1997). All statistical analyses were done using Stata version 12.0 software (StataCorp, College Station, TX).
3. Results
3.1. Study characteristics
A total of 142 potentially relevant studies were identified. A flowchart of the detail selection and exclusion process is shown in Fig. 1. From this process, twenty-three articles were included for the meta-analysis, of which, seven papers (Hao, 2014, Li, 2012, Huang et al., 2013, Wang, 2011, Xu, 2010, Zhang et al., 2011, Zhou, 2014) were identified through the CNKI database and Wanfang database (five papers were from theses that have not been published (Table 1)), with others identified through PubMed, EMBASE and the Cochrane Library. There were eleven papers (Kim et al., 2012, Zhang et al., 2013, Hao, 2014, Li, 2012, Huang et al., 2013, Xu, 2010, Zhang et al., 2011, Chu et al., 2014, Kou et al., 2014, Qi et al., 2014, Zhou et al., 2014b) overlapping with both miR-146a and miR-196a-2, thus resulting in nineteen studies for miR-146a with 7170 cases and 9443 controls (Table 1) and fifteen studies for miR-196a-2 with 6417 cases and 7627 controls (Table 2), respectively. For miR-146a, one study used a Turkish population (Akkiz et al., 2011b); one study used a Korean population (Kim et al., 2012); and seventeen studies used a Chinese population (Cong et al., 2014, Zhang et al., 2013, Xu et al., 2008, Hao, 2014, Li, 2012, Huang et al., 2013, Wang, 2011, Xu, 2010, Zhang et al., 2011, Zhou, 2014, Chu et al., 2014, Kou et al., 2014, Qi et al., 2014, Zhou et al., 2014b, Zhou et al., 2012, Shan et al., 2013, Xiang et al., 2012). For miR-196a-2, one study used a Turkish population (Akkiz et al., 2011a); one used a Korean population (Kou et al., 2014); and thirteen studies used a Chinese population (Li et al., 2010, Zhang et al., 2013, Han et al., 2013, Hao, 2014, Li, 2012, Huang et al., 2013, Xu, 2010, Zhang et al., 2011, Chu et al., 2014, Kou et al., 2014, Qi et al., 2014, Zhou et al., 2014b, Qi et al., 2010).
Table 1.
Characteristics of studies included in the meta-analysis for miR-146a rs2910164.
| Study | Year | Country | Ethnicity | Genotyping method | Source of controls | Journal | GG1 | CG1 | CC1 | GG0 | CG0 | CC0 | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xu et al. | 2008 | China | Asian | PCR-RFLP | PB | Carcinogenesis | 80 | 241 | 158 | 58 | 249 | 197 | 0.119 |
| Xu | 2010 | China | Asian | PCR-RFLP | PB | [Thesis] | 86 | 237 | 177 | 87 | 238 | 197 | 0.296 |
| Akkiz et al. | 2011 | Turkey | Caucasian | PCR-RFLP | PB | Gene | 137 | 75 | 10 | 144 | 67 | 11 | 0.384 |
| Wang | 2011 | China | Asian | MALDI-TOF | PB | [Thesis] | 212 | 561 | 343 | 272 | 924 | 673 | 0.115 |
| Zhang et al. | 2011 | China | Asian | PCR-RFLP | PB | [CJPM] | 156 | 450 | 319 | 291 | 725 | 577 | 0.149 |
| Kim et al. | 2012 | Korean | Asian | PCR-RFLP | PB | Gene | 14 | 88 | 57 | 24 | 103 | 74 | 0.19 |
| Li | 2012 | China | Asian | AS-PCR | PB | [Thesis] | 124 | 302 | 134 | 92 | 288 | 180 | 0.196 |
| Xiang et al. | 2012 | China | Asian | PCR-RFLP | HB | Molecular biology report | 27 | 45 | 28 | 21 | 46 | 33 | 0.506 |
| Zhou et al. | 2012 | China | Asian | PCR-RFLP | HB | DNA and cell biology | 33 | 86 | 67 | 71 | 254 | 158 | 0.056 |
| Huang et al. | 2013 | China | Asian | MADLI-TOF | HB | [CJOPT] | 12 | 58 | 40 | 15 | 41 | 54 | 0.122 |
| Shan et al. | 2013 | China | Asian | PCR-RFLP | HB | GMR | 28 | 62 | 82 | 36 | 71 | 78 | 0.009 |
| Zhang et al. | 2013 | China | Asian | MADLI-TOF | PB | APJCP | 163 | 503 | 331 | 156 | 475 | 367 | 0.911 |
| Chu et al. | 2014 | China | Asian | PCR-RFLP | HB | Plos One | 22 | 82 | 84 | 50 | 146 | 141 | 0.23 |
| Cong et al. | 2014 | China | Asian | PCR-RFLP | HB | Tumor Biology | 27 | 85 | 94 | 17 | 84 | 117 | 0.723 |
| Hao | 2014 | China | Asian | PCR-RFLP | PB | [Thesis] | 23 | 133 | 70 | 30 | 154 | 97 | 0.007 |
| Kou et al. | 2014 | China | Asian | PCR-RFLP | HB | Oncology Letter | 25 | 147 | 99 | 56 | 297 | 179 | < 0.001 |
| Qi et al. | 2014 | China | Asian | HRM | PB | BMC cancer | 0 | 165 | 149 | 3 | 244 | 159 | < 0.001 |
| Zhou et al. | 2014 | China | Asian | PCR-RFLP | HB | Tumor Biology | 40 | 153 | 73 | 30 | 154 | 97 | 0.007 |
| Zhou | 2014 | China | Asian | PCR-RFLP | HB | [Thesis] | 26 | 86 | 61 | 14 | 15 | 12 | 0.088 |
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism, MALDI-TOF: matrix assisted laser desorption/ionization-time of flight mass spectrometry, AS-PCR: allele specific-polymerase chain reaction, HRM: high resolution melting, PB: population based studies, HB: hospital based studies, []: published in Chinese, [Thesis]: thesis published in Chinese, [CJPM]: Chinese Journal of preventive medicine, [CJOPT]: Chinese Journal of Oncology Prevention and Treatment, GMR: Genetics and Molecular Research, APJCP: Asian Pacific Journal of Cancer Prevention, HWE: Hardy–Weinberg equilibrium in control samples, GG1, CG1 and CC1: genotype frequency in cases, GG0, CG0 and CC0: genotype frequency in controls.
Table 2.
Characteristics of studies included in the meta-analysis for miR-196a-2 rs11614913.
| Study | Year | Country | Ethnicity | Genotyping |
Source of |
Journal | CC1 | CT1 | TT1 | CC0 | CT0 | TT0 | HWE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Controls | ||||||||||||
| Li et al. | 2010 | China | Asian | PCR-RFLP | HB | Pathology | 78 | 150 | 82 | 42 | 102 | 78 | 0.402 |
| Qi et al. | 2010 | China | Asian | PCR-LDR | HB | Human immunology | 82 | 179 | 100 | 92 | 197 | 102 | 0.869 |
| Xu | 2010 | China | Asian | PCR-RFLP | PB | [Thesis] | 115 | 247 | 130 | 100 | 251 | 144 | 0.621 |
| Akkiz et al. | 2011 | Turkey | Caucasian | PCR-RFLP | PB | Journal of Viral Hepatitis | 77 | 86 | 22 | 58 | 87 | 40 | 0.492 |
| Zhang et al. | 2011 | China | Asian | PCR-RFLP | PB | [CJPM] | 208 | 449 | 277 | 328 | 817 | 477 | 0.972 |
| Kim et al. | 2012 | Korea | Asian | PCR-RFLP | PB | Gene | 34 | 84 | 41 | 45 | 107 | 49 | 0.356 |
| Li | 2012 | China | Asian | AS-PCR | PB | [Thesis] | 148 | 194 | 218 | 98 | 246 | 216 | 0.057 |
| Han et al. | 2013 | China | Asian | PCR-RFLP | HB | Plos One | 227 | 505 | 305 | 220 | 485 | 304 | 0.31 |
| Huang et al. | 2013 | China | Asian | MADLI-TOF | HB | [CJOPT] | 25 | 52 | 32 | 30 | 53 | 26 | 0.784 |
| Zhang et al. | 2013 | China | Asian | MADLI-TOF | PB | APJCP | 214 | 488 | 294 | 165 | 502 | 328 | 0.245 |
| Chu et al. | 2014 | China | Asian | PCR-RFLP | HB | Plos one | 41 | 81 | 66 | 70 | 167 | 100 | 0.986 |
| Hao | 2014 | China | Asian | PCR-RFLP | PB | [Thesis] | 77 | 126 | 32 | 67 | 160 | 55 | 0.022 |
| Kou et al. | 2014 | China | Asian | PCR-RFLP | HB | Oncology Letter | 84 | 150 | 37 | 125 | 304 | 103 | 0.001 |
| Qi et al. | 2014 | China | Asian | HRM | PB | BMC cancer | 45 | 209 | 60 | 71 | 214 | 121 | 0.156 |
| Zhou et al. | 2014 | China | Asian | PCR-RFLP | HB | Tumor biology | 93 | 139 | 34 | 66 | 160 | 55 | 0.018 |
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism, PCR-LDR: polymerase chain reaction-ligase detection reaction, MALDI-TOF: matrix assisted laser desorption/ionization-time of flight mass spectrometry, AS-PCR: allele specific-polymerase chain reaction, HRM: high resolution melting, PB: population based studies, HB: hospital based studies, []: published in Chinese, [Thesis]: thesis published in Chinese, [CJPM]: Chinese Journal of Preventive Medicine, [CJOPT]: Chinese Journal of Oncology Prevention and Treatment, APJCP: Asian Pacific Journal of Cancer Prevention, HWE: Hardy–Weinberg equilibrium in control samples, CC1, CT1 and TT1: genotype frequency in cases, CC0, CT0 and TT0: genotype frequency in controls.
3.2. Association between miRNA polymorphism and HCC susceptibility
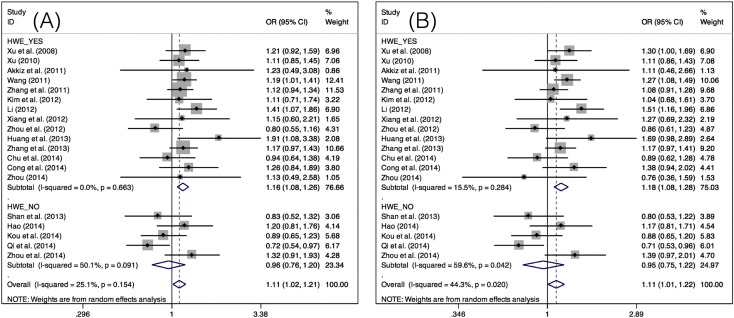
For the miR-146a rs2910164 polymorphism, significantly increased risks for HCC development were observed when all studies were pooled under two models (CG vs CC: OR = 1.11, 95% CI = 1.02–1.21, P = 0.021; and GG + CG vs CC: OR = 1.11, 95% CI = 1.01–1.22, P = 0.035). In the subgroup analyses by ethnicity, a significantly increased risk for HCC was found in the Asian population (GG + CG vs CC: OR = 1.11, 95% CI = 1.00–1.23, P = 0.041). Subgroup analysis by control source showed population-based studies to have a significantly increased risk for HCC (GG vs CC: OR = 1.25, 95% CI = 1.04–1.50, P = 0.02; CG vs CC: OR = 1.14, 95% CI = 1.03–1.25, P = 0.01; and GG + CG vs CC: OR = 1.15, 95% CI = 1.02–1.29, P = 0.017). Subgroup analysis by HWE status showed a significantly increased risk for HCC for studies consistent with HWE (G vs C: OR = 1.10, 95% CI = 1.01–1.19, P = 0.022; CG vs CC: OR = 1.16, 95% CI = 1.08–1.26, P < 0.001; and GG + CG vs CC: OR = 1.18, 95% CI = 1.08–1.28, P < 0.001) (Table 3, Fig. 2).
Table 3.
Meta-analysis for the two miRNAs polymorphism and HCC susceptibility.
| Allele model |
Homozygous model |
Heterozygote model |
Dominant model |
Recessive model |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P/Ph | OR (95% CI) | P/Ph | OR (95% CI) | P/Ph | OR (95% CI) | P/Ph | OR (95% CI) | P/Ph | |
| miR-146a rs2910164 (G/C) | ||||||||||
| G vs C | GG vs CC | CG vs CC | GG + CG vs CC | GG vs CG + CC | ||||||
| Overall | 1.06 (0.98, 1.15) | 0.124/0.001 | 1.15 (0.98, 1.36) | 0.090/0.003 | 1.11 (1.02, 1.21) | 0.021/0.154 | 1.11 (1.01, 1.22) | 0.035/0.020 | 0.97 (0.86, 1.10) | 0.663/0.047 |
| S1-Caucasian | 0.92 (0.67, 1.27) | 0.619/NA | 1.05 (0.43, 2.54) | 0.920/NA | 1.23 (0.49, 3.08) | 0.657/NA | 1.11 (0.46, 2.66) | 0.823/NA | 0.86 (0.59, 1.27) | 0.452/NA |
| S1-Asian | 1.07 (0.99, 1.15) | 0.103/0.001 | 1.15 (0.97, 1.37) | 0.097/0.002 | 1.11 (1.01, 1.21) | 0.027/0.119 | 1.11 (1.00, 1.23) | 0.041/0.014 | 0.98 (0.86, 1.12) | 0.765/0.037 |
| S2-HB | 1.02 (0.88, 1.18) | 0.772/0.024 | 1.02 (0.75, 1.39) | 0.880/0.032 | 1.05 (0.89, 1.25) | 0.562/0.213 | 1.05 (0.88, 1.26) | 0.597/0.106 | 0.94 (0.76, 1.17) | 0.599/0.273 |
| S2-PB | 1.09 (1.00, 1.19) | 0.062/0.008 | 1.25 (1.04, 1.50) | 0.020/0.024 | 1.14 (1.03, 1.25) | 0.010/0.214 | 1.15 (1.02, 1.29) | 0.017/0.047 | 0.99 (0.84, 1.16) | 0.858/0.024 |
| S3-HWE_Yes | 1.10 (1.01, 1.19) | 0.022/0.011 | 1.20 (1.00, 1.43) | 0.053/0.005 | 1.16 (1.08, 1.26) | 0.000/0.663 | 1.18 (1.08, 1.28) | 0.000/0.284 | 0.99 (0.86, 1.14) | 0.879/0.029 |
| S3-HWE_No | 0.96 (0.82, 1.14) | 0.667/0.057 | 0.99 (0.66, 1.49) | 0.975/0.126 | 0.96 (0.76, 1.20) | 0.702/0.091 | 0.95 (0.75, 1.22) | 0.702/0.042 | 0.90 (0.68, 1.10) | 0.445/0.352 |
| miR-196a-2 rs11614913 (C/T) | ||||||||||
| C vs T | CC vs TT | TC vs TT | CC + TC vs TT | CC vs TC + TT | ||||||
| Overall | 1.14 (1.06, 1.23) | 0.001/0.009 | 1.31 (1.12, 1.53) | 0.001/0.006 | 1.10 (0.96, 1.25) | 0.166/0.006 | 1.16 (1.03, 1.31) | 0.018/0.005 | 1.14 (1.00, 1.30) | 0.043/0.006 |
| S1-Caucasian | 1.52 (1.13, 2.04) | 0.006/NA | 2.41 (1.30, 4.50) | 0.005/NA | 1.80 (0.99, 3.27) | 0.055/NA | 2.04 (1.16, 3.60) | 0.013/NA | 1.34 (0.87, 2.06) | 0.185/NA |
| S1-Asian | 1.12 (1.05, 1.21) | 0.002/0.021 | 1.28 (1.10, 1.49) | 0.002/0.014 | 1.08 (0.95, 1.22) | 0.267/0.009 | 1.13 (1.01, 1.28) | 0.038/0.013 | 1.13 (0.99, 1.29) | 0.072/0.004 |
| S2-HB | 1.10 (0.96, 1.28) | 0.178/0.005 | 1.24 (0.92, 1.67) | 0.157/0.005 | 1.06 (0.89, 1.27) | 0.488/0.170 | 1.11 (0.90, 1.38) | 0.315/0.026 | 1.12 (0.95, 1.32) | 0.170/0.202 |
| S2-PB | 1.16 (1.07, 1.25) | 0.000/0.176 | 1.36 (1.15, 1.61) | 0.000/0.139 | 1.13 (0.93, 1.38) | 0.216/0.003 | 1.20 (1.02, 1.41) | 0.029/0.019 | 1.15 (0.94, 1.40) | 0.165/0.003 |
| S3-HWE_Yes | 1.09 (1.02, 1.18) | 0.016/0.050 | 1.21 (1.04, 1.40) | 0.013/0.047 | 1.05 (0.91, 1.22) | 0.474/0.005 | 1.10 (0.97, 1.25) | 0.138/0.012 | 1.09 (0.94, 1.26) | 0.248/0.007 |
| S3-HWE_No | 1.14 (1.06, 1.23) | 0.000/0.809 | 1.31 (1.12, 1.53) | 0.000/0.857 | 1.38 (1.05, 1.80) | 0.019/0.994 | 1.16 (1.03, 1.31) | 0.001/0.956 | 1.38 (1.12, 1.70) | 0.003/0.606 |
S1: subgroup by ethnicity, S2: subgroup by source of controls, S3: Subgroup by HWE, PB: population based, HB: hospital based, P: P values of association, Ph: P values of heterogeneity, OR: odds ratio, CI: confidence intervals, P < 0.05 are in bold text.
Fig. 2.
Forest plots of the OR for the association of miR-146a rs2910164 with HCC risk in subgroup analysis by HWE status under A) the heterozygote model (CG vs CC), and B) the dominant model (GG + CG vs CC).
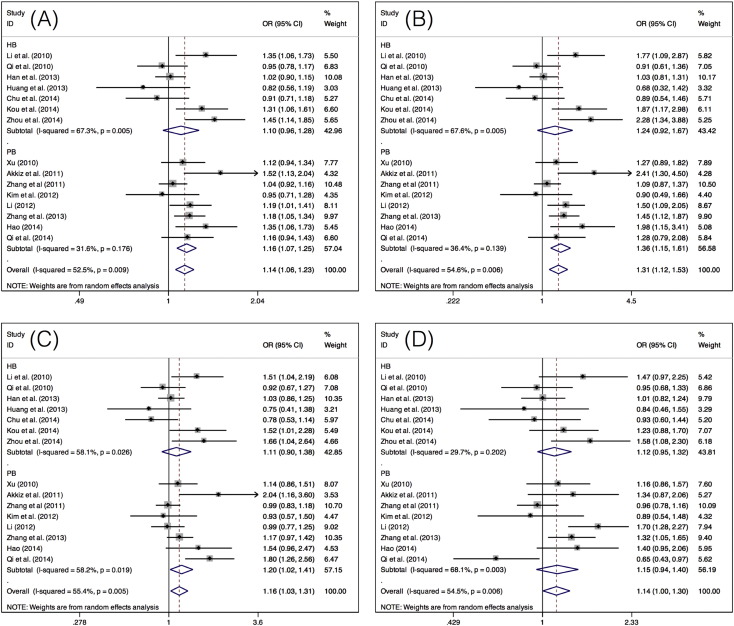
For the miR-196a-2 rs11614913 polymorphism, significantly increased risks for HCC development were observed when all studies were pooled under four models (C vs T: OR = 1.14, 95% CI = 1.06–1.23, P = 0.001; CC vs TT: OR = 1.31, 95% CI = 1.12–1.53, P = 0.001; CC + TC vs TT: OR = 1.16, 95% CI = 1.03–1.31, P = 0.018; and CC vs TC + TT: OR = 1.14, 95% CI = 1.00–1.30, P = 0.043). In the subgroup analysis for studies with the Asian population and population-based studies, significantly increased risks for HCC development were consistent between the three models (C vs T, CC vs TT and CC + TC vs TT) (Table 3, Fig. 3).
Fig. 3.
Forest plots of the OR for the association of miR-196a-2 rs11614913 with HCC risk in subgroup analysis by source of control under A) the allele model (C vs T), B) homozygous model (CC vs TT), C) dominant model (CC + TC vs TT), and D) recessive model (CC vs TC + TT).
3.3. Heterogeneity analysis
Between-study heterogeneity was tested for all the genetic models using the Q-test. For the miR-146a rs2910164 polymorphism, significant heterogeneity between studies was observed for the models (G vs C: Ph = 0.001; GG vs CC: Ph = 0.003; GG + CG vs CC: Ph = 0.020; and GG vs CG + CC: Ph = 0.047) (Table 3). To investigate the sources of heterogeneity, we first carried out meta-regression analyses. These showed that ethnicity, source of controls, genotyping method, HWE in controls and publication year were not effect modifiers (P > 0.05, data not shown). We then performed subgroup analyses by ethnicity, source of controls and HWE in controls. However, we still observed significant heterogeneity in the subgroup for the models (G vs C, GG vs CC and GG vs CG + CC). Lastly, we did the Galbraith plot analysis to identify outliers that might be causing the heterogeneity. For model (G vs C), the plot showed four studies — Li (2012), Wang (2011)), Zhou (2014)) and Qi et al. (2014) — as the outliers in the overall population (Fig. S1). For model (GG vs CC), the plot showed Li (2012), Wang (2011) and Zhou (2014) as the outliers. For model (GG vs CG + CC), the plot showed Zhang et al. (2011) as the outlier (Fig. S1). After excluding these outliers in each model, we did not observe any significant heterogeneity in overall results (G vs C: Ph = 0.17; GG vs CC: Ph = 0.17; and GG vs CG + CC: Ph = 0.24). We also did not observe any significant association for HCC risk between these models.
Regarding the miR-196a-2 rs11614913 polymorphism, it showed significant heterogeneity between studies for all the models (C vs T: Ph = 0.009; CC vs TT: Ph = 0.006; TC vs TT: Ph = 0.006; CC + TC vs TT: Ph = 0.005; and CC vs TC + TT: Ph = 0.006) (Table 3). Meta-regression analysis revealed that the HWE in controls was the major heterogeneity source (P = 0.026), while ethnicity, source of controls, genotyping method, and publication year were not effect modifiers (P > 0.05, data not shown). For models (C vs T) and (CC vs TT), Galbraith plot analysis showed Akkiz et al. (2011a) and Zhou et al. (2014b)as the outliers contributing to the heterogeneity. For models (TC vs TT) and (CC vs TC + TT), the plot showed Qi et al. (2014) and Li (2012) as outliers. For model (CC + TC vs TT), the plot showed Akkiz et al. (2011a) and Qi et al. (2014) as outliers (Fig. S2). However, after excluding these studies in each model, there still remained some degree of heterogeneity for the following models (C vs T: Ph = 0.058; CC vs TT: Ph = 0.043; and CC + CT vs TT: Ph = 0.082). After excluding the outliers, only the CC vs TC + TT model remained significant as showing a link between this SNP and HCC risk (OR = 1.13, 95% CI = 1.02–1.26, P = 0.018, Ph = 0.200). In the subgroup analysis by HWE in controls after excluding the outliers in each model, no heterogeneity was observed (Ph > 0.05) for the group consistent with HWE (P > 0.05) in controls.
3.4. Sensitivity analyses and assessment of publication bias
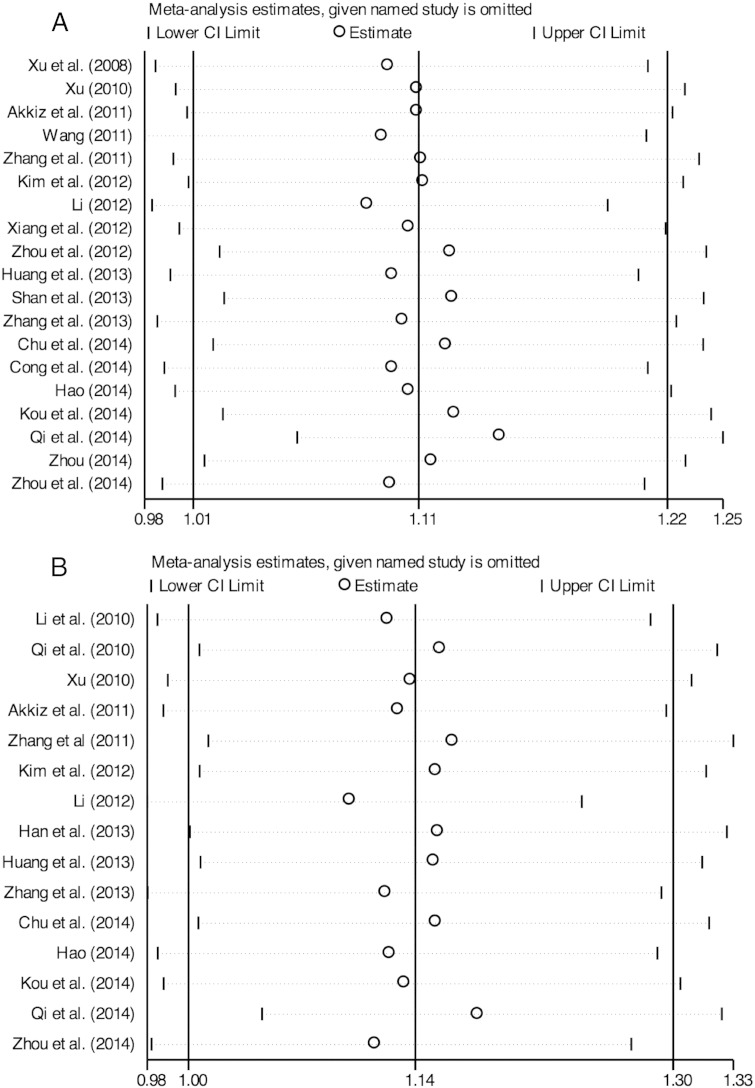
Sensitivity analyses were performed to evaluate the influence of each study on the pooled ORs by sequentially omitting studies. No individual study significantly altered the significance of the pooled ORs, suggesting that the results of the meta-analysis were not being driven by single data points (Fig. 4).
Fig. 4.
Sensitivity analysis of miRNA polymorphism with HCC. A) miR-146a rs2910164 model (GG + CG vs CC), and B) miR-196a-2 rs11614913 model (CC vs TC + TT).
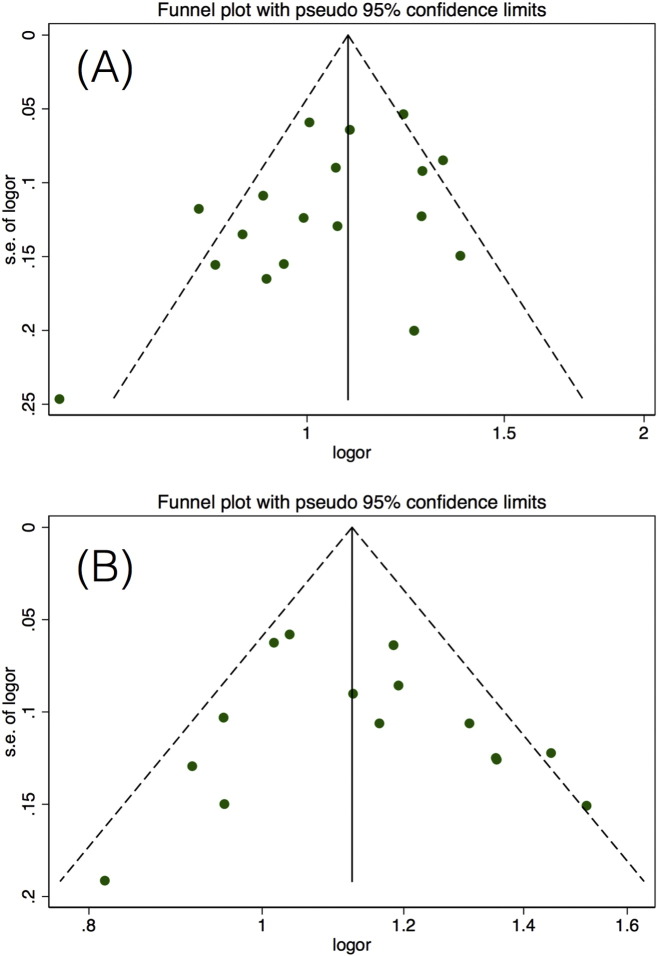
Begg's funnel plot and Egger's linear regression test were performed to evaluate potential publication bias. No evidence of obvious asymmetry for the two SNPs was found based on the shapes of the funnel plots (Fig. 5). The statistic generated using Egger's test (t = − 0.80, P = 0.433 for miR-146a rs2910164 model (GG + CG vs CC); t = 0.03, P = 0.980 for miR-196a-2 rs11614913 model (CC vs TC + TT)) also showed no evidence of publication basis.
Fig. 5.
Begg's funnel plot for publication bias. A) Funnel plot for miR-146a rs2910164 with HCC risk in overall analysis for model (GG + CG vs CC), and B) funnel plot for miR-196a-2 rs11614913 with HCC risk in overall analysis for model (CC vs TC + TT).
4. Discussion
Recent studies have suggested an important role for miRNAs' function as a tumor suppressor influencing both pro- and anti-proliferative cascades (Fabbri et al., 2007). Deregulated miRNA and its associated post-transcriptional gene silencing or gene expression comprise an important part in the pathogenesis of HCC (Yin et al., 2015). Apart from the role of hepatitis viruses which could alter the expression of miRNA (Sidhu et al., 2015), variation within the miRNA genetic code itself may also impact the structure of miRNA and thus its expression. To date, nearly 2000 different miRNAs have been identified in humans and about 2200 SNPs within pre-miRNA regions discovered (Gong et al., 2012). Of these, the two most studied SNPs are the miR-146a rs2910164 and miR-196a-2 rs11614913. However, results for the two SNPs are still controversial. Therefore, we performed this meta-analysis using all the available studies with the largest sample size (N = 16,613 for miR-146a; N = 14,044 for miR-196a-2) to systematically assess the associations and guide future studies. Our results indicate that both polymorphisms are significantly associated with increased HCC risk. In line with other studies, the miR-146a rs2910164 and miR-196a-2 rs11614913 polymorphisms are associated with increased risk for other cancers like colorectal cancer (Xu & Tang, 2015) and gastric cancer (Ni et al., 2015) in the Asian population. The increased risk estimates found in our analysis are somewhat modest (OR < 1.6). However, the magnitudes found are consistent with those from traditional GWAS studies, which have found many common variants (MAF > 5%) with modest effects (OR < 2) (McCarthy et al., 2008), but which collectively account for a substantial proportion of risk for developing complex disease.
Functional studies (Zhang et al., 2015) have revealed that miR-146a expression in hepatoma cells and hepatoma tissues is significantly downregulated compared to related normal tissues; this then correlated with liver cancer metastasis. Further evidence has shown that by restoring miR-146a expression, HCC cell invasion and metastasis were significantly suppressed. This inhibition of cell dysfunction by miR-146a was mediated by miR-146a inhibition of VEGF expression through a dual signal pathway model via beta-catenin and NF-kB. Our results support this role for miR-146a in HCC, showing that for the rs2910164 SNP, CG vs CC and GG + CG vs CC increased risk for HCC. As the C to G change for rs2910164 led to reduced expression of miR-146a, carrying the allele G could result in higher HCC risk. However, in the allele model (G vs C), we did not observe a significant association, potentially due to between-study heterogeneity. When we did the subgroup analysis by HWE status, significant association was observed, although some degree of heterogeneity still existed. When outlier studies in each genetic model were excluded, no significant heterogeneity was found. These outlier studies were mostly of relatively small sample size and thus their exclusion is not a great weakness.
Our results indicated the miR-196a-2 rs11614913 SNP as significantly predictive of HCC under all genetics models. However, caution should be taken when interpreting our results, as high heterogeneity existed in these models. The major heterogeneity source was the HWE status in controls as revealed by meta-regression analysis. Another source of heterogeneity came from outliers. The outliers were predominantly either of a Caucasian population (Akkiz et al., 2011a) or a study by Qi et al. (2014). After excluding the outliers in each genetic model, no heterogeneity was found for groups that were consistent with HWE (P > 0.05) in controls. In line with the functional studies, deregulated miR-196a-2 has been shown to target the downstream genes homobox (HOX) and annexin A1 (ANXA1) (Chen et al., 2011), which both played an important role in the carcinogenesis and malignant transformation of HCC (Kanai et al., 2010).
Several meta-analyses (Wang et al., 2012, Xu et al., 2013, Liu et al., 2014, Wang et al., 2014, Peng et al., 2014) have systematically summarized the potential association of the two SNPs with susceptibility to HCC. Compared with these previous results, for miR-146a rs2910164, our results differ from the first meta paper published by Wang et al. (2012), in that they did not observe any significant association between miR-146a and HCC risk under any genetic model. This may be because that study only included five studies, with relatively small sample sizes. This is compared with the latest meta-analysis published by Peng et al. (2014), in which they only found such association in the Dominant model (GG + CG vs CC), but not in the heterozygote model (CG vs CC).
For miR-196a-2 rs11614913, Xu et al. (2013) only showed the polymorphism as a risk for HCC in Caucasians, not in Asians. Later work by Wang et al. (2014) found a risk for HCC by the miR-192a SNP to be present in both Caucasians and Asians. However, the latest meta-analysis by Peng et al. (2014) failed to observe any association for this SNP. Our results using the largest samples so far, found the rs11614913 polymorphism as a significant risk factor for HCC in both Caucasians and Asians. One thing in common for previous meta-analyses is the existence of between-study heterogeneity, which can hinder meta-analysis. Therefore, in our analysis, we thoroughly examined the sources of heterogeneity by doing subgroup analysis, meta-regression and Galbraith plot analysis.
There are some limitations in our meta-analysis. Firstly, despite our efforts, some degree of heterogeneity still existed for miR-196a-2 rs11614913. Therefore, it should be acknowledged that the potential heterogeneity might influence these results. Second, we only examined the polymorphism in Caucasians and Asians, while the role of the two SNPs in other ethnicities remains unknown. Lastly, as we do not have the access to the original data, the results could not be adjusted for any other covariates, such as age, gender, etc.
Our findings may provide guidance for further studies. We propose that further studies should try to focus on the role of these two SNPs in predicting HCC clinical course using a prospective cohort study design. Beyond epidemiological studies, laboratory studies attempting to explore the role of miRNA in cellular dysfunction and aberrant proliferation could not only aid in our understanding of these SNPs' roles, but potentially improve our understanding neoplasia more generally. Taken together, these epidemiological and other analytical methods could aid in the development of targets for intervention in preventing HCC and moderating its clinical course.
In summary, we performed the largest meta-analysis of miR-146a rs2910164 and miR-196a-2 rs11614913 in predicting HCC. Our results provided strong evidence that SNPs within miR-146a and miR-196a-2 contribute to HCC risk in the Asian population. However, further studies validating the results in this and other ethnicities are essential.
Acknowledgments
We greatly thank Dr. Steve Simpson, Jr. (Menzies Institute for Medical Research) for revising and providing valuable comments to the paper.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mgene.2015.11.002.
Appendix A. Supplementary data
Supplementary figures.
References
- Akkiz H., Bayram S., Bekar A., Akgollu E., Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case–control study. J. Viral Hepat. 2011;18(7):e399–e407. doi: 10.1111/j.1365-2893.2010.01414.x. [DOI] [PubMed] [Google Scholar]
- Akkiz H., Bayram S., Bekar A., Akgollu E., Uskudar O., Sandikci M. No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case–control study. Gene. 2011;486(1–2):104–109. doi: 10.1016/j.gene.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Chen C., Zhang Y., Zhang L., Weakley S.M., Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J. Cell. Mol. Med. 2011;15(1):14–23. doi: 10.1111/j.1582-4934.2010.01219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.H., Hsieh M.J., Chiou H.L., Liou Y.S., Yang C.C., Yang S.F. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0089930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong N., Chen H., Bu W.Z., Li J.P., Liu N., Song J.L. miR-146a G > C polymorphisms and risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2014;35(6):5669–5673. doi: 10.1007/s13277-014-1750-2. [DOI] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Tong Y., Zhang H.M., Wang K., Hu T., Shan G. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum. Mutat. 2012;33(1):254–263. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- Han Y., Pu R., Han X., Zhao J., Zhang Y., Zhang Q. Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0058564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoll H.H. Systematic reviews on rehabilitation interventions. Arch. Phys. Med. Rehabil. 2006;87(6):875. doi: 10.1016/j.apmr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Hao Y. 2014. The Effect and Mechanism of miR196a SNP in Hepatocellular Carcinoma: Shanxi Medical University. [Google Scholar]
- Huang Q.H., Li J.L., Wang C.K., Wei Z.L., Li Y.X., Zhang C.Y. Correlation between microRNA-146a polymorphism and primary liver carcinoma in the Guangxi Zhuang population. Chin. J. Oncol. Prev. Treat. 2013;5:100–104. [Google Scholar]
- Jazdzewski K., Murray E.L., Franssila K., Jarzab B., Schoenberg D.R., de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA. 2008;105(20):7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D.K., Sun J., Cao G., Liu Y., Lin D., Gao Y.Z. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat. Genet. 2013;45(1):72–75. doi: 10.1038/ng.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M., Hamada J., Takada M., Asano T., Murakawa K., Takahashi Y. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol. Rep. 2010;23(3):843–851. [PubMed] [Google Scholar]
- Kim W.H., Min K.T., Jeon Y.J., Kwon C.I., Ko K.H., Park P.W. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504(1):92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Kou J.T., Fan H., Han D., Li L., Li P., Zhu J. Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol. Lett. 2014;8(3):1255–1260. doi: 10.3892/ol.2014.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y. 2012. MicroRNA Related SNPs and Genetic Susceptibility to Hepatocellular Carcinoma: Zhengzhou University. [Google Scholar]
- Li X.D., Li Z.G., Song X.X., Liu C.F. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology. 2010;42(7):669–673. doi: 10.3109/00313025.2010.522175. [DOI] [PubMed] [Google Scholar]
- Liu J., Xie B., Chen S., Jiang F., Meng W. Association study of two inflammation-related polymorphisms with susceptibility to hepatocellular carcinoma: a meta-analysis. BMC Med. Genet. 2014;15:92. doi: 10.1186/s12881-014-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7) doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q., Ji A., Yin J., Wang X., Liu X. Effects of two common polymorphisms rs2910164 in miR-146a and rs11614913 in miR-196a2 on gastric cancer susceptibility. Gastroenterol. Res. Pract. 2015;2015:764163. doi: 10.1155/2015/764163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Li S., Lao X., Chen Z., Li R., Deng Y. The association of common functional polymorphisms in mir-146a and mir-196a2 and hepatocellular carcinoma risk: evidence from a meta-analysis. Medicine. 2014;93(29) doi: 10.1097/MD.0000000000000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P., Dou T.H., Geng L., Zhou F.G., Gu X., Wang H. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum. Immunol. 2010;71(6):621–626. doi: 10.1016/j.humimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Qi J.H., Wang J., Chen J., Shen F., Huang J.T., Sen S. High-resolution melting analysis reveals genetic polymorphisms in microRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer. 2014;14:643. doi: 10.1186/1471-2407-14-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y.F., Huang Y.H., Chen Z.K., Huang K.T., Zhou M.T., Shi H.Q. miR-499A > G rs3746444 and miR-146aG > C expression and hepatocellular carcinoma risk in the Chinese population. Genet. Mol. Res. 2013;12(4):5365–5371. doi: 10.4238/2013.November.7.11. [DOI] [PubMed] [Google Scholar]
- Sidhu K., Kapoor N.R., Pandey V., Kumar V. The “macro” world of microRNAs in hepatocellular carcinoma. Front Oncol. 2015;5:68. doi: 10.3389/fonc.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venook A.P., Papandreou C., Furuse J., de Guevara L.L. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15(Suppl. 4):5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- Wang W. 2011. Association of MiR-146a Single Nucleotide Polymor- Phism with Susceptibility to Hepatocellular Carcinoma and the Microarray Analysis of Tumor Related MicroRNAs: Fudan University. [Google Scholar]
- Wang Z., Cao Y., Jiang C., Yang G., Wu J., Ding Y. Lack of association of two common polymorphisms rs2910164 and rs11614913 with susceptibility to hepatocellular carcinoma: a meta-analysis. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang L., Shi X., Xu H., Wang T., Bian J. Association between two common polymorphisms and risk of hepatocellular carcinoma: evidence from an updated meta-analysis. Bio. Med. Res. Int. 2014;2014:468605. doi: 10.1155/2014/468605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Fan S., Cao J., Huang S., Zhang L.P. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol. Biol. Rep. 2012;39(6):7019–7023. doi: 10.1007/s11033-012-1532-0. [DOI] [PubMed] [Google Scholar]
- Xu Y. 2010. Association Study of Polymorphisms in MiRNAs Genes with the Susceptibility of Hepatocellular Carcinoma: Nanjing Medical University. [Google Scholar]
- Xu L., Tang W. Associations of polymorphisms in mir-196a2, mir-146a and mir-149 with colorectal cancer risk: a meta-analysis. Pathol. Oncol. Res. 2015 doi: 10.1007/s12253-014-9843-1. [DOI] [PubMed] [Google Scholar]
- Xu T., Zhu Y., Wei Q.K., Yuan Y., Zhou F., Ge Y.Y. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29(11):2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- Xu Y., Li L., Xiang X., Wang H., Cai W., Xie J. Three common functional polymorphisms in microRNA encoding genes in the susceptibility to hepatocellular carcinoma: a systematic review and meta-analysis. Gene. 2013;527(2):584–593. doi: 10.1016/j.gene.2013.05.085. [DOI] [PubMed] [Google Scholar]
- Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat. Rev. Gastroenterol. Hepatol. 2010;7(8):448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Zhao Y., Ji Y.J., Tong L.P., Liu Y., He S.X. Serum/plasma microRNAs as biomarkers for HBV-related hepatocellular carcinoma in China. Bio. Med. Res. Int. 2015;2015:965185. doi: 10.1155/2015/965185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhai Y., Hu Z., Wu C., Qian J., Jia W. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat. Genet. 2010;42(9):755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- Zhang X.W., Pan S.D., Feng Y.L., Liu J.B., Dong J., Zhang Y.X. Relationship between genetic polymorphism in microRNAs precursor and genetic predisposition of hepatocellular carcinoma. Chin. J Prev. Med. 2011;45(3):239–243. [PubMed] [Google Scholar]
- Zhang J., Wang R., Ma Y.Y., Chen L.Q., Jin B.H., Yu H. Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac. J. Cancer Prev. 2013;14(11):6427–6431. doi: 10.7314/apjcp.2013.14.11.6427. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Sun X.X., Ma X., Chen Z.N. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol. Cancer. 2015;14(1):5. doi: 10.1186/1476-4598-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. 2014. The Association of the SNPs in miRNA-146a and in miRNA-122 with Hereditary Predisposition and the Earlier Recurrence After Resection for Hepatocellular Carcinoma: Southern Medical University. [Google Scholar]
- Zhou J., Lv R., Song X., Li D., Hu X., Ying B. Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 2012;31(4):524–530. doi: 10.1089/dna.2011.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Simpson S., Jr., Holloway A.F., Charlesworth J., van der Mei I., Taylor B.V. The potential role of epigenetic modifications in the heritability of multiple sclerosis. Mult. Scler. 2014;20(2):135–140. doi: 10.1177/1352458514520911. [DOI] [PubMed] [Google Scholar]
- Zhou B., Dong L.P., Jing X.Y., Li J.S., Yang S.J., Wang J.P. Association between miR-146aG > C and miR-196a2C > T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 2014;35(8):7775–7780. doi: 10.1007/s13277-014-2020-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.