Abstract
We compare the historical demographies of two Müllerian comimetic butterfly species: Heliconius erato and Heliconius melpomene. These species show an extensive parallel geographic divergence in their aposematic wing phenotypes. Recent studies suggest that this coincident mosaic results from simultaneous demographic processes shaped by extrinsic forces over Pleistocene climate fluctuations. However, DNA sequence variation at two rapidly evolving unlinked nuclear loci, Mannose phosphate isomerase (Mpi) and Triose phosphate isomerase (Tpi), show that the comimetic species have quite different quaternary demographies. In H. erato, despite ongoing lineage sorting across the Andes, nuclear genealogical estimates showed little geographical structure, suggesting high historical gene flow. Coalescent-based demographic analysis revealed population growth since the Pliocene period. Although these patterns suggest vicariant population subdivision associated with the Andean orogeny, they are not consistent with hypotheses of Pleistocene population fragmentation facilitating allopatric wing phenotype radiation in H. erato. In contrast, nuclear genetic diversity, θ,in H. melpomene was reduced relative to its comimic and revealed three phylogeographical clades. The pattern of coalescent events within regional clades was most consistent with population growth in relatively isolated populations after a recent period of restricted population size. These different demographic histories suggest that the wing-pattern radiations were not coincident in the two species. Instead, larger effective population size (Ne) in H. erato, together with profound population change in H. melpomene, supports an earlier hypothesis that H. erato diversified first as the model species of this remarkable mimetic association.
The neotropical butterflies Heliconius erato and Heliconius melpomene are an exceptional example of Müllerian mimicry. These distantly related (1, 2), unpalatable species have both radiated into almost 30 wing phenotypic races. However, in any one location across their sympatric ranges, they exhibit nearly perfect convergence in their warningly colored wing patterns. This parallel diversification argues forcefully for the adaptive nature of the mimicry between the two species, and strong stabilizing selection on local patterns has been demonstrated (3, 4).
Nonetheless, the origins of this coincident mosaic of divergence and convergence remain highly controversial (5, 6). According to Müllerian mimicry theory, strong frequency-dependent selection should promote convergence to one common wing pattern at both intraspecific and interspecific levels. One explanation is allied closely with the “Pleistocene refugium model” for the evolution of diversity in Amazonia (7). The so-called “Brown–Sheppard–Turner” model (8, 9) proposes that the two species underwent simultaneous population constrictions during the climatic perturbations of the Pleistocene period, with each mimetic racial pair originating in response to stochastic changes in the biotic environment in isolated populations. Based on cladograms of the two radiations constructed from allelic changes in major wing-patterning loci, Sheppard et al. (8) further suggested that similar wing-pattern races within a species were most closely related, despite often being separated geographically by other, phenotypically distinct races. Matching pattern-gene cladograms may, however, simply reflect similar underlying developmental pathways, rather than a coevolutionary process (10). A recent study using mtDNA sequences (11, 12) challenged some of the earlier conclusions drawn from parsimony analysis of pattern loci. Sequence variation in both species was structured into major geographical regions. Thus, adjacent, often distinct wing-pattern races within both species were more closely related than allopatric but convergent races, indicating that similar patterns within each species had originated independently. Furthermore, the expected phylogenetic signature of coevolution, that of matching phylogenies between the two species, was not seen. H. erato showed a single divide across the Andes, whereas H. melpomene was subdivided into four major biogeographic regions: west of the Andes, the Amazon basin, southeast Brazil, and the Guyanan Shield. Given these phylogenetic patterns, Brower (12) concluded that the contemporary wing-pattern mosaic arose from several independent mimetic origins. Also, based on similar and low levels of mtDNA divergence within the comimics, Brower argued that the time frame of the phenotypic diversification in both species was consistent with the Pleistocene refuge theory.
Recently, there has been renewed interest in the dynamics of Müllerian mimicry and, in particular, the conditions in which coevolution is expected. Müller's original mathematical formulation (13) demonstrated clearly that, given unequal abundances of two similarly distasteful comimics, the rarer species gains a much larger benefit from mimicry than the more abundant species. Thus, in contrast to the mutual convergence implicit in strict coevolution, the rarer species may evolve, or “adverge” (14), toward the more common species. To truly understand the relative roles of each species in the evolution of this parallel diversification, we first need to have a clear picture of the historical demographies of the comimics. Here, we present a comparative historical demographic analysis of H. erato and H. melpomene by using sequence data from two highly variable, unlinked nuclear loci: the autosomal Mannose phosphate isomerase (Mpi) and the sex-linked Triose phosphate isomerase (Tpi) loci. These data complement the previous mtDNA study of the two species (11, 12) and permit more detailed insights into the population history of this mimetic association. Genealogical examination across multiple loci is essential for distinguishing population processes, such as growth or subdivision, from the evolutionary forces that are specific to a single locus.
Methods
We sequenced intron regions of Mpi and Tpi from 10 wing-pattern races of H. erato and nine wing-pattern races of H. melpomene, encompassing three major biogeographical regions: West Andes, East Andes, and the Guyanan Shield (see Table 3, which is published as supporting information on the PNAS web site). To place the history of racial diversification within a broader evolutionary context, we also sampled the sister species of both radiations. Heliconius himera is a geographic replacement of H. erato in the dry, elevated forest regions of southern Ecuador and northern Peru. Heliconius cydno occurs sympatrically with its sister species, H. melpomene, in the northern Andes and Central America. Although these sister-species pairs are known to hybridize in the wild at low (H. cydno and H. melpomene) to moderate (H. himera and H. erato) levels, significant behavioral, ecological, and genetic differentiation warrant species-level designation (15–17). Nonetheless, the exact phylogenetic relationships remain problematic (1). We also sampled the following outgroup species: Heliconius clysonimus, Heliconius telesiphe, and Heliconius hecale (see Table 3).
DNA sequence data were obtained from Mpi and Tpi according to Beltrán et al. (1). For each individual, 3–10 clones were sequenced to identify distinct alleles. Sequences chosen for inclusion generally were represented by at least two identical clones. High substitution rates restricted sequence comparison between closely related species (1); thus, data were compiled into four distinct alignments (see Fig. 3, which is published as supporting information on the PNAS web site). Because of the possibility of recombination in our data sets, we coestimated nucleotide diversity, θ, and recombination, r, by using the genealogical maximum-likelihood (ML) approach in the program recombine (version 0.4) (18). We used an unweighted pair group method with arithmetic mean (UPGMA) tree generated with paup* (version 4.0b8) (19) for the initial tree. recombine was then implemented by using 10 short-chain runs, with sampling every 40 steps for 2,000 steps, followed by a single long-chain run, with sampling every 20 steps for 40,000 steps. To facilitate comparisons, these parameters were also estimated also from a comparable subset of Brower's original mtDNA data (11, 12).
Phylogenetic relationships were estimated by using ML under the heuristic search process in paup* (version 4.0b8) (19). The best models of DNA substitution were identified by using modeltest 3.06 (20). To identify optimal topologies, an initial ML tree was estimated by using a tree bisection-reconnection (TBR) branch-swapping routine on the neighbor-joining tree. This tree seeded a second search using nearest-neighbor-interchange (NNI) branch swapping while reestimating the most-likely substitution parameters. ML topologies were compared with maximum parsimony (MP). The MP was generated by using the heuristic search option and TBR reconstruction. The tree used to seed this run was chosen from the shortest-length trees generated during 100 replicates (each running for 2 min) of a TBR branch-swapping routine run on a tree obtained by random stepwise sequence addition. Confidence within each node was assessed by 500 bootstrap replicates by using the heuristic search with TBR branch swapping. To include the information in insertion and/or deletion events, each length polymorphism was coded in the MP analysis as a single binary character (0 or 1) added to the end of the sequence. To test specific phylogenetic relationships between the sister species, the shortest trees representing alternative a priori scenarios were generated by using macclade (version 3.07) (21), starting with the ML trees shown in Fig. 1. These topologies were then compared with the ML tree by using the method of Shimodaira and Hasegawa (22), implemented in paup*.
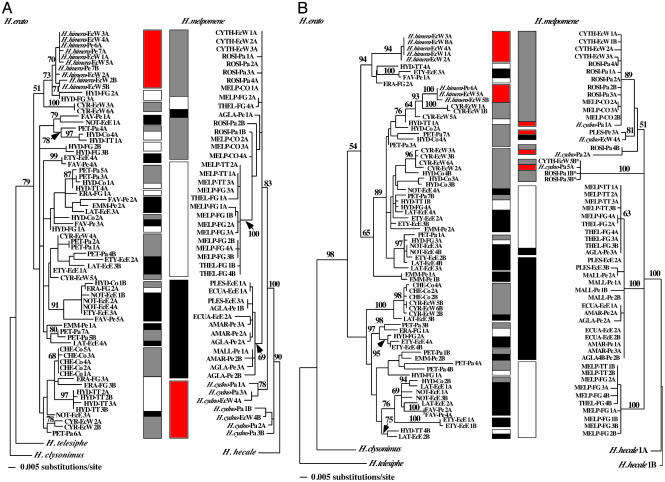
Fig. 1.
ML genealogies for H. erato and H. melpomene of Tpi (A) and Mpi (B) alleles. The following models of evolution were the best: TrN+I+G, P(I) = 0.3402, a = 0.8850, for Tpi H. erato; TrN+I, P(I) = 0.6301, for Tpi H. melpomene; GTR+G, a = 0.6013, for Mpi H. erato; and TrN+I, P(I) = 0.4376, for Mpi H. melpomene. MP genealogies were qualitatively and quantitatively similar. Numbers on the branches are parsimony bootstrap values for the equivalent nodes on the ML tree. Sequences are labeled with a racial code, individual number, allele identity (A or B), and country code (see Table 3 for additional information on phylogenetic analysis). The three main geographic regions are indicated by bars. Racial identity for H. erato is given as follows: CHE, H. erato chestertonii; CYR, H. erato cyrbia; EMM, H. erato emma; ERA, H. erato erato; ETY, H. erato etylus; FAV, H. erato favorinus; HYD, H. erato hydara; LAT, H. erato latavitta; NOT, H. erato notabilis; PET, H. erato petiverana; and HIM, H. himera. Racial identity for H. melpomene is given as follows: AGLA, H. melpomene aglaope; MALL, H. melpomene malleti; AMAR, H. melpomene amaryllis; CYTH, H. melpomene cythera; ECUA, H. melpomene ecuadoriensis; MELP, H. melpomene melpomene; PLES, H. melpomene plessini; ROSI, H. melpomene rosina; and THEL, H. melpomene thelxiopeia. Pe, Peru; EcW, Ecuador, western slope of Andes; EcE, Ecuador, eastern slope of Andes; Pa, Panama; Co, Colombia; TT, Trinidad and Tobago; FG, French Guyana. In B, H. melpomene alleles marked with an asterisk are hypothesized to be of hybrid origin based a larger sampling of alleles (V. Bull, M. Beltrán, E. Bermingham, C. Jiggins, W.O.M., and J. Mallet, unpublished data).
We explored the partitioning of genetic variation across geographic regions (East Andes, West Andes, and Guyana) by using two approaches. First, the hierarchical components of sequence variation were estimated at the racial, regional (as defined from mtDNA; ref. 12), and species level by using an analysis of molecular variance, implemented in arlequin (version 2001) (23). Distance matrices were calculated by using the best model of DNA substitution. Probabilities were calculated by using 20,000 permutations of the data matrix. Second, we compared the most parsimonious number of changes needed to explain the geographic location of each allele with the number expected under the null hypothesis of panmixis by using the program macclade (version 3.07) (21). The expected distribution for the null model was calculated by randomizing geographic location 1,000 times.
Finally, we investigated the historical demography of each species by using a graphical nonparametric estimate of effective population size, known as the generalized skyline plot (24), and we evaluated alternative parametric demographic models by using genie (version 3.0) (25). Input consisted of ML genealogies that were first subjected to a nonparametric rate-smoothing method (26), with rate differences weighted by the mean at all nodes, as implemented in the program treeedit (version 1.0α 10; available at http://evolve.zoo.ox.ac.uk). ML parameter estimation was performed by using the “differential evolution” optimizer, and coalescent intervals were grouped by using the ε parameter (24), with ε set between 0.001 and 0.0025.
Results
We sampled 58 and 64 alleles from Tpi and Mpi, respectively, in H. erato and 42 and 51, respectively, in H. melpomene. The two loci showed high intraspecific sequence variation, and patterns of variation were consistent with an interspecific study (1). Estimated levels of r, the ratio between per-site recombination rate and per-site mutation rate (18), at Mpi and Tpi were low in both species and comparable with those in mtDNA data sets (Table 1). This finding suggests that the effect of recombination is unlikely to strongly affect phylogenetic or demographic analysis. Consistent with this conclusion, we observed that levels of homoplasy were similar within Mpi, Tpi, and mtDNA genealogies when the number of taxa was kept constant (ref. 1, and data not shown).
Table 1.
Estimates of genetic diversity and recombination (18) for mtDNA, Tpi, and Mpi in H. erato and H. melpomene with ≈95% confidence intervals
| Species | Locus | n | θ (95% CI) | r (95% CI) |
|---|---|---|---|---|
| H. erato | mtDNA | 31 | 0.05 (0.03-0.08) | 0.00 (0.00-0.14) |
| H. erato | Tpi | 58 | 0.27 (0.19-0.38) | 0.01 (0.00-0.04) |
| H. erato | Mpi | 64 | 0.38 (0.28-0.53) | 0.00 (0.00-0.01) |
| H. melpomene | mtDNA | 22 | 0.02 (0.01-0.04) | 0.00 (0.00-0.00) |
| H. melpomene | Tpi | 42 | 0.06 (0.03-0.09) | 0.00 (0.00-0.04) |
| H. melpomene | Mpi | 51 | 0.08 (0.05-0.11) | 0.00 (0.00-0.04) |
The two comimics possessed large differences in levels of nucleotide variation at all three loci. This difference was most striking at the two nuclear genes for which estimates of nucleotide diversity were nearly five times higher for H. erato than for H. melpomene (Table 1). In H. melpomene, estimated values of θ were concordant across loci when adjusted for differences in allelic copy number. Thus, as expected by assuming similar evolutionary rates among loci (1), estimates of θ based on the haploid mtDNA were one-third of the estimates of θ for the sex-linked Tpi locus and one-fourth of the estimate of θ for the autosomal Mpi locus. In contrast, for H. erato, there was some discrepancy between the expected and observed levels of nucleotide diversity among loci. In particular, the observed level of nucleotide diversity at mtDNA was 70–80% lower than expected, given the levels of variation at both Tpi and Mpi.
Genetic variation was also partitioned in fundamentally different ways within the species. In H. erato, the vast majority of the diversity at both loci was found within the sampled races, and only a very small amount was due to regional differences (92.85% versus 2.35% for Tpi, and 94.56% versus 3.18% for
Mpi). In contrast, in H. melpomene, among-regional differences explained approximately one-half of the total variance (51.34% for Tpi and 44.18% for Mpi), and geographic race explained most of the remaining variance.
The differences in the levels and distribution of Tpi and Mpi diversity were clearly evident in the allelic genealogies of the two species. Within H. erato, very similar alleles were distributed broadly across major phenotypic and biogeographic boundaries at both loci (Fig. 1). At the sex-linked Tpi locus, alleles sampled from the sister species H. himera and alleles from the geographic race Heliconius erato chestertonii, occurring in the isolated Cauca Valley of Colombia, formed two separate, nested monophyletic lineages. Paraphyly of H. erato alleles with respect to H. himera was significantly more likely than reciprocal monophyly of alleles from each sister species (Shimodaira and Hagegawa test: D = 14.05; P = 0.033). At Mpi, H. erato and H. himera alleles were clearly polyphyletic, with H. himera alleles falling into two distinctive lineages within the larger H. erato genealogy. Despite the lack of a clear phylogeographic signal within H. erato, genetic variation at both loci was not distributed randomly, and there were differences in the distribution of alleles across the Andes (Mpi, P = 0.003; Tpi, P = 0.007, based on randomization tests); this population structure remains significant with H. erato chestertonii alleles excluded.
In contrast, in H. melpomene variation in both nuclear loci coalesced broadly into the following three major biogeographic regions delimited previously by the mtDNA data (12): west of the Andes, Amazonia, and the Guyanan Shield (Fig. 1). At the Tpi locus, H. melpomene alleles were monophyletic, with H. cydno alleles being basal and paraphyletic. Although paraphyly of H. cydno alleles relative to H. melpomene was the most likely model, neither reciprocal monophyly or polyphyly could be rejected (ref. 1, and data not shown). At Mpi, the genealogical pattern was more complex, with H. melpomene and H. cydno sharing very similar alleles. Nonetheless, three major regional clades in H. melpomene were clearly identifiable (Fig. 1B). Within both nuclear genealogies, there were several exceptions to this geographic structure. For example, at the Mpi locus, alleles sampled from the Guyanan Shield fell into two clades. One clade consisted only of alleles found on the Guyanan Shield, and the other clade formed a single derived lineage within a clade of alleles sampled from the Amazonian region. These inconsistencies, however, may be explained by a few migration events.
The pattern of coalescent events in the Tpi and Mpi genealogies further highlighted differences in the evolutionary history of the two comimics. In both species, genealogies were most consistent with a history of population growth rather than with constant effective population size (Table 2 and Fig. 2). However, the processes shaping the demographic histories operated over very different time scales. In H. erato, assuming approximately clock-like evolution and a evolutionary rate comparable with the mitochondrial cytochrome c oxidase subunit I/II (COI/COII) region [1.1–1.2% divergence per lineage per 1 million years (11)] (1), the time to most-recent common ancestor at both loci occurred within the Pliocene period (Fig. 2). Since that time, the genealogies of both nuclear loci were most consistent with population growth across the species and within major biogeographic regions (Table 2). In contrast, for H. melpomene, assuming the same substitution rate the time to most-recent common ancestor was much more recent at both Tpi and Mpi, occurring near the Pliocene–Pleistocene boundary. Nonetheless, there was similar evidence for population growth (Table 2). In this case, regional population growth was likely to have occurred within the latter half of the Pleistocene; average pairwise differences within regional clades ranged from 0.52–0.55% (uncorrected) at Mpi and 0.53–1.1% (uncorrected) at Tpi, suggesting that these independent population expansions began ≈250,000–500,000 years ago.
Table 2. Evaluation of alternative parametric demographic models for the genealogies of H. erato and H. melpomene and their subpopulations at Mpi and Tpi.
|
H. melpomene
|
H. erato
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Mpi
|
Tpi
|
Mpi
|
Tpi
|
|||||||||||
| Guyana | East | West | All | Guyana | East | West | All | East | West | All | East | West | All | |
| n | 19 | 16 | 16 | 51 | 16 | 13 | 13 | 42 | 37 | 24 | 61 | 34 | 19 | 53 |
| Highest-likelihood model | Log | Log/expo | Log | Log | Log | Log | Log | Log | Log | Log | Log | Log | Log | Log |
| Constant model rejected | Yes** | Yes** | No | Yes* | Yes* | Yes** | Yes** | Yes** | Yes** | Yes** | Yes** | Yes** | Yes** | Yes** |
| Expo model rejected | Yes** | No | Yes* | Yes* | Yes* | No | Yes** | No | No | Yes** | No | Yes** | Yes** | Yes** |
The most likely model was compared with constant and exponential models. An expansion growth model never gave a better fit, and results are not presented. Genealogies of H. erato represented by this data do not include H. erato chestertonii. *, P = 0.05; **, P = 0.01. Log, logistic; expo, exponential.
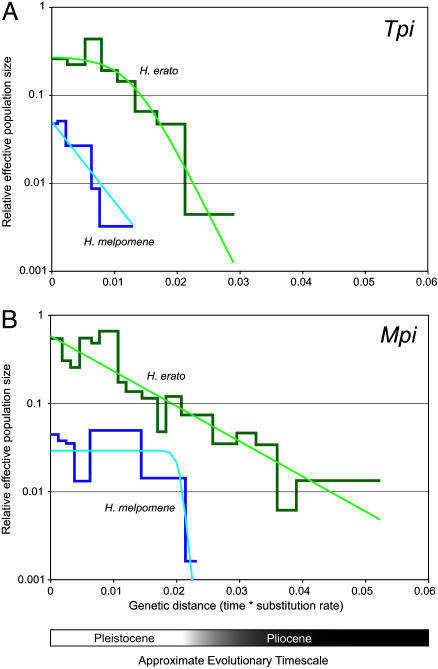
Fig. 2.
Estimated demographic histories of the H. erato and H. melpomene radiations, for Tpi (A) and Mpi (B). The thicker, piecewise plots are the generalized skyline plots. The approximate timescale assumes an evolutionary rate of ≈1.1–1.2% per lineage per 1 million years (1). Thus, the approximate Pleistocene–Pliocene boundary corresponds to a genetic distance of ≈0.023 substitutions per site. The thinner, smooth curves represent the parametric ML demographic history (see Table 2). Plots for H. erato (green) and H. melpomene (blue) are shown on the same graph.
Discussion
Our understanding of the evolutionary history of the parallel mimetic radiations within H. erato and H. melpomene is enhanced significantly by the addition of high-resolution genealogical information for two unlinked nuclear loci. Consistent with the mtDNA study described in refs. 11 and 12, the phylogeographic patterns at the nuclear loci were not concordant between the comimics. In contrast to the mtDNA data, however, genealogical and coalescent-based analyses of nuclear loci revealed that the comimics have had very different demographic histories, and they suggest that racial diversification did not occur simultaneously in response to extrinsic forces over the Pleistocene period (8, 12).
Evolutionary and Demographic History of H. erato. The nuclear data argue for a significantly different population history in H. erato relative to that previously envisioned from mtDNA, highlighting the importance of using multilocus comparisons to infer the population and demographic history of a species (27). Mitochondrial DNA variation in H. erato was partitioned into two regional clades separated by the Andes (11). Very low levels of intraclade mtDNA variation suggested that racial diversification had evolved recently (within the last 200,000 years) and independently in the two areas, which is broadly consistent with a Pleistocene refugium hypothesis for wing-pattern radiation (11, 12).
In contrast, concordant patterns of variation at the two nuclear loci in H. erato (Table 1) are not consistent with the refugium hypothesis, and they do not provide phylogenetic support for independent color-pattern radiations. There was evidence for some genetic differences at both nuclear loci between H. erato populations separated by the Andes. However, both eastern and western populations retained high levels of genetic diversity (Table 1), which would not have been expected had population contractions been experienced. In fact, the genealogical patterns at both loci revealed population growth beginning in the Pliocene period and continuing into the Pleistocene period (Table 2 and Fig. 2). These patterns, concordant across two unlinked nuclear loci, point to an evolutionary history of these allelic samples dating back into the Pliocene period. The similarity of alleles sampled from either side of the Andes (Fig. 1) likely reflects high levels of historical gene flow before the establishment of the Andes as a significant dispersal barrier in these lowland forest insects.
Some of the phylogenetic discrepancies between the nuclear and mtDNA data may reflect differences in expected coalescence times for the different loci. However, mtDNA diversity within H. erato was much lower than expected based on extant levels of variation at both Tpi and Mpi (Table 1). It is unclear whether low mtDNA diversity is a sampling artifact, whether it reflects stochastic lineage extinction, or whether it is the result of purifying selection on mtDNA. Nonetheless, the agreement between the two unlinked nuclear loci suggests that genealogical patterns at these loci better reflect historical demographics within H. erato. Furthermore, lack of evidence for Pleistocene population constrictions in H. erato is concordant with accumulating palynological data. Contrary to the hypothesis of allopatric forest refugia, recent paleoecological studies (28, 29) indicate that continuous tropical forest is likely to have persisted in the Amazon region across the climatic fluctuations of the Pleistocene period.
The nuclear data also help to clarify our understanding of the evolutionary relationship between H. himera and H. erato and shed additional light onto the age of the H. erato lineage. The behavioral, ecological and genetic differences between H. himera and H. erato have been documented (15–17); however, their phylogenetic relationship has remained unresolved (1, 11). Our data significantly reject reciprocal monophyly of H. erato and H. himera; Tpi alleles of H. erato are paraphyletic relative to H. himera, and Mpi alleles of the two species are polyphyletic. These phylogenetic patterns, combined with the restricted, elevated distribution of H. himera, suggest that this species evolved from a peripherally isolated wing-pattern variant within the geographically widespread H. erato lineage by means of a classical “peripatric” speciation event. The slight genetic differences between the two sister species suggest that peripheral speciation occurred ≈2 million years ago (1, 11). Interestingly, the race, H. erato chestertonii displays a similar phylogenetic and biogeographic pattern and may represent another example of peripatric divergence within H. erato. In this scenario, sequence differences between the H. erato and H. himera do not reflect the origin of the H. erato wing-pattern radiation. The age of the H. erato lineage is better estimated relative to divergence between H. erato and its most closely related sister species, Heliconius hecalesia (1). These species are reciprocally monophyletic, and they show a corrected maximum pairwise divergence at mtDNA of 11.3% (mean, 11.0%; SD, 0.5%) (1). Assuming a rate of 1.1–1.2% per lineage per 1 million years (11), this divergence dates the origin of the H. erato lineage back into the Pliocene period, consistent with the levels of extant variation at both Mpi and Tpi.
Evolutionary and Demographic History of H. melpomene. In H. melpomene, the mtDNA (12), Tpi, and Mpi gene genealogies were largely concordant and supported a very different population history for this species relative to its comimic. Variation at all three loci was lower than in H. erato (Table 1), indicating a much lower effective population size in this species. Additionally, in contrast to H. erato, the time to most-recent common ancestor of extant variation (Fig. 2) and estimates of divergence time between H. melpomene and H. cydno (1) place the origin of the H. melpomene lineage within the Pleistocene period.
Furthermore, all three loci demonstrate clear population fragmentation over the Pleistocene period. Genetic variation was divided into major biogeographic regions, but the phylogenetic relationships among-regional clades varied between loci. For example, the Guyana Shield lineage was basal to the rest of the H. melpomene radiation for mtDNA (12) but was the most derived lineage at Tpi (Fig. 1A). At Mpi, the relationship between the three lineages was unresolved (Fig. 1B). These inconsistencies are likely to reflect the stochastic fixation of ancestral variation within emerging regional clades, and they suggest that the clades were formed roughly simultaneously, early in the evolution of H. melpomene lineage. Both of the two main color-pattern types, “rayed” and “banded,” occur in more than one of the geographical lineages identified in H. melpomene, supporting Brower's contention that similar color patterns have evolved independently within each biogeographic region (12).
This Pleistocene subdivision was associated with changes in the population dynamics of the emerging geographic lineages. The individual clades at both nuclear loci for H. melpomene were characterized by low levels of variation relative to that of the entire species sample (Fig. 1), suggesting a loss of variation (possibly as a consequence of population constrictions in each region). Also, the star-like nature of the genealogies of the H. melpomene clades, resulting from a high frequency of singleton mutations between these alleles, is highly characteristic of recent population expansion. Furthermore, despite small sample sizes and low levels of sequence variation, models of historical population growth were almost always supported over constant population size in three geographical regions at both nuclear loci (Table 2). Sampling at the Mpi locus in the western Andean population yielded a logistic growth model with the highest likelihood; however, a model of constant population size could not be rejected. However, at Mpi, demographic analysis was complicated by polyphyly of H. melpomene and H. cydno (Fig. 1B). A more extensive sampling in areas of sympatry suggests high levels of introgressive hybridization between these species (V. Bull, M. Beltrán, E. Bermingham, C. Jiggins, W.O.M., and J. Mallet, unpublished data). Indeed, we observed several H. melpomene alleles that were nearly identical to extant H. cydno alleles. Removing probable introgressed alleles (asterisk in Fig. 1B) provides support for population expansion in H. melpomene west of the Andes (constant size versus logistic growth with hybrid alleles removed: -2Δln = 6.36, P = 0.04, df = 2).
Interestingly, these strong genealogical and demographic trends are coupled with the evolution of significant postzygotic hybrid sterility between geographical regions of H. melpomene (30) in contrast to the lack of postzygotic reproductive isolation among races of H. erato (8).
Advergence and the Evolution of Mimicry Between H. erato and H. melpomene. We found no evidence that the two comimics shared similar demographic histories, as predicted by the Brown–Sheppard–Turner coevolution model. Instead, our data strongly suggest that the phenotypic diversity in H. erato arose first and that H. melpomene radiated subsequently to mimic H. erato, a scenario first proposed by Eltringham (31) in the early 20th century. Eltringham noted that H. erato was almost always much more common than H. melpomene, an observation borne out by 90 years of subsequent field research (6). Based on differences in relative abundances, he argued that H. erato was the model for the mimetic association and that H. melpomene had evolved unilaterally to mimic H. erato. As Eltringham recognized, the number-dependent selection proposed by Müller (13) provides a potent driving force for this mimetic advergence. The relative gain due to Müllerian mimicry between similarly unpalatable species is proportional to the square of the ratio of their relative abundances. Thus, the rarer species gains considerably more from mimicry than the common species. Our data support Eltringham's mimetic advergence hypothesis in three ways. First, the much higher levels of variation in H. erato relative to H. melpomene (Table 1) are consistent with a larger historical effective population size in H. erato. The plots of the effective population size through time indicate that this demographic difference has persisted over the histories of both species (Fig. 2). Second, our analyses indicate that H. erato is almost twice as old as the H. melpomene lineage. Finally, the likely peripatric differentiations of both H. himera and H. erato chestertonii suggest that the phenotypic radiation in H. erato may have evolved quite early in the history of the species.
Although Eltringham (31) did not articulate how he envisioned the initial wing-pattern radiation in H. erato to have occurred, the evolution of novel patterns would not necessarily have required allopatry (5). Recent empirical research (32) has shown that the selection against novel wing patterns is extremely sensitive to changes in density. Thus, even a relatively small number of novel-patterned, aposematic butterflies can educate potential predators. If the novel phenotype reaches a critical density or occupies a critical radius, selection will cause the two warning patterns to be separated by a steep cline (5). These clines are unstable and may spread (33), but they can become trapped in regions of low density, at boundaries between ecotones, or at other physical barriers to gene flow (5, 33). Hybrid zones between races will permit introgression at most loci not closely linked to the genes determining wing-pattern differences; the high levels of gene flow detected across contemporary hybrid zones between parapatric races (J. Mallet and N.S.F., unpublished data) is consistent with this prediction. The establishment of novel pattern variation in H. erato in this manner could then easily drive mimetic advergence in H. melpomene. Indeed, with the 5-fold difference in abundances suggested by the nuclear data and often observed in real populations (6), H. melpomene would receive 25 times more benefit from mimicry than H. erato. A newly formed H. melpomene pattern would spread quickly to occupy the range of its model, generating the parallel color-pattern mosaic that we observe today (33).
The differing evolutionary and demographic histories that have been revealed by analysis of nuclear data provide strong evidence against simultaneous diversification of H. erato and H. melpomene wing patterns. Instead, the older origin, greater abundances, and earlier population expansion in H. erato suggest that the wing-pattern radiation occurred first in this species and that H. melpomene diverged later to mimic the preexisting patterns in H. erato. Nonetheless, concerted evolution of wing patterns in these butterflies cannot be rejected unequivocally. The wing patterns of Heliconius butterflies are remarkably labile, as evidenced by the rapid, independent diversification within the regional clades of H. melpomene. Extremely strong selection acts on the relatively small number of loci that control wing-pattern variation (10), providing the opportunity for continual mimetic evolution under some conditions. Additional comparative demographic analysis incorporating variation at the color-pattern loci themselves will clarify further the population conditions under which new wing patterns evolve.
Supplementary Material
Acknowledgments
We thank Margarita Beltran, Chris Jiggins, and Jim Mallet for sharing samples; Chris Jiggins, Tupac Otero, and Shannon Bennett for valuable discussions; and two reviewers for constructive criticisms. We also thank Germania Estévez (Charles Darwin Research Station, Puerto Ayora, Islas Galapagos, Ecuador), Courtney Rooks and the Paria Springs Eco Community (Maraval, Trinidad) for help with collections, the Autoridad Nacional del Ambiente (Panama City, Panama), and the Ministerio del Ambiente (Quito, Ecuador) for permission to collect butterflies. This work was supported by the National Science Foundation (W.O.M. and D.H.), the Puerto Rican Experimental Program to Stimulate Competitive Research (N.S.F. and W.O.M.), and the Wellcome Trust (O.G.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mpi, Mannose phosphate isomerase; Tpi, Triose phosphate isomerase;ML, maximum likelihood.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF413719, AF413720, AF413723–AF413725, AF413728, AF413726, AF413739, AF413743, AF413744–AF413746, AF413758, AF413752–AF413755, AF413758, AF413764, AF413770, AF4123774–AF413775, AF413782, AF413783, AF413785–AF413787, AF413789, AF413790–AF413792, AF516220, AY319192–AY319254, AY319856-AY319883, AY319885–AY319923, AY329801–AY329812, AY329817–AY329843, AY332412–AY332432, and AY332435–AY332464).
References
- 1.Beltrán, M., Jiggins, C. D., Bull, V., Linares, M., Mallet, J., McMillan, W. O. & Bermingham, E. (2002) Mol. Biol. Evol. 19, 2176-2190. [DOI] [PubMed] [Google Scholar]
- 2.Brower, A. V. Z. (1994) Mol. Phylogenet. Evol. 3, 159-174. [DOI] [PubMed] [Google Scholar]
- 3.Benson, W. W. (1972) Science 176, 936-939. [DOI] [PubMed] [Google Scholar]
- 4.Mallet, J. & Barton, N. H. (1989) Evolution (Lawrence, Kans.) 43, 421-431. [DOI] [PubMed] [Google Scholar]
- 5.Turner, J. R. G. & Mallet, J. L. B. (1996) Philos. Trans. R. Soc. London B 351, 835-845. [Google Scholar]
- 6.Mallet, J. (2001) Evol. Ecol. 13, 777-806. [Google Scholar]
- 7.Haffer, J. (1969) Science 165, 131-137. [DOI] [PubMed] [Google Scholar]
- 8.Sheppard, P. M., Turner, J. R. G., Brown, K. S., Benson, W. W. & Singer, M. C. (1985) Philos. Trans. R. Soc. London B 308, 433-613. [Google Scholar]
- 9.Brown, K. S., Sheppard, P. M. & Turner, J. R. G. (1974) Philos. Trans. R. Soc. London B 187, 369-387. [Google Scholar]
- 10.Nijhout, H. F. (1991) The Development and Evolution of Butterfly Wing Patterns (Smithsonian Institute, Washington).
- 11.Brower, A. V. Z. (1994) Proc. Natl. Acad. Sci. USA 91, 6491-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brower, A. V. Z. (1996) Evolution (Lawrence, Kans.) 50, 195-221. [DOI] [PubMed] [Google Scholar]
- 13.Müller, F. (1879) Philos. Trans. R. Soc. London B 1879, xx-xxix. [Google Scholar]
- 14.Brower, L. P., Brower, J. V. Z. & Collins, C. T. (1972) in Ecological Essays in Honour of G. Evelyn Hutchinson, ed. Deevey, E. (Connecticut Academy of Arts and Sciences, New Haven), pp. 57-67.
- 15.McMillan, W. O., Jiggins, C. D. & Mallet, J. (1997) Proc. Natl. Acad. Sci. USA 94, 8628-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiggins, C., McMillan, W. O., Neukirchen, W. & Mallet, J. (1997) Biol. J. Linn. Soc. 59, 221-242. [Google Scholar]
- 17.Jiggins, C. D., King, P., McMillan, W. O. & Mallet, J. (1997) Heredity 79, 495-505. [Google Scholar]
- 18.Kuhner, M. K., Yamato, J. & Felsenstein, J. (2000) Genetics 156, 1393-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swofford, D. L. (2000) (Sinauer, Sunderland, MA).
- 20.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 21.Maddison, W. P. & Maddison, D. R. (1997) (Sinauer, Sunderland, MA).
- 22.Shimodaira, H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 1114-1116. [Google Scholar]
- 23.Schneider, S., Roessli, D. & Excoffier, L. (2000) arlequin, A Software for Population Genetics Data Analysis (Genetics and Biometry Laboratory, Univ. of Geneva, Geneva), Version 2000.
- 24.Strimmer, K. & Pybus, O. G. (2001) Mol. Biol. Evol. 18, 2298-2305. [DOI] [PubMed] [Google Scholar]
- 25.Pybus, O. G. & Rambaut, A. (2002) Bioinformatics 18, 1404-1405. [DOI] [PubMed] [Google Scholar]
- 26.Sanderson, M. J. (1997) Mol. Biol. Evol. 14, 1218-1231. [Google Scholar]
- 27.Edwards, S. V. & Beerli, P. (2000) Evolution (Lawrence, Kans.) 54, 1839-1854. [DOI] [PubMed] [Google Scholar]
- 28.Colinvaux, P. A. & Oliveira, P. E. D. (2001) Palaeogeogr. Palaeoclimatol. Palaeoecol. 166, 51-63. [Google Scholar]
- 29.Knapp, S. & Mallet, J. (2003) Science 300, 71-72. [DOI] [PubMed] [Google Scholar]
- 30.Jiggins, C. D., Naisbit, R. E., Coe, R. L. & Mallet, J. (2001) Nature 411, 302-305. [DOI] [PubMed] [Google Scholar]
- 31.Eltringham, H. (1916) Trans. Entomol. Soc. London 1916, 101-148. [Google Scholar]
- 32.Kapan, D. D. (2001) Nature 409, 338-340. [DOI] [PubMed] [Google Scholar]
- 33.Sasaki, A., Kawaguchi, I. & Yoshimori, A. (2002) Theor. Popul. Biol. 61, 49-71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.