Abstract
Painting of fourth (POF) is a chromosome-specific protein in Drosophila and represents the first example of an autosome-specific protein. POF binds to chromosome 4 in Drosophila melanogaster, initiating at the proximal region, followed by a spreading dependent on chromosome 4-specific sequences or structures. Chromosome-specific gene regulation is known thus far only as a mechanism to equalize the transcriptional activity of the single male X chromosome with that of the two female X chromosomes. In Drosophila, a complex including the male-specific lethal proteins, “paints” the male X chromosome, mediating its hypertranscription, explained to some extent by the acetylation of lysine 16 on histone H4. Here, we show that Pof is essential for viability in both sexes and for female fertility. POF binding to an autosome, the F element, is conserved in genus Drosophila, indicating functional conservation of the autosome specificity. In three of nine studied species, POF binds to the male X chromosome. When bound to the male X, it also colocalizes with the dosage compensation protein male-specific lethal 3, suggesting a relationship to dosage compensation. The chromosome specificity is determined at the species level and not by the amino acid sequence. We argue that POF is involved in a chromosome-specific regulatory function.
Chromosome-specific gene regulation has so far been identified only as the mechanism for dosage compensation of sex chromosomes. Dosage compensation equalizes the transcriptional activities of the two X chromosomes in the homogametic sex with that of the single X chromosome in the heterogametic sex (1, 2). Dosage compensation strategies vary widely between species (3); in female mammals, one X chromosome is inactivated and forms the Barr body, whereas in Drosophila melanogaster, dosage compensation is achieved by increasing transcription of the single male X chromosome 2-fold (1, 3).
In D. melanogaster, two noncoding RNAs, roX1 and roX2, are essential components of the dosage compensation system (4). These noncoding RNAs, together with the five MSL (male-specific lethal) proteins, “paint” the dosage-compensated male X chromosome (5). The MSL complex mediates acetylation of H4 at lysine 16 on the male X chromosome (6), which in part explains the subsequent hypertranscription. The role played by the roX RNAs in Drosophila has suggested parallels between the fly and the mammalian dosage-compensating systems. In female mammals, a large noncoding RNA, the product of the Xist gene, plays a critical role both in the choice of which X chromosome remains active and in the initial spread and establishment of silencing on the inactivated X chromosome (7). A connection between BRCA1, a breast and ovarian tumor suppressor, and Xist RNA has been reported (8). BRCA1 associates with Xist RNA and supports X inactivation, and is thus the first identified protein factor in the mammalian dosage-compensating system. A Polycomb group protein, Eed, has been shown to play a role in imprinted X inactivation; i.e., in the extra embryonic tissues (9), and is localized on the inactive X chromosome (10).
The mammalian Xist RNA, like the Drosophila roX1 and roX2 RNAs, paints the dosage-compensated X chromosome. Although the way compensation for chromosome dosage is achieved differs between mammals and Drosophila, the basic features of the process display striking similarities. In both systems, chromatin complexes containing spliced, polyadenylated, noncoding RNAs become associated with the compensated chromosome and paint it by spreading along the chromosome. The painting initiates at “nucleation” site(s) and spreads from them to involve most of the chromosome. Our discovery of Painting of fourth (POF) (11), provides the first example of a chromosome-specific protein for an autosome. Unlike BRCA1, Eed, and the MSL proteins, POF paints an autosome, the fourth chromosome of D. melanogaster. Chromosome translocation analysis showed that the binding depends on an initiation site in the proximal region of chromosome 4 and spreads in cis to involve the entire chromosome. The spreading depends on sequences or structures specific to chromosome 4 and cannot extend to parts of other chromosomes translocated to the fourth. Spreading can also occur in trans to a paired homologue that lacks the initiation region. In the related species Drosophila busckii, POF paints the entire X chromosome exclusively in males, suggesting a relationship between the fourth chromosome and the X and between POF complexes and dosage compensation complexes (11). The fourth chromosome is the smallest chromosome, ≈5 Mb (12), and consists of a centromeric, highly condensed region that is underreplicated in polytenic tissues, and a banded, polytenized region, corresponding to cytological sections 101E-102F. Chromosome 4 is in many respects an atypical autosome. Many features indicate that the fourth chromosome of D. melanogaster is largely heterochromatic in nature. The heterochromatin protein HP1 is found associated with much of chromosome 4 (13), and certain histone modifications, identifying heterochromatin are enriched, e.g., H3K9-methylated (14). Repetitive elements normally confined to heterochromatin are distributed throughout the banded region of chromosome 4 (15). Furthermore, reporter genes inserted at many sites in this chromosome often display a variegated, partially repressed expression typical of heterochromatic position-effect variegation (16, 17). Chromosome 4 is also the only autosome that survives in a haploid condition (18). The majority of Drosophila species have a corresponding microchromosome (19) also called the F element (20). In some Drosophila species, such as D. busckii, the microchromosome appears to form the proximal part of the X chromosome (21).
Here, using genetic and molecular analysis of the Pof gene and deduced protein, we show that Pof is essential for female fertility and viability of both sexes. Immunostainings of chromosomes in different species show that the F element specificity is conserved in evolution and that POF is connected to dosage compensation. Based on this finding we conclude that the F element specificity supports a function and we speculate that this function may be a first example of chromosome-specific regulation of an autosome.
Materials and Methods
Fly Strains and Crosses. Fly stocks from different species were provided from the Tucson and Umeå stock centers; for D. busckii see ref. 11. Established mutants and fly lines used in this study have been described (11, 22). Flies were cultivated in vials with potato mash-yeast-agar medium, except for D. busckii, which was raised in Formula 4-24 Instant medium (Carolina Biological Supply).
Generation of Pof Mutants by P Element Excision. We excised the P element inserted close to the transcription start of Pof in the fly line EP (2)2285 to create short deletions in the Pof region. Single white-eyed, non-Sco males from the cross (yw/yw; Sco/CyO × w1118; EP (2)2285/CyO; mus309D3, Δ2-3/mus309D2) were crossed to yw; Sco/CyO females. After 4 days, the single male was removed from the cross, DNA was prepared (23), and PCR was used to screen for short imprecise excisions. The primers used were: 5′-TCCGCATCAGAATCCTCCC-3′ and 5′-GATCATCGGTGGTATCAAGG-3′ to screen for promoter deletions and 5′-ATTTAACACTTCGTAAGAGGGC-3′ and 5′-CGACGAGTAATTTGGTACACTG-3′ to screen for deletions in the transcribed region. Crosses from males with excisions were continued to establish balanced stocks. The mus309 trans-heterozygous genotype increases the yield of imprecise excisions (24).
Transgenic Flies. The P[white+ Pof] construct was prepared as follows: A genomic Pof fragment was amplified by using primers 5′-TATCTCGAGCTGATCGGCAAAATACCCAAAATGAAG-3′ and 5′-AATCTAGAGCCATATGCAAGGAATTGGAGAGAA-3′, wild-type DNA and pfuTURBO polymerase. The PCR product was cut with XhoI/XbaI and ligated into P{CaSpeR-4} (25). The P[white+ Pof.EYFP] construct expressing the POF complete protein with a yellow fluorescent protein (YFP) tag at the C terminus was made as follows: Primers 5′-TATCTCGAGCTGATCGGCAAAATACCCAAAATGAAG-3′ and 5′-GCAGAATTCGAGGATCAGGATCGC-3′ were used to amplify the Pof fragment. The PCR product was cut with XhoI/EcoRI. The enhanced YFP (EYFP)-encoding fragment was excised from the pEYFP-N1 plasmid (Clontech) by using EcoRI/NotI. The two fragments were purified and ligated into a XhoI/NotI cut P{CaSpeR-4} vector. The P[white+ Drosophila ananassaePof.EYFP] construct: The DaPof fragment was amplified from a DaPof cDNA clone by using primers 5′-GCTAGGAACTCGAGTAGGACAGTAT-3′ and 5′-CGCGGTACCGTCTCGGCATCTGATTCCAATTGAGCATAT-3′ and cut with XhoI/KpnI. The EYFP fragment was excised from the pEYFP-N1 plasmid by using KpnI/XbaI. The two fragments were purified and ligated into a XhoI/XbaI cut pUAST vector (26). Salivary gland overexpression of DaPOF.EYFP was achieved by combining P[white+ D. ananassaePof.EYFP] with the salivary gland-specific driver P[w+ GawB]1. The constructs were sequenced to confirm the absence of mutations resulting from errors during PCR. Germ-line transformation of the construct was completed according to described methods (27), by using the Df (1)w67c23, yw, or w1118 strain as host.
Molecular Biology. To clone the D. ananassae Pof cDNA a 350-bp DaPof fragment, isolated by PCR, 32P-labeled by random priming was used to probe a D. ananassae cDNA third-instar larvae library (kindly provided by Kiyohito Yoshida, Hokkaido University, Sapporo, Japan). Inserts from two positive colonies were subcloned into pBluescript II KS(+) and sequenced. An 1,897-bp cDNA was isolated and the encoded amino acid sequence was deduced (GenBank accession no. AY545996). RT-PCR was used to determine transcript levels in the induced Pof mutants. The primer pair 5′-CCACCATCTGCGACATAAACAG-3′ and 5′-CGACGAGTAATTTGGTACACTG-3′ will amplify only the endogenous Pof transcript, 5′-CCACCATCTGCGACATAAACAG-3′ and 5′-CGTCGCCGTCCAGCTCGACCA-3′ only the Pof.EYFP transcript, and 5′-AAAGACGATGCCAAGGACTCAC-3′ and 5′-GCTTGGCGGCCGTGCCGACA-3′ will amplify the neighboring gene CG4806. Poly(A)+ RNA was isolated as described (28). One microliter of poly(A)+ RNA, pd(T)12-18 first-strand primer, primer pairs indicated above, and Ready-to-Go RT-PCR beads (Amersham Biosciences, Piscataway, NJ) was used in each RT-PCR. The amplified products were separated on agarose gels, were visualized, and were quantified by using a Fluor-S MultiImager (Bio-Rad). Sequence alignments were performed by using bioedit and clustalw (29).
Antibodies. Polyclonal antibodies were raised in hen and rabbit by using a full-length bacterially expressed protein as antigen (AgriSera, Vännäs, Sweden). A construct expressing a GST-POF fusion protein was generated by cloning a Pof cDNA into the pGEX5X-3 vector. The fusion protein was produced and purified according to the manufacturer's instruction (Amersham Pharmacia Biosciences) by using Factor Xa cleavage to elute the untagged POF protein from a GSTrap (5 ml) column. The eluted protein was concentrated and used for immunization. The rabbit sera and the purified hen IgY were affinity-purified on an UltraLink Iodoacetyl column as described by the manufacturer (Pierce).
Immunostainings and Immunofluorescence. Polytene chromosomes were prepared and stained from the salivary glands of third-instar larvae essentially as described (11). As primary antibodies against POF a rabbit or hen polyclonal anti-POF antibody raised against a synthetic peptide (11) diluted 1:100, and rabbit or hen antibodies raised against full-length POF protein (see above, 1:400) were used. For double staining, a goat anti-MSL3 antisera, diluted 1:1,000 (kindly provided by M. Kuroda, Harvard Medical School, Boston), or rabbit anti-H4K16ac (Serotec, 1:250) were used. Anti-GFP (Living Colors Av peptide antibody, Clontech) were used to detect EYFP, diluted 1:40. As secondary antibodies, donkey anti-rabbit, donkey anti IgY and donkey anti-goat conjugated with Cy3 or FITC were used (The Jackson Laboratory), diluted 1:400. Schneider 2 (S2) cells were grown and fixed according to standard methods and were stained as described above. Live ovaries and salivary glands were dissected and mounted in halocarbon oil 27 as described (30). Preparations were analyzed by using a Zeiss Axiophot microscope equipped with a KAPPA DX30C charge-coupled device camera. Images were assembled, contrasted, and merged electronically by using photoshop (Adobe Systems, San Jose, CA). The filter set 41028 (Chroma Technology, Rockingham, VT) was used to detect EYFP.
Results
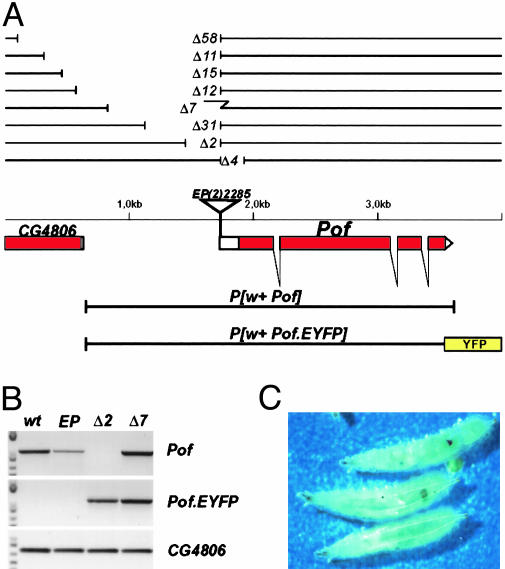
Pof Mutants Were Isolated by P Element Excision. To learn about the in vivo function of Pof besides its binding to chromosome 4, short deletion mutants were created by imprecise excision of the EP element located at the transcription start point of Pof in line EP (2)2285 (Fig. 1A). The EP insertion itself decreases the amount of transcript as seen in Fig. 1B but causes no mutant phenotype. Eight induced Pof-deletion alleles are shown in Fig. 1 A. PofΔ4 deletes the normal translation start point of Pof, but a truncated protein is formed that restores POF function (results not shown). Deletions removing the promoter region of Pof (PofΔ2; PofΔ31; PofΔ7) cause recessive lethality, which is completely rescued by a genomic copy of Pof carried by a transposon, i.e., P[w+ Pof] or P[w+ Pof.EYFP]. By using a sensitive RT-PCR method, we detect no transcript from the endogenous Pof in P[w+ Pof.EYFP]; PofΔ2/PofΔ2 rescued females (Fig. 1B) nor in P[w+ Pof.EYFP]; PofΔ31/PofΔ31 females (results not shown). The PofΔ7 allele still expresses Pof RNA despites its lethality. The residual P element sequence seen in this allele (Fig. 1 A), containing part of the hsp70 control region, probably drives this expression, which does not restore Pof function. Previously (11), we have shown that only a small amount of transcript is detected in females compared to males (20- to 30-fold less). However, the high-level expression seen in males originates from the testes (results not shown). A low expression is seen in both sexes in most tissues, whereas a very high amount of transcript is found in male testes (results not shown). PofΔ2 and PofΔ31 abolish the low overall expression and cause lethality but do not affect the testes specific expression. Because the P[w+ Pof.EYFP] construct still shows correct testes expression (results not shown), we conclude that a testes-specific regulatory element is located in the 472-bp fragment between the upstream end of P[w+ Pof.EYFP] and the PofΔ31 break point. The longer deletion alleles, i.e., PofΔ58, PofΔ11, PofΔ15, and PofΔ12, which also affect expression from CG4806, are not trans-complemented by a transgenic Pof construct. Homozygous PofΔ2 or PofΔ31 are lethal in third-instar larvae, no pupae form. By picking non-GFP homozygous first-instar larvae from a PofΔ2/CyO, GFP stock we could see that PofΔ2/PofΔ2 hatch as first-instar larvae without significant delay. Their development is then progressively slowed down. Crawling third-instar larvae are found after 14 days, compared with 4-5 days in wild type. After 31 days, >20% of these larvae are still moving. In a majority of third-instar PofΔ2/PofΔ2 larvae melanotic masses of different sizes are found (Fig. 1C). However, whether these “pseudotumours” are a direct consequence of the mutant or caused by the developmental delay is not known. As expected, no POF staining is seen on polytene chromosomes from PofΔ2/PofΔ2 larvae. To investigate the role of maternal Pof mRNA to embryo development, we removed Pof in germ-line clones by using FLP/FRT-induced recombination. However, PofΔ31/PofΔ31 germ cells arrest at an early stage of oogenesis and no eggs are produced. The effect of homozygous Pof mutation on male fertility has not been tested. We conclude that the Pof gene function is essential for female fertility and proper development in both sexes.
Fig. 1.
Map of the Pof gene. (A) The exon-intron structure of the Pof gene is shown below the genomic DNA line. Filled boxes represent coding regions and open boxes represent untranslated sequences. The EP element insertion in the homozygous viable EP (2)2285 stock and the extent of induced deletions and transgenic constructs are shown. The broken line in the PofΔ7 deletion symbolizes a 600-bp residual piece from the P element. The EYFP fusion sequence is indicated as a yellow box. (B) RT-PCR analysis on wild-type, EP (2)2285/EP (2)2285, P[w+ Pof.EYFP]; PofΔ2/PofΔ2 and P[w+ Pof.EYFP]; PofΔ7/PofΔ7 female poly(A)+ RNA. The primer pairs amplify only endogenously transcribed Pof (Top), Pof.EYFP (Middle), and CG4806 (Bottom). (C) Sixteen-day-old PofΔ2/PofΔ2 third-instar larvae with melanotic masses.
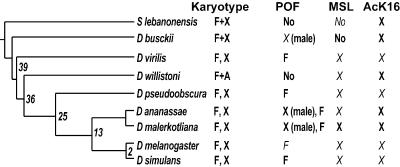
F Element-Specific Binding Is Conserved in Evolution. POF binds specifically to chromosome 4 in D. melanogaster (11) and is essential. This finding is controversial because it implies that chromosome 4 might be under a chromosome-wide control. However, it is not clear that the binding of the fourth chromosome has a function in vivo. To test this hypothesis, we used an evolutionary approach. We raised two new polyclonal antibodies against the whole POF protein sequence to examine different species for presence and specificity of POF. By using four different POF antibodies, we stained eight species from the genus Drosophila and one species from the genus Scaptodrosophila (Scaptodrosophila lebanonensis; Fig. 2). Strikingly, we find evolutionary conservation of the binding of POF to the F element. Distantly related species, such as Drosophila virilis and Drosophila pseudoobscura, still show a specific binding of POF to the F element (Fig. 3A). In D. virilis, POF also decorates the nucleolus. We believe that this staining is also specific because we find it in the close relative species Drosophila americana americana as well (results not shown). The F element-specific binding is always seen in both sexes in the tested species. The conservation through evolution suggests that POF binding to the F element confers some selective advantage in vivo. The binding results are summarized in Fig. 2.
Fig. 2.
Phylogenetic relationship among the species studied based on ref. 33. All species belong to the Drosophila genus, except S. lebanonensis from the Scaptodrosophila genus. Karyotypes are indicated, F element fused to X (F+X), F element as a unique dot chromosome (F, X), and F element fused to an autosome (F+A). POF bindings are tested in both sexes and indicated if sex-specific. The F element specificity is always seen in both sexes. No, No staining is detected. The indicated stainings for MSL and H4K16ac are specific for the male X. Results in italics have been shown (11, 31, 45).
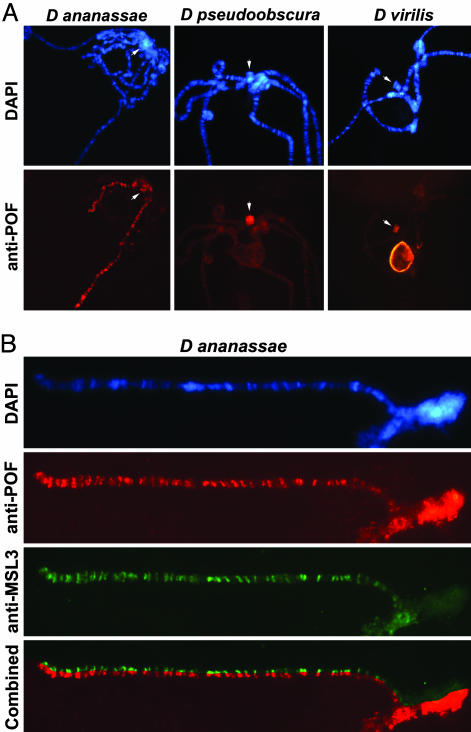
Fig. 3.
Localization of POF on salivary gland chromosomes from different species. (A) POF binds to the male X chromosome and to the F element in D. ananassae (Left) and to the F element in D. pseudoobscura and D. virilis. In D. virilis, the nucleolus is also decorated by POF. An arrow indicates the F element. (B) Higher magnification of a stretched male X chromosome (right arm) and the F element in D. ananassae. The combined image (Bottom) shows that POF colocalizes perfectly with MSL3 on the X chromosome, whereas only POF decorates the F element.
Male X-Specific Binding Is Conserved in Evolution. Our earlier results showing POF staining on the male X-chromosome in D. busckii supported the suggested relationship between the F element and the X chromosome and suggested that POF may be part of an ancient dosage compensation system (11). No MSL binding has been found on D. busckii male X chromosome (ref. 31 and results not shown; MSL1, 2, 3, maleless, males absent on the first tested); however, the H4K16 acetylation is conserved and colocalizes with POF (Fig. 6, which is published as supporting information on the PNAS web site). We found that POF binds to both arms of the male X chromosome also in D. ananassae (Fig. 3A) as well as to the F element in both sexes. Also in the D. ananassae relative Drosophila malerkotliana a specificity to the male X chromosome is seen (Fig. 2, and Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, on the X chromosome, POF perfectly colocalizes with the MSL complex, as shown by double staining with MSL3 in Figs. 3B and 7. This finding indicates that POF binds to the same chromosomal loci as the dosage-compensation complex and may be included in the complex. We did not observe binding of POF to any chromosome in two species. There was no signal in Scaptodrosophila lebanonensis, which is the form farthest away from D. melanogaster. We tested S. lebanonensis because it has been suggested to have the F element fused to the X chromosome in the fashion of D. busckii (32). We did not see any signal in Drosophila willistoni either. It is interesting to note that D. willistoni is the only tested species that has the F element fused to one of the major autosomes (32). It is also the only Drosophila where we fail to detect POF with any of the four used antibodies.
The Chromosome-Specific Property Is Species-Specific. We were interested to test the determinant for the chromosome specificity. We therefore cloned the Pof gene from D. ananassae, in which POF binds the male X. We fused the DaPof gene to EYFP, put it under control of a UAS promoter, and introduced it into D. melanogaster, P[w+ DaPof.EYFP]. A genomic D. melanogaster Pof was also fused to EYFP to be used as a control of the system, P[w+ Pof.EYFP] (Fig. 1 A). Salivary glands dissected from P[w+ Pof.EYFP] transgenic larvae show a bright fluorescence of a chromosomal part in the nuclei corresponding to the fourth chromosome (Fig. 4A). This finding tells us that POF binds the fourth chromosome specifically also in vivo and that the formaldehyde fixation used for preparing chromosomes does not alter the in vivo situation. The P[w+ DaPof.EYFP] transgenic larvae have a clearly fluorescent fourth chromosome, indicating that also the DaPOF protein binds specifically to the F element when introduced in D. melanogaster, seen both in vivo (Fig. 4B) and in formaldehyde-fixed squashes (Fig. 4C). DaPOF is found preferentially in interbands rather than in condensed chromatin, which is similar to the DmPOF. The fourth chromosome specificity is seen in three tested independent P[w+ DaPof.EYFP] lines without a GAL4 driver. We have not succeeded in rescuing Pof mutants by using the UAS-driven DaPOF transgene. Whether this outcome is caused by a dysfunction of the DaPOF protein or a failure to achieve proper expression is not known. When overexpressed in salivary glands, in contrast to DmPOF (11), the DaPOF protein binds to all chromosomes. The results suggest that the chromosome specificity of POF is at the species level and is not determined by the amino acid sequence per se. This finding is confirmed by sequence comparison where the D. pseudoobscura POF protein (F binding) is not more similar to the D. melanogaster sequence (46% identical) than to the D. ananassae sequence (51%) (Fig. 8, which is published as supporting information on the PNAS web site).
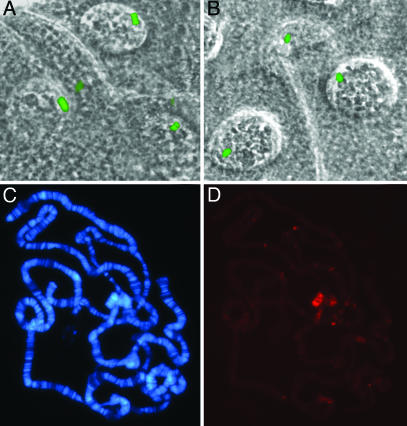
Fig. 4.
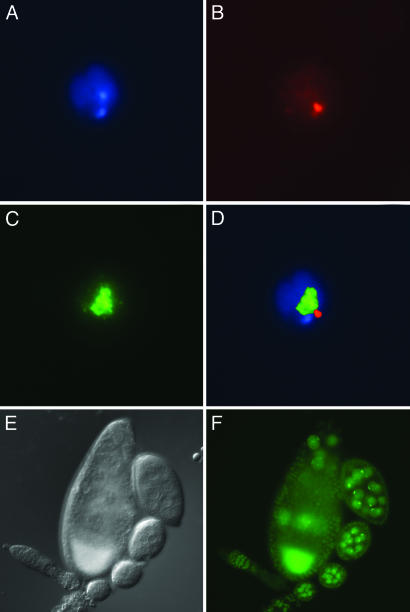
Localization of DmPOF and DaPOF in D. melanogaster salivary glands. (A)Unfixed salivary gland showing DmPof.EYFP fluorescence of chromosome 4 in the nuclei. The phase contrast and the fluorescent images are merged. (B) Unfixed salivary gland showing DaPof.EYFP fluorescence of chromosome 4. (C and D) Localization of DaPOF.EYFP on D. melanogaster salivary gland chromosomes as detected by anti-EYFP antibody. DAPI (C) and anti-EYFP visualized by Cy3 (D).
Chromosome 4 Specificity Is Seen in Different Cell Types. We next wanted to see whether POF binding to the fourth chromosome is unique to polytene chromosomes or polyploid tissues. We therefore examined antibody staining and POF.EYFP fluorescence in different cells and tissues. Antibody staining of S2 cells shows that POF is localized at specific foci in the nuclei (Fig. 5 A-D). These foci are separable from the male X visualized by anti-MSL3 staining. By comparing the sizes, we suggest that POF decorates the fourth chromosome also in S2 cells. The staining is seen as one or two clear foci, suggesting that in some nuclei the two fourth chromosomes are paired or are colocalized. During mitosis, POF is not seen on the fourth chromosome, which is in contrast to MSL3 that is still detectable at the metaphase X chromosome (results not shown). By using the P[w+ Pof.EYFP] transgenic line, we also tested other tissues. We detect POF, presumably on chromosome 4, in a variety of cell types, including ovary nurse cells (Fig. 5 E and F) and follicle cells (data not shown). The presence of POF in these cell types may explain the sterility of PofΔ31/PofΔ31 germ-line clones.
Fig. 5.
POF localization in S2 cells and in ovaries. (A-D) S2 cells: DAPI (A), anti-POF (B), anti-MSL3 (C), and merged (D). The merged image shows that POF staining is separable from the MSL3-labeled X chromosome. Immunofluorescence of POF.EYFP as detected in live ovary preparations. (E) Nomarski image. (F) EYFP fluorescent image. The strong signal in the oocyte is caused by autofluorescence.
Discussion
Chromosome-specific binding of proteins has so far been described only for the dosage-compensated X chromosome and the binding of POF to the entire F element is to our knowledge a unique characteristic. Although heterochromatin proteins, e.g., HP1 (13), have been shown to bind extensively to chromosome 4 and certain histone modifications are enriched, e.g., H3K9-methylated (14), this finding is accompanied by association to other heterochromatic regions, whereas POF is strictly specific for chromosome 4 and is not detectable in the general centromeric region. By inducing deletions in the promoter region of Pof, presumably null mutations according to RT-PCR, we show that Pof is essential for oogenesis in females and viability in both sexes. The phenotypic manifestation of mutated Pof, i.e., prolonged larval development, melanotic masses, and failure to form pupae, is hard to interpret. At this point, we cannot distinguish the direct consequences of the Pof mutation from indirect effects. We conclude that Pof is essential, which raises the controversial question whether the F element may be under a chromosome-specific control. To link the binding of POF to the F element with a function, we undertook an evolutionary study, arguing that conservation of POF binding to the F element indicates function. Our study of different species within the genus Drosophila shows this conclusion to be the case. The F element is found to be decorated by POF in both D. pseudoobscura and D. virilis, species that diverged from D. melanogaster 25 and 39 million years ago, respectively (33), and we conclude that the F element-binding property serves a function. We have previously shown that the binding of POF in D. melanogaster depends on an initiation site in the proximal region of chromosome 4 and spreads in cis to involve the entire chromosome. The spreading depends on sequences or structures specific to chromosome 4 and cannot extend to parts of other chromosomes translocated to the fourth (11). In both D. virilis and Drosophila simulans, the whole banded region of chromosome 4 has been inverted relative to e.g., D. melanogaster (34), yet POF binds to their respective F elements. This discovery shows that the gene order of the chromosome is not involved in the spreading process.
Previously, we have shown that in D. busckii, POF paints the entire X chromosome exclusively in males, suggesting a relationship between the fourth chromosome and the X and between POF complexes and dosage compensation complexes (11). Although none of the MSL proteins has so far been detected on the male D. busckii X chromosome, by using the available antibodies, the fact that the H4K16 acetylation is conserved argues that a similar dosage compensation system is used. Our finding of POF binding to the male X chromosome in D. ananassae strengthens the suggested relationship between POF and dosage compensation. The colocalization of POF and MSL3 in these species further suggests this finding to be the case. When introduced into D. melanogaster, the DaPOF binds to the fourth chromosome and not to X, in neither male nor female. This finding indicates that the chromosome specificity of POF is determined at the species level rather than at protein sequence level, and may include interactions with other proteins or include noncoding RNAs in similarity to the dosage compensation systems in Drosophila and mammals (1).
Considering this association, we suggest that the function of POF binding to the F element is of regulatory character. Chromosome 4 is a unique chromosome in several aspects. One is its heterochromatic nature, including binding of the heterochromatic protein HP1 (35), and late replication of the chromosome as a whole (36). It has been suggested that heterochromatic and euchromatic regions are interspersed along the banded region of the chromosome (37). Another aspect is that similar to the X chromosome but in contrast to the autosomes, haplo-4 and triplo-4 flies are viable and fertile, although haplo-4 females breed poorly (19), suggesting a mechanism of dosage compensation (18). Alternatively, the small size of chromosome 4 may account for the haplosufficiency. In general, deletions that extend over more than one of Bridges numbered chromosomal divisions are lethal (38). However, exceptions exist as exemplified by the large deletions Df(2L)H and Df(3L)Vn (19). The numbered divisions are estimated to be in the range of 800-1,500 kb with few exceptions (39) and the Df(2L)H and Df(3L)Vn deletions span <2.8 and <1.7 Mb, respectively. Although the banded (and sequenced) region of chromosome 4 is 1,230 kb, the chromosome as a whole is estimated to 4.5-5.2 Mb (12). This finding means that chromosome 4 is the only haplosufficient autosome and is also by far the longest region of haplosufficiency among autosomes in D. melanogaster. In this respect, it is interesting to note that in the only species tested within the genus Drosophila where POF is not detected, D. willistoni, the F element is fused to one of the autosomes, element E corresponding to 3R in D. melanogaster (32). We speculate that the potential POF-supported regulatory mechanism is not needed when the F element is part of one of the major autosomes. So far, there are to our knowledge, no proof of chromosome specific regulation on autosomes. Chromosome number differences between related species must be considered as an argument against this possibility. Extreme examples of different diploid numbers between related species exist, as exemplified by the barking deers, muntjacs (40). The extremes among these, the Indian muntjac (Muntiacus muntjac) with 2n = 6 and the Chinese muntjac (Muntiacus reevesi) with 2n = 46. Despite the karyotypic differences, these species are morphologically similar, and can hybridize, although the hybrids are sterile. It is argued that the correct somatic development of these hybrids implies that it is irrelevant if the genes underlying the developmental circuit are on 6 or 46 chromosomes (40). Although, in general, diploid species within a genus show a narrow range of chromosome numbers, examples of high karyological differences exist. Cytogenetic studies of titi monkeys (Callicebus) have provided a similar example, with diploid numbers ranging from 2n = 50 (Callicebus hoffmannsii; ref. 41) to 2n = 16 (Callicebus lugens; ref. 42). However, in this case, detailed cytogenetic analysis by using FISH shows that despite the diversity and the rearranged karyotype, the synteny of 11 human chromosomes is maintained intact in Callicebus callicebus (43). At this point, it is therefore hard to conclude on the potential generality of chromosome-specific regulatory functions.
Our hypothesis is that POF is involved in a chromosome 4-specific gene regulation mechanism. This control may include the stimulation of gene expression in a highly heterochromatinized environment. Such a mechanism may explain, to some extent, the haplosufficiency of chromosome 4. Recently, a systematic analysis of the Caenorhabditis elegans genome, by using the RNAi technique, has shown that genes of similar function cluster in distinct regions of individual chromosomes, suggesting that different chromosomes and regions of the genome may be specialized for particular functions (44). In this context, it should be noted that similar observations were made also in Drosophila by Hochman (18), who concludes that both the X and the fourth chromosome are highly enriched in mutations, resulting in pupal lethality. Further work on POF has the potential to contribute to our understanding of chromosome regulatory complexes, their composition, function, and evolution.
Supplementary Material
Acknowledgments
We thank K. Yoshida for providing the D. ananassae cDNA library; M. Kuroda for anti-MSL antibodies; S. Åström for the mus309 mutant stocks; I. Dacklin for injecting the P[w+ DaPof.EYFP] construct; and V. Pirrotta, D. Roberts, A. Saura, and S. Tuck for comments on the manuscript. This work was supported by grants from the Swedish Research Council and the Åke Wiberg, Carl Tryggers, and Philip Sörenssen Foundations (to J.L.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: POF, Painting of fourth; MSL, male-specific lethal; YFP, yellow fluorescent protein; EYFP, enhanced YFP; S2, Schneider 2.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY545996).
References
- 1.Akhtar, A. (2003) Curr. Opin. Genet. Dev. 13, 161-169. [DOI] [PubMed] [Google Scholar]
- 2.Park, Y. & Kuroda, M. I. (2001) Science 293, 1083-1085. [DOI] [PubMed] [Google Scholar]
- 3.Pannuti, A. & Lucchesi, J. C. (2000) Curr. Opin. Genet. Dev. 10, 644-650. [DOI] [PubMed] [Google Scholar]
- 4.Franke, A. & Baker, B. S. (1999) Mol. Cell 4, 117-122. [DOI] [PubMed] [Google Scholar]
- 5.Stuckenholz, C., Kageyama, Y. & Kuroda, M. I. (1999) Trends Genet. 15, 454-458. [DOI] [PubMed] [Google Scholar]
- 6.Smith, E. R., Pannuti, A., Gu, W., Steurnagel, A., Cook, R. G., Allis, C. D. & Lucchesi, J. C. (2000) Mol. Cell. Biol. 20, 312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plath, K., Mlynarczyk-Evans, S., Nusinow, D. A. & Panning, B. (2002) Annu. Rev. Genet. 36, 233-278. [DOI] [PubMed] [Google Scholar]
- 8.Ganesan, S., Silver, D. P., Greenberg, R. A., Avni, D., Drapkin, R., Miron, A., Mok, S. C., Randrianarison, V., Brodie, S., Salstrom, J., et al. (2002) Cell 111, 393-405. [DOI] [PubMed] [Google Scholar]
- 9.Wang, J., Mager, J., Chen, Y., Schneider, E., Cross, J. C., Nagy, A. & Magnuson, T. (2001) Nat. Genet. 28, 371-375. [DOI] [PubMed] [Google Scholar]
- 10.Mak, W., Baxter, J., Silva, J., Newall, A. E., Otte, A. P. & Brockdorff, N. (2002) Curr. Biol. 12, 1016-1020. [DOI] [PubMed] [Google Scholar]
- 11.Larsson, J., Chen, J. D., Rasheva, V., Rasmuson Lestander, A. & Pirrotta, V. (2001) Proc. Natl. Acad. Sci. USA 98, 6273-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke, J. & McDermid, H. (1993) Chromosoma 102, 718-723. [DOI] [PubMed] [Google Scholar]
- 13.Eissenberg, J. C., Morris, G. D., Reuter, G. & Hartnett, T. (1992) Genetics 131, 345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czermin, B., Melfi, R., McCabe, D., Seitz, V., Imhof, A. & Pirrotta, V. (2002) Cell 111, 185-196. [DOI] [PubMed] [Google Scholar]
- 15.Miklos, G. L. G., Yamamoto, M. T., Davies, J. & Pirrotta, V. (1988) Proc. Natl. Acad. Sci. USA 85, 2051-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallrath, L., Guntur, V., Rosman, L. & Elgin, S. (1996) Chromosoma 104, 519-527. [DOI] [PubMed] [Google Scholar]
- 17.Wallrath, L. & Elgin, S. (1995) Genes Dev. 9, 1263-1277. [DOI] [PubMed] [Google Scholar]
- 18.Hochman, B. (1976) in The Genetics and Biology of Drosophila, eds. Ashburner, M. & Novitski, E. (Academic, San Diego), pp. 903-928.
- 19.Ashburner, M. (1989) Drosophila: A Laboratory Handbook (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Muller, H. J. (1940) in The New Systematics, ed. Huxley, J. (Clarendon Press, Oxford), pp. 185-268.
- 21.Krivshenko, J. D. (1955) Proc. Natl. Acad. Sci. USA 41, 1071-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsley, D. L. & Zimm, G. G. (1992) The Genome of Drosophila melanogaster (Academic, New York).
- 23.Gloor, G. B., Preston, C. R., Johnson-Schlitz, D. M., Nassif, N. A., Phillis, R. W., Benz, W. K., Robertson, H. M. & Engels, W. R. (1993) Genetics 135, 81-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beall, E. L. & Rio, D. C. (1996) Genes Dev. 10, 921-933. [DOI] [PubMed] [Google Scholar]
- 25.Pirrotta, V. (1988) Bio/Technology 10, 437-456. [DOI] [PubMed] [Google Scholar]
- 26.Brand, A. H., Manoukian, A. S. & Perrimon, N. (1994) in Methods in Cell Biology, eds. Goldstein, L. S. B. & Fyrberg, E. A. (Academic, San Diego), pp. 635-654.7707973
- 27.Spradling, A. C. (1986) in Drosophila, a Practical Approach, ed. Roberts, D. B. (IRL, Oxford), pp. 175-197.
- 28.Svensson, M. J., Chen, J. D., Pirrotta, V. & Larsson, J. (2003) Chromosoma 112, 133-143. [DOI] [PubMed] [Google Scholar]
- 29.Hall, T. A. (1999) Nucleic Acids Symp. Ser. 41, 95-98. [Google Scholar]
- 30.Hazelrigg, T. (2000) in Drosophila Protocols, eds. Sullivan, W., Ashburner, M. & Hawley, R. S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 313-344.
- 31.Marin, I., Franke, A., Bashaw, G. J. & Baker, B. S. (1996) Nature 383, 160-163. [DOI] [PubMed] [Google Scholar]
- 32.Papaceit, M. & Juan, E. (1998) Chromosome Res. 6, 49-54. [DOI] [PubMed] [Google Scholar]
- 33.Russo, C. A., Takezaki, N. & Nei, M. (1995) Mol. Biol. Evol. 12, 391-404. [DOI] [PubMed] [Google Scholar]
- 34.Podemski, L., Ferrer, C. & Locke, J. (2001) Chromosoma 110, 305-312. [DOI] [PubMed] [Google Scholar]
- 35.James, T. C., Eissenberg, J. C., Craig, C., Dietrich, V., Hobson, A. & Elgin, S. C. (1989) Eur. J. Cell Biol. 50, 170-180. [PubMed] [Google Scholar]
- 36.Barigozzi, C., Dolfini, S., Fraccaro, M., Raimondi, G. R. & Tiepolo, L. (1966) Exp. Cell Res. 43, 231-234. [DOI] [PubMed] [Google Scholar]
- 37.Sun, F. L., Cuaycong, M. H., Craig, C. A., Wallrath, L. L., Locke, J. & Elgin, S. C. (2000) Proc. Natl. Acad. Sci. USA 97, 5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsley, D. L., Sandler, L., Baker, B. S., Carpenter, A. T., Denell, R. E., Hall, J. C., Jacobs, P. A., Miklos, G. L., Davis, B. K., Gethmann, R. C., et al. (1972) Genetics 71, 157-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorsa, V. (1988) Chromosome Maps of Drosophila (CRC, Boca Raton).
- 40.John, B. & Miklos, G. B. G. (1988) The Eukaryote Genome in Development and Evolution (Allen & Unwin, London).
- 41.Rodrigues, L. R., Barros, R. M., Pissinati, A., Pieczarka, J. C. & Nagamachi, C. Y. (2001) Cytobios 105, 137-145. [PubMed] [Google Scholar]
- 42.Bonvicino, C. R., Penna-Firme, V., do Nascimento, F. F., Lemos, B., Stanyon, R. & Seuánez, H. N. (2003) Folia Primatol. (Basel) 74, 141-149. [DOI] [PubMed] [Google Scholar]
- 43.Stanyon, R., Bonvicino, C. R., Svartman, M. & Seuánez, H. N. (2003) Chromosoma 112, 201-206. [DOI] [PubMed] [Google Scholar]
- 44.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- 45.Bone, J. R. & Kuroda, M. I. (1996) Genetics 144, 705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.