Abstract
Phenotypic differences between individuals and species are controlled in part through differences in expression of a relatively conserved set of genes. Genes expressed in the immune system are subject to especially powerful selection. We have investigated the evolution of both gene expression and candidate enhancers in human and mouse macrophages exposed to glucocorticoid (GC), a regulator of innate immunity and an important therapeutic agent. Our analyses revealed a very limited overlap in the repertoire of genes responsive to GC in human and mouse macrophages. Peaks of inducible binding of the glucocorticoid receptor (GR) detected by ChIP-Seq correlated with induction, but not repression, of target genes in both species, occured at distal regulatory sites not promoters, and were strongly enriched for the consensus GR binding motif. Turnover of GR binding between mouse and human was associated with gain and loss of the motif. There was no detectable signal of positive selection at species-specific GR binding sites, but clear evidence of purifying selection at the small number of conserved sites. We conclude that enhancer divergence underlies the difference in transcriptional activation after GC treatment between mouse and human macrophages. Only the shared inducible loci show evidence of selection and therefore these loci may be important for the subset of responses to GC that is shared between species.
Keywords: enhancer, evolution, glucocorticoid, macrophage
Introduction
The gene complements of different mammals are remarkably similar (1), which implies that phenotypic variation is driven mainly by differences in transcriptional regulation (2–4) . Expression of orthologous genes can differ between closely related primates (5) and even between individuals within a species (6), with genes involved in extracellular processes and the immune response being the most divergent (7). The most highly-conserved, and highly-inducible promoters are, paradoxically, the most sensitive to variation in expression across species (8). Much of this divergence is driven by the evolution of cis-regulatory elements (9), such as enhancers (10).
Deep evolutionary conservation has been used to identify candidate enhancers (11, 12). Nevertheless, complete gain and loss of enhancers between species is also common (13–15). In mammals, at least, turnover of cis-regulatory elements can occur rapidly enough to be identified between strains of a single species (16). Most evolutionarily labile enhancers can be aligned to the genomes of distantly-related species (17), suggesting that the acquisition of novel enhancers can occur through mobilization of existing sequences, including transposable elements (18). Species-specific regulatory elements display a level of nucleotide diversity consistent with a relaxation in evolutionary constraint (14). Since not all transcription factor-binding sites have a direct effect on gene expression (18–20), these sites may represent functionally neutral sequence.
The profound differences in the immune systems of mouse and man have long been recognised (8, 21–23), and are accompanied by both high levels of gene expression divergence and cis-regulatory element turnover (7). This divergence is likely driven by the evolutionary pressure of host-pathogen interactions, alongside changes in the expressed protein-coding sequences driven by positive natural selection (24).
Glucocorticoids (GC) are powerful metabolic hormones that are released in response to stress (25) and that provide natural feedback regulation of immune function. Exogenous GC are widely-used as anti-inflammatory therapy (26). Accordingly, their actions on immune cells are likely to be affected by evolutionary selection. GC act by binding to an intracellular nuclear hormone receptor - the glucocorticoid receptor (GR). Nuclear GR may bind directly to DNA, classically as a homo-dimer (27), to a canonical glucocorticoid response element (GRE), or may act indirectly by binding other transcription factors such as NFκB and AP-1 (28), as well as by recruiting coregulators, for example GRIP1 (29). Gene repression by GC in inflammation has been linked to binding of negative GR binding elements (nGRE) (30, 31), that are distinct from the consensus GRE. Aside from their ability to repress the actions of proinflammatory stimuli, GC alone act directly on macrophages, producing changes in cell survival, proliferation, morphology and phagocytosis (32–35). In other cellular systems, GR binds DNA mainly at cis-regulatory elements (36) and alters chromatin organization (37–39).
We sought evidence of the evolutionary pressure on GC actions by comparing the responses of human and mouse macrophages. Only limited expression data for macrophages responding to GC has been generated previously (40–42). We confirmed that the majority of genes with a significant shift in expression in response to GC are upregulated (36), but few target genes were shared between the two species. GR binding sites were enriched near to inducible genes in both species and species-specific binding was associated with species-specific upregulation of genes in the same genomic region. However, these species-specific sites do not appear to be experiencing a selective pressure and the only selection we could detect was for the preservation of the small set of GR-binding sites that were shared between human and mouse.
Methods
Ethics
Procedures involving human volunteers were approved by the South East Scotland National Health Service Research Ethics Committee. All volunteers gave informed consent. Animals were cared for and managed within the Roslin Institute’s guidelines for animal safety and welfare.
Cell culture
8-10 week male wild type C57BL/6 mice were culled by cervical dislocation. Bone marrow was flushed from hind limbs and then cultured in RPMI supplemented with Penicillin/Streptomycin, Glutamax (Invitrogen), and 10% Foetal Calf Serum for 7 days in the presence of rhCSF-1 at 104U/ml. Human peripheral blood monocytes were isolated from blood samples by Ficoll gradient separation of buffy coats followed by MACS CD14+ve selection (Milteny). They were then cultured as above for 7 days before being treated as indicated with 100nM dexamethasone (Sigma) or ethanol vehicle.
RNA extraction and processing
RNA was prepared using RNeasy column based extraction with on column DNase treatment (Qiagen). RNA quality was checked using a 2100 Bioanalyzer (Agilent). For RT-qPCR, cDNA was prepared using SuperscriptIII (Invitrogen). Relative expression was determined using SyBR green on a LightCycler480 (Roche) and compared with GAPDH as a reference. Primer sequences are given in Table S1G. For expression microarrays, RNA was prepared using standard Affymetrix protocols and applied to the HT-MG430PM (mouse), or HT-U33plusPM (human) chip by Edinburgh Genomics.
Chromatin Immunoprecipitation
Antibodies used for chromatin immunoprecipitation of mouse GR were BuGR2 1ug/106 cells (ThermoFisher / Pierce) and normal rabbit IgG sc-2025 (Santa Cruz). For human GR ChIP we used Sigma Imprint anti-GR, 1ug/106 cells and mouse IgG.
To prepare antibody-bound beads, 20ul of Protein A Dynabeads (Invitrogen) per immunoprecipitation (IP) were washed once then diluted to 200ul in block solution (1xPBS, 0.5% BSA, +2 l 0.1M PMSF). Antibody was added and rotated for 3h at 4°C.
Cells were washed gently once with phosphate buffered saline (PBS) then cross-linked in tissue culture plates with 1% formaldehyde/RPMI at room temperature for 10 min (mouse) or 7.5 min (human) and then quenched with 0.125M glycine. Cells were detached by scraping in PBS then spun down (400g, 5 min, 4°C), resuspended and counted. For mBMDM, 106 cells per IP were then lysed for 15 minutes on ice in 1%SDS, 10mM EDTA, 50mMTris-HCl pH 8.1 supplemented with Protease Inhibitors (Calbiochem), 1mM DTT and 0.2mM PMSF (Sigma). The solution was diluted in IP dilution buffer (0.1% Triton-X100, 2mM EDTA, 150mM NaCl, 20mM Tris-HCl pH8.1) and sonicated using a Soniprep 150 to produce an average fragment size 300-500 bp. Chromatin was spun for 10min at 10,000 g 4°C then supplemented with 20% Triton-X100 to 1% and Bovine Serum Albumin (BSA) (Sigma) to 50ug/ml. Input aliquots were removed and stored at −20°C. Chromatin was then added to the antibody-bound Protein A Dynabeads (Life Technologies) and rotated overnight at 4°C. Beads were washed 3 times for 10 minutes each in 1 - 1% IP dilution buffer, 2 - 1%Triton-X100/0.1%Na-deoxycholate/0.1%SDS, 50mM Hepes pH7.9, 500mM NaCl, 1mM EDTA and 3 - 0.5%Na-deoxycholate/0.5%NP-40, 20mM Tris-HCl pH 8, 1mM EDTA, 250mMLiCl. Chromatin was extracted at 37°C for 15min on a vibrating platform in 100ml extraction buffer (0.1M NaHCO3, 1%SDS). To reverse crosslinks, samples were supplemented to 300mM with NaCl, treated with RNaseA 20mg (Roche) then incubated for ~8h at 65°C. Proteinase K 40ug (Genaxxon) was added and samples incubated at 55°C for 1h. DNA was purified using the QIAquick PCR purification kit (Qiagen). Real-time qPCR analysis to determined percent input bound at known GR target loci was carried out on a LightCycler 480 System using SYBR Green Master Mix (Roche). Primers used are presented in Table S1G. For sequencing ChIP DNA was prepared and amplified using Illumina adapters and Tru-Seq multiplex primers then sequenced using a HiSeq2500 by Edinburgh Genomics.
For hMDM the same protocol was followed with the following differences. Due to constraints on cell number for sequencing material was prepared from 4 volunteers, treated, fixed for 7.5 minutes and lysed as above. Consistency of the assay was assessed by biological replicates of ChIP qPCR for a known GR target in the FKBP5 locus (Figure S2A). Chromatin was sonicated to a fragment size of 400-600 bp and the chromatin pooled, to give 25 × 106 cells in total. This was split into 3 for the IP step and recombined at extraction. DNA was then isolated as above, split into 3 aliquots and blunt ended with Klenow (Roche), PNK (NEB) and T4 DNA polymerase (Roche). An overhanging A base was added using Klenow (−exo) (NEB) and Illumina adapters ligated overnight at 16°C with T4 DNA ligase (NEB). The IP samples were recombined after ligation and then split again into 7 aliquots. Libraries were amplified from each of these aliquots using Illumina Tru-seq multiplex primers and Phusion high-fidelity DNA polymerase (NEB) and the resulting material pooled and sequenced by Edinburgh Genomics on a Hiseq-2500.
Data Analysis
Expression data
Analysis was performed using R/Bioconductor packages ‘arrayQualityMetrics’, ‘affy’ and ‘limma’ (64–66). Expression values were generated using rma. Further exploratory expression analysis used unlogged expression values prefiltered for low expressed probesets as input for the graphical correlation based tool Biolayout Express3D (67). A range of correlation coefficients and MCL values was used to determine an optimal graph structure from which clusters of genes were then read. Clusters were then manually curated to remove artefacts. Genes from these lists were selected across a range of fold-changes for analysis by RT-qPCR and a threshold drawn at log2 fold change = 1, where all tested genes were confirmed (Figure S1A & S1B). To limit loss of genes with extreme profiles – and hence less likely to cluster −genes reaching log2 fold change >1.5 using a conventional analysis by log fold change were also retained if the corresponding expression profile was consistent with a response across all replicates. Orthologues were identified using the HGNC Comparison of Orthology Predictions (HCOP) tool (68).
Promoter Analysis
Promoters were defined as −300, +100bp of the transcription start sites (TSS) described by the FANTOM5 consortium (49). Where multiple TSSs are known, any overlaps were concatenated. Average sequence conservation scores (phastCons) for promoter regions were extracted and enriched motifs identified using HOMER (51).
Comparison to GWAS results and inflammatory genes
The GWAS catalogue (48) ( http://www.genome.gov/gwastudies/) was manually edited to retain only hits with association to inflammatory / immune conditions (Table S1H, 1408 unique SNPs). The intersection of reported genes was assessed by fold change above a background distribution generated using permutation (100,000) of random gene sets. Significance of the difference was assessed using Pearson’s chi-squared test. The intersection of risk SNPs and promoters was ascertained using BEDtools (69).
Functional Annotation
Lists of functional terms from multiple publically available databases were generated using HOMER (51), filtered using a threshold of −log p-value 6.5, then manually curated to remove duplicate terms.
ChIP-sequencing
Sequencing quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and sequence from adapters removed using trimmomatic (70). Paired end reads were aligned to mm9 or hg19 by Bowtie2 (71) using default options (−D 15 −R 2 −L 22 −i S,1,1.15). Downstream analysis was performed using HOMER (51), including creation of bedGraph files for visualization, peak calling and annotation. Peaks were called by comparison with the sequenced input sample for each experiment as a measure of background. For the mBMDM data after confirming congruence (86% peak overlap), data from two independent replicates were combined. Publically available sequencing data for comparisons was accessed via NCBI GEO for Uhlenlaut et al (GSE31796) and Ostuni et al. (GSE38379).
We compared the number of observed intersections of our GR bound sites with sites of PU.1 binding, H3K4me1, H3K4me3, H3K27ac and FAIRE-seq reported in unstimulated mBMDM (57) to the median intersection that occurred in 1,000 genome permuted GR peak locations. Counts of intersections for all marks were generated using HOMER (mergePeaks −cobound −d given).
To compare the locations of GC-regulated genes with GR peaks we calculated the proportion of peaks within a given genomic interval from the TSS of a regulated gene (Figure 3A & B). We calculated the enrichment of GR peaks near to regulated genes as the ratio of the proportion of regulated genes near to a GR peak to the proportion of unregulated genes with a GR peak within the same genomic interval. The significance of both these results was estimated by comparing them to the 95% confidence interval of 1,000 replicates of genome permuted GR peak locations.
We compared peaks between species using liftOver provided by UCSC to get the coordinates of those peaks falling within syntenic blocks. Peaks with > 1bp overlap were assigned as shared. Insertions and deletions were called by comparison with dog (CanFam2), horse (EquCab2), cow (BosTau6) and pig (SuScr3) genomes. If the sequence underlying a peak could be aligned to at least one of these species the peak was defined as being deleted, if not then it was called as a insertion. Human GR sites were assigned as deletions in along the mouse lineage (Figure 2). Where multiple orthologues were present this analysis was run ‘all to all’.
The role of species-specific GR binding was assessed by calculating the proportion of genes with GR binding within 1 Mb that were upregulated to the proportion of all genes that were upregulated. These ratios were calculated separately for each combination of shared, mouse-specific and human-specific GR peaks and upregulated genes (Figure 4). Significance of the difference between mouse and human specific binding was assessed using Pearson’s chi squared.
To compare the enrichment of motifs in shared vs. aligned non-bound peaks we performed motif finding using HOMER as above, using the non-bound as background.
GERP scores for the locations of bound GR motifs in both human and mouse were extracted from the UCSC genome browser. These scores have been calculated by running the GERP++ algorithm on the 36-way mammalian genome alignments (72).
Data Access
Microarray and sequencing data presented in this paper have been submitted to NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/), accession GSE61881. Publically available sequencing data for comparisons was accessed via NCBI GEO for Uhlenlaut et al (40)(GSE31796) and Ostuni et al. (57) (GSE38379).
Results
Glucocorticoid induced gene expression in macrophages
The response to GC was determined by gene expression profiling, using the most commonly used models of macrophage biology, bone marrow-derived macrophages in mouse (mBMDM) and monocyte-derived macrophages in human (hMDM), both cultivated in vitro using the macrophage growth factor, CSF1 (43–45), as used in a previous comparison of the response to LPS (8).
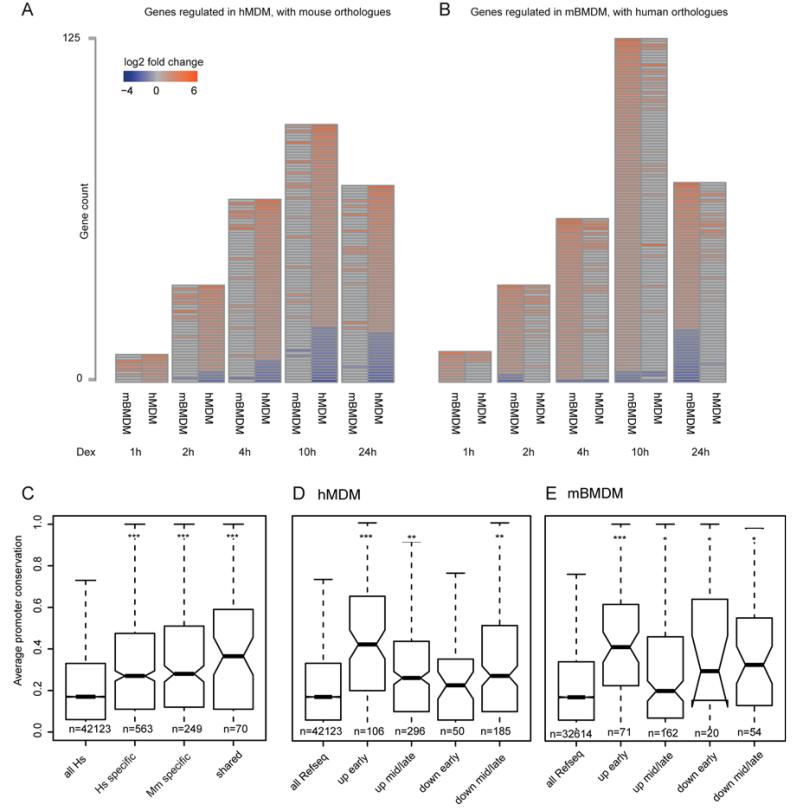
To identify both initial direct targets and downstream secondary consequences, gene expression was measured at six time points over 24 hours following treatment with dexamethasone. After filtering for low expressed and low variance probes, lists of regulated genes were confirmed by RT-qPCR (see methods and Figure S1A and S1B) to produce a high confidence set. For mBMDM there were 160 induced and 50 repressed genes over the full 24h time series. In the hMDM 225 genes were induced and 125 repressed (Fig.1A and 1B, full lists are presented in supplementary Tables S1A & S1B). In both species, induced genes responded more quickly than repressed genes: in mBMDM 10%, 32% and 62% within 1, 2 and 4 hours respectively, compared to the repressed gene set (0%, 4% and 14% at the same time points). The equivalent figures for hMDM were 11, 30 and 70% induced within 1, 2 and 4 hours respectively (2, 14, and 47% for the repressed set at the same time points).
Figure 1. The expression response of macrophages to dexamethasone is divergent despite target gene promoter sequence conservation.
(A) For each time point studied heatmaps of genes regulated by 100nM dexamethasone in hMDM alongside the orthologous genes from mouse and their expression values in mBMDM with height proportional to the number of genes changing (red = induced, blue = repressed). (B) The inverse analysis to that in (A) showing genes regulated in mBMDM by 100nM dexamethasone alongside their human orthologues. (C) Box plots showing average sequence conservation scores (phastCons) of promoters for; (all Hs) all human genes, genes responding only in either hMDM (Hs specific) or mBMDM (Mm specific) and genes with a shared expression response (shared). (D & E) as in (C) but for all genes regulated by GC in (D) mBMDM and (E) hMDM, categorised by response and kinetics. All Refseq promoters are shown as a measure of background. (*** p <1×10−10, ** p <1×10−4, * p <0.05, Wilcoxon rank sum; n = number of promoters; median = horizontal bar, whiskers = 1.5x interquartile range, statistical comparisons are made to background).
For mBMDM and hMDM the robust induced gene set included several known GR targets (e.g. Dusp1, Tsc22d3, Fkbp5 (40) and Per1(36)). As expected from earlier studies of mBMDM (46) both repressed gene lists contained urokinase plasminogen activator (Plau) (47). Since Plau is a target of sustained MAP kinase signaling (47) its repression may be an indirect consequence of the induction of the MAPK inhibitor Dusp1. In mBMDM eight annotated transcription factors were amongst the induced gene set, including four (Fos, Hivep2, Klf4, Ncoa5) that were induced within two hours and that could contribute to the downstream regulatory cascade. Similarly, 10 transcription factors were induced and two repressed within two hours in hMDM (Table S1C). Functional annotation revealed a number of terms shared by the induced sets in both species, including stimulus response, immune/wounding response and regulation of transcription (Fig. S1C). The terms nuclear processes, apoptosis and development were enriched in the early-induced set; cell surface immune response, phagocytosis, migration and cytoskeleton were found amongst the late responders in mBMDM. Functional annotation of induced genes from hMDM revealed terms absent from the mouse dataset including: adipogenesis, FOXO and insulin signalling, MAPK cascade. The terms immune pathways, IL-10 production, NFκB, TNF, NOD-like receptor and rheumatoid arthritis were enriched in the hMDM repressed set (Fig. S1C).
Genes regulated by GC in human macrophages are candidates for involvement in inflammatory and metabolic disease. Indeed, among GC regulated genes in hMDM there was a 3.7-fold enrichment of genes reported to have a genetic association with an inflammatory condition or metabolic disease (n = 48, chi-squared p =1.1×10−5, Tables S1D & S1H) in the GWAS catalogue (48). In mBMDM, only 3 of these genes were upregulated following GC treatment (Fos, Mertk and Tlr7).
GC gene induction in hMDM differs from mBMDM
228 mouse orthologues for the 225 GC-induced hMDM genes and 131 orthologues for 125 repressed genes were identified (see methods). The reciprocal analysis found 157 human orthologues of 160 mBMDM induced and 55 orthologues of 50 mBMDM repressed genes. From amongst this set of robustly-regulated genes, 33 induced and three repressed genes were shared by the two species (Table S1E), a small but significant overlap (p=4×10−15, cumulative binomial probability). The magnitude of expression change between genes upregulated in both species compared to the complete set of upregulated genes was only marginally different, being greater for the mouse genes (1.4-fold enrichment, Wilcoxon rank sum p = 9×10−4).
Promoters of genes induced by GC in hMDM and mBMDM are conserved
Despite their discordant regulation between the species, and in contrast to the situation for the response of macrophages to LPS (8), the promoter regions of GC-inducible genes were conserved (Fig. 1C-E). The genes that had a shared response did not have significantly higher promoter conservation (p=0.166 and p=0.3117 compared to human specific and mouse specific respectively, Wilcoxon rank sum) (Figure 1C). In each species, promoters of genes that were induced the most rapidly to GC were more highly conserved than those with a slower response or than for genes whose expression was repressed in response to GC (Fig. 1D and 1E).
Analysis of the promoters (−300bp, +100bp of the TSS defined previously by CAGE analysis) (49) of GC-regulated genes for transcription factor binding motifs provided no evidence of enrichment for GREs and few motifs were even marginally enriched in the induced gene sets (analysis not shown).
GR binding occurs at canonical GRE sites in distal enhancers
To identify the sites involved in the GC response in the two species, chromatin immunoprecipitation for GR and sequencing (ChIP-seq) was performed two hours after dexamethasone treatment in both mBMDM and hMDM. Representative UCSC browser tracks for GR binding in mBMDM and hMDM are shown in Fig. 2A and 2C.
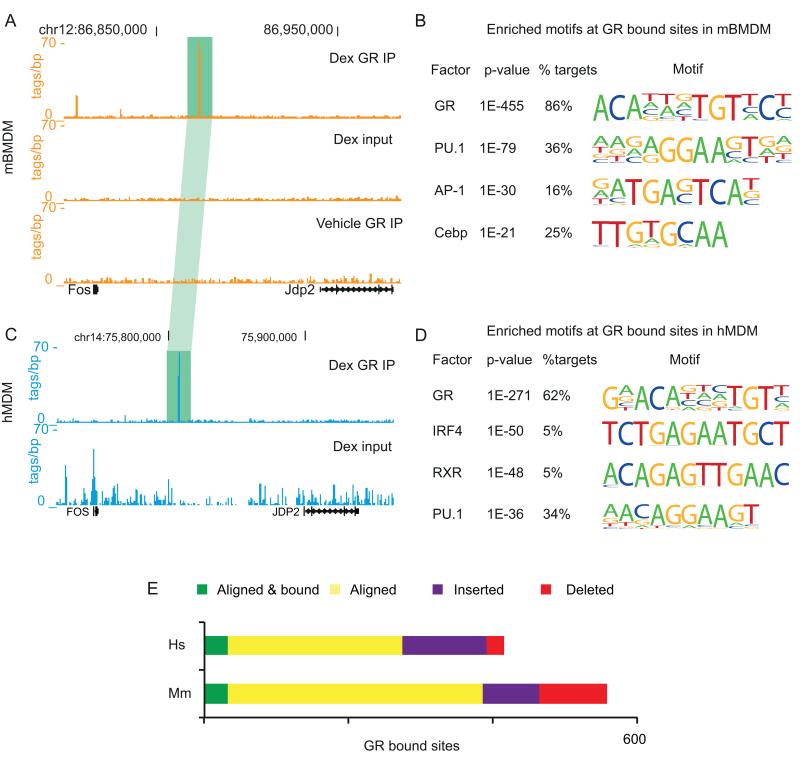
Figure 2. GR binding in mBMDM and hMDM occurs at sites with the canonical GRE.
(A) ChIP-seq data tracks from the UCSC browser for the Fos – Jdp2 region, for GR binding in mBMDM. Data from ChIP with anti-GR antibodies after treatment with 100nM dexamethasone for 2h (Dex GR IP), input material (Dex input) and immunoprecipitated material from a vehicle treated control (Vehicle GR IP) are shown. (C) As in (A) but for FOS – JDP2 in hMDM. Vehicle GR IP data was not generated in hMDM due to constraints on available cell numbers. A GR bound site that aligns between mBMDM and hDMD is highlighted in green. Alignments are to mm9 and hg19 versions of the mouse and human genomes, respectively. (B and D) Enriched motifs found de novo within GR bound sites in mBMDM (B) and hMDM (D). (E) Evolutionary outcomes for GR peaks in human. Aligned sites where the orthologous region is bound by GR in mouse are shown in green and in yellow if the site is not bound by GR in mouse. Sites that could not be aligned are defined as either insertions (purple) or deletions (red) by comparison with dog (CanFam2), horse (EquCab2), cow (BosTau6) and pig (SuScr3) genomes (see methods), Human GR sites were assigned as deletions in the mouse lineage.
There were 488 high confidence GR binding peaks in chromatin from mBMDM, most (474) of which were induced by dexamethasone. These peaks lie away from promoters, in intergenic regions and introns (Fig. S2B). Based upon de novo motif finding, the majority (78%) of the GR peaks contained a motif, or motifs, closely resembling the GRE within +/− 25bp of the peak centre. This figure rises to 86% when considering a region +/−100bp from the peak centre (Fig. 2B). There were no matches under the GR peaks for the nGRE (CTCC(n)0-2GGAGA, where (n)0-2 indicates flexibility in spacing) (30). A more permissive search revealed 59/488 peaks containing a weak match for nGRE. Of these, only 4 have a repressed gene within 1Mb (Plau, Egr2, Rgs2, Cy2s1), one of which has a prominent canonical GRE-containing peak at 74kb (Rgs2).
In hMDM treated with dexamethasone, 484 high confidence GR peaks were detected (Fig. 2C). As in the mouse data, these were remote from promoters in non-coding regions of the genome (Fig. S2B) and were highly enriched for a motif closely matching the consensus GRE (52% within +/−25bp, 62% within +/−100bp) (Fig. 2D and S2B) but not for the nGRE. None of the inflammation-associated SNPs from the GWAS catalogue identified above (Table S1G) directly overlap with GR bound peaks in hMDM, although two lie within 350bp (rs10499197, near TNFAIP3 linked to SLE and systemic sclerosis and rs12466022, linked to multiple sclerosis).
Inducible transcription factors often bind to regions that are already in open chromatin. The GR peaks in mBMDM were clearly associated with sites marked by various enhancer-associated histone marks in unstimulated cells (Fig. S2C). Overall, 462/488 (94%) of our sites overlap with one or more enhancer marks (Fig. S2C), and individual marks also showed enrichments above genome-wide expectations (H3K27ac (70.3%, 24.5-fold enriched, p<2.2×10−16), H3K4me1 (90.4%, 18.4-fold enriched, p<2.2×10−16)) and open chromatin (43.4%, 53.9-fold enriched, p<2.2×10−16). The ETS factor PU.1 is a master regulator of macrophage transcription, and other stimulus-induced transcription factors have been shown to bind at enhancers marked by PU.1 in activated macrophages (50, 51). The sites recruiting GR after dexamethasone treatment also show significant overlap with PU.1- binding sites in unstimulated mBMDM (72.3%, 32.1-fold enriched, p<2.2×10−16 Pearson’s chi squared, Fig. S1C). Overall, a close match to the PU.1 consensus motif was found in 36% of GR-bound sites in mBMDM (Fig. 2B, p=1×10−79) and 76% had an ETS motif. Similarly, 34% of GR peaks in treated hMDM had a PU.1 motif (Fig. 2D, p=1×10−36) and 62% an ETS factor binding motif. The GRE and ETS sites were closely spaced (average of 40bp between the motifs in mouse and 43bp in human), suggestive of cooperativity between ETS factors and GR binding. Aside from PU.1, there was little similarity in other transcription factor motifs enriched around the GR peaks in the two species. For the mouse, other enriched motifs include those that can bind Cepb, AP-1, c-Jun, and Runx1 (Fig. 2B). For the hMDM data set, IRF4 and RXR sites were the most significantly enriched (Fig. 2D).
GR binding differs in hMDM and mBMDM
The majority of GR bound sites can be aligned between human and mouse (n = 274 (56.6%) and 354 (79.2%) in human and mouse, respectively) but only a minority of these (n = 32) are bound by GR in both (Fig. 2E; Table S1F). There was no difference between the rate of turnover of GR binding in evolution at these sites in human and mouse (chi-squared, p = 0.18). Sites that could not be aligned between human and mouse were defined as de novo insertions if they could not be aligned to any of several outgroup species, or deletions if they could be (see methods). As previously reported (52), there was an increased rate of deletions along the mouse lineage (3.8-fold, chi-squared, p = 2.6×10−11) and a smaller, but significant, 1.6-fold increased rate of insertions along the human lineage (chi-squared, p = 2.0×10−3). There have been several reports that transposable elements are the source of species-specific sequences (14, 53). Indeed, transposable elements were the source of almost all inserted GR-binding sequence (96.6% and 75.5% in human and mouse, respectively), which represent 1.8- and 1.4-fold enrichments (chi-squared, p < 7.6×10−3) over the deleted sequences in human and mouse, respectively.
GR binding is associated with induced genes in both human and mouse macrophages but the target loci are not conserved
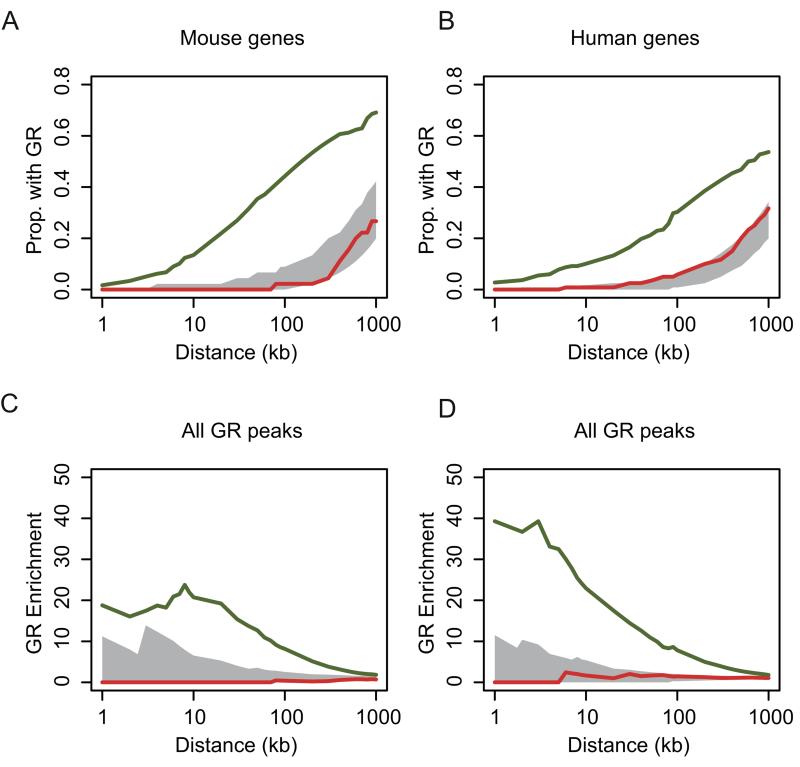
There was a clear association between GR binding peaks and the loci encoding GC-induced genes in both human and mouse macrophages. This enrichment was greatest at 10kb from the TSS, but still marginally detectable at 1Mb in both species (Fig. 3). In A549 cells, GR has been reported to be bound closer to induced genes than to repressed genes (36), while in our macrophage data, there was no detectable relationship between GR-binding peaks and repressed genes (Fig. 3).
Figure 3. Induced genes are associated with GR binding.
(A and B) The proportion of GR ChIP peaks with induced (green line) or repressed (red line) gene promoters within a given genomic distance for (A) mBMDM and (B) hMDM. The 95% confidence intervals from matched genome permuted distributions of GR peaks are shown in grey. (C and D) Enrichment of the proportion of induced (green line) gene promoters with a GR peak within a given interval versus the proportion of induced genes without a GR peak (red line) within that interval for (C) mBMDM and (D) hMDM. No enrichment is seen for repressed genes (shown in red) The 95% confidence interval from a genome permuted distribution of GR peaks is shown in grey.
The expression response to GC was stronger where there were multiple GR peaks within 200kb of the TSS of an induced gene than if there was only a single site (mBMDM 1.7-fold median log2 fold change p = 0.0018, hMDM 1.4-fold median log2 fold change p = 0.031, Wilcoxon rank sum). Early (<2h) induction was not associated with greater proximity to a GR peak (Wilcoxon rank sum p = 0.20 and p = 0.16, for human and mouse, respectively).
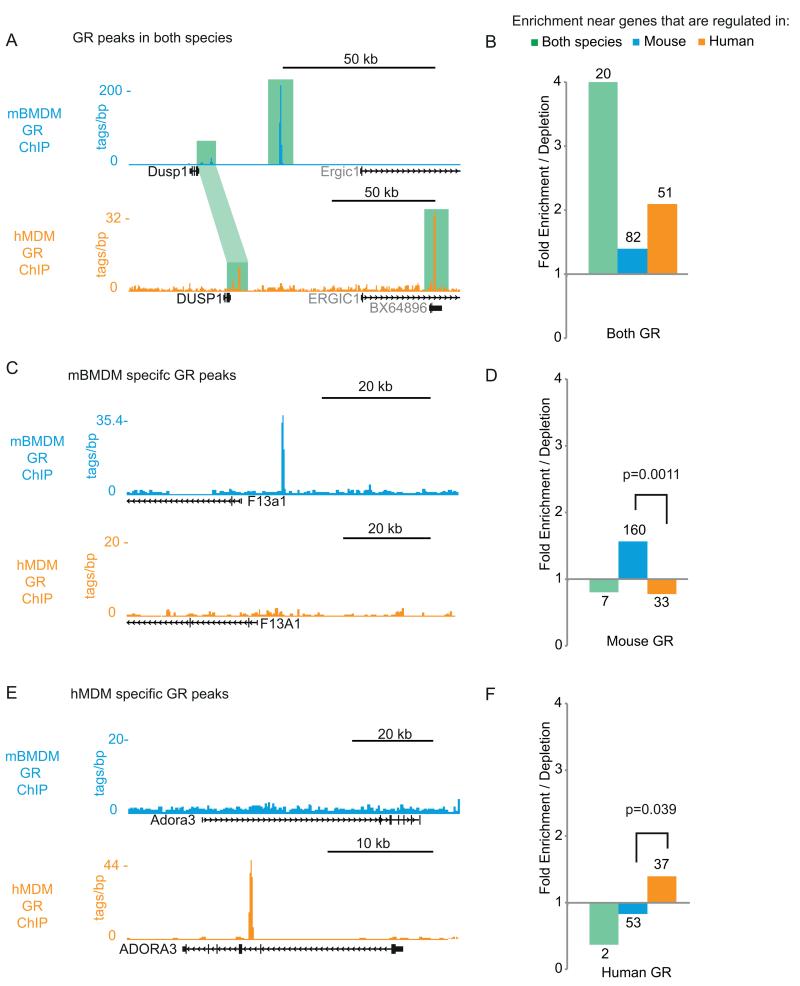
As stated above, amongst the set of GR bound sites from hMDM and mBMDM that can be assigned to regions of conserved DNA, only a minority (32) were clearly conserved in binding (Table S1F). Sixteen of the conserved GR sites are adjacent to genes that were induced in both species and that encode known regulators of the inflammatory response (e.g. DUSP1, FKBP5, MAP3K6, TSC22D3, FOS (Fig. 2A&C), KLF4). Even at these conserved loci GR binding differed: a previously described proximal peak at the DUSP1/Dusp1 locus was retained (54) but the strongest binding site was not shared (Fig. 4A). Overall, genes which were induced in both species were enriched for having GR bound within 1 Mb – but not necessarily at orthologous positions – in both species (4.0-fold enrichment, chi-squared p = 2.9×10−7 ; Fig. 4A).
Figure 4. GR binding sites are minimally conserved between mouse and man and this is linked to the divergent transcriptional response to GC.
(A) GR ChIP-seq data from mBMDM (orange) and hMDM (cyan) showing conserved GR binding (green highlight) at a locus (DUSP1/Dusp1) whose expression is rapidly induced by GC in both mouse and human macrophages. GR bound sites aligned between species are linked by light green highlight: the most prominent sites are bound in only one species. (B) Enrichment / depletion for mouse/human shared GR binding within 1Mb for GC-responsive genes that are; shared between mouse and human (green), mouse-specific (cyan), human-specific (orange). Numbers give raw counts for each category. (C) As in (A) but showing mouse-specific GR binding at F13a1/F13A1 which is induced in mBMDM but not hMDM. (D) As for B, but for mouse-specific GR binding sites. The chi squared p value for the difference between mouse and human specific sites is given. (E) As in (A) but showing human-specific GR binding at ADORA3/Adora3 which is induced in hMDM but not mBMDM. (F) As for (D) but for human-specific GR-binding sites.
Expanded windows for F13A1 and Adora3 are shown in Figures S1D.
The more common pattern was for GR binding at regulated loci to be divergent between the species; for example the GR peak upstream of F13a1, a component of the coagulation cascade, is mouse-specific (Fig. 4C, expanded view in Fig. S2D). Genes like F13a1 that responded to GC only in mouse were 2.0-fold enriched over human-specific genes for GR binding within 1Mb in the mouse (chi-squared p = 0.0011; Fig. 4D). Similarly, human specific GR binding was enriched adjacent to human-specific GC regulated genes. For example ADORA3, which has a known role in driving the human macrophage phenotype (55), has an intronic GR peak that is not present in mouse. As for the mouse, there was an overall 1.7-fold enrichment for a human-specific GR peak within 1Mb of genes that were specifically upregulated in human macrophages (chi-squared, p = 0.034; Fig. 4E and 4F, expanded view in Fig. S2D). These data highlight the strong correlation between divergent GR binding and divergent gene expression response to dexamethasone across species (Figures 1 and 2), and the shared enrichment for GR binding in the vicinity of genes that were induced in both species (Fig. 3). We conclude that the turnover of GR binding sites between human and mouse is the primary driver of the divergent transcriptional response to GC.
Most of the species-specific GR sites are in genomic regions that can be aligned between mouse and human (Fig. 2E), suggesting that the changes that cause motif loss occurred as a result of nucleotide substitutions and deletions, rather than insertion or deletion of sequence. Loss of GR binding was associated with loss of the GRE motif. Amongst aligning sequence, there was significant enrichment for the GRE motif in the subset that is bound in both species compared to those that are species-specific (Fig. 5A and 5B). This is also true for PU.1, although less strongly, reflecting the lower enrichment for this motif in the baseline dataset. Consistent with the loss of GR binding, there was depletion for the GRE motif in locations that were not bound by GR, even when the orthologous location in the opposite species was bound (Fig. S3A and S3B). There was some enrichment for PU.1 in human sequence orthologous to mouse specific sites, which suggests there may be some residual regulatory function in macrophages at these sites.
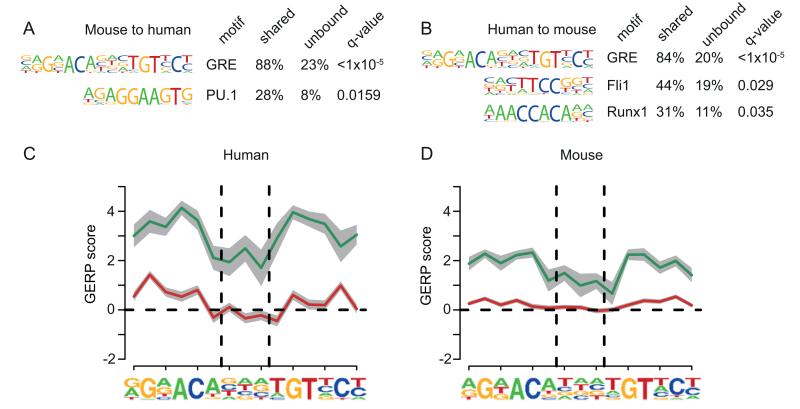
Figure 5. Conserved GR binding is linked to conservation of the GRE.
(A) Motif enrichment for the sites bound by GR in mBMDM that aligned and were also bound in hMDM, using as background the mouse sites that could be aligned to human but were not bound in hMDM (q values shown, Benjamini-Hochberg). (B) Analogous to (A) but for hMDM sites bound in mBMDM vs sites that could be aligned but were not bound in mBMDM. (C) Mean per base constraint scores calculated using GERP (72) across the GRE in shared (green) and species-specific (red) peaks found in hMDM, where the grey bars represents the standard error of the mean. Vertical dashed lines delineate the centre NNN for the GRE, as derived de novo from our hMDM data. (D) Analogous to (C) for GR bound peaks and GRE motif found in mBMDM.
The turnover of the GRE motif at species-specific GR peaks could be driven by a selective pressure on the immune response to modulate host-pathogen interactions (8, 23). Evolutionary constraint was detectable around the shared sites in each species (Fig. 5C and 5D), extending to approximately 100 bp beyond the motif (Fig. S3C and S3D). This implies that purifying selection has acted to preserve this motif between human and mouse. As predicted by the degeneracy of the GRE motif, evidence of selection was reduced in the centre of the motif. However, within the species-specific GR binding sites, there was no signature consistent with substantial selection across the motif (Fig. 5C and 5D). Indeed, there was some slight constraint at the non-degenerate sites in the motif. This suggests that turnover of the GRE motif at individual loci is driven by nucleotide substitutions at approximately the genome-wide mutation rate and not by positive selection driving the gain or loss of new binding sites (Fig. 4).
Discussion
This study shows that the divergence of the transcriptional response to GC in mouse and human macrophages is associated with evolutionary turnover of candidate enhancers. These enhancers contain canonical inverted repeat GR-response elements (GRE) and are bound by GR in glucocorticoid-treated macrophages. Although GC have commonly been studied as repressors of inflammatory gene expression, the data indicate that, in the context studied here, GC act on macrophages primarily as inducers of gene expression when measured at the level of stable mRNA. The induced targets are largely distinct between human and mouse (Fig. 1). This adds to the weight of evidence that caution must be exercised when translating mouse findings to man (8, 21, 22).
Repressed genes have been shown to lie further from GR bound sites than induced genes at candidate loci (56) and in previously published ChIP-seq data from A549 lung epithelial cancer cell line (36). Based upon genome-wide comparison, the genes repressed after GC-treatment in macrophages had no significant genomic association with direct GR-DNA binding in either species (Fig. 3), nor was there any support for the existence of negative GR binding elements (nGRE) (30, 31) in either mouse or human GR ChIP-seq datasets. Lower binding affinity and faster turnover time could compromise their detection under the conditions we employed. Alternatively, there may be context-dependent use of different types of regulatory element. For example, the nGRE may only be relevant in macrophages when GR acts to suppress inflammatory gene induction, (40) rather than in the basal CSF1-dependent state we have examined. The GC-responsive GR elements we identified were often associated with binding motifs for the macrophage master transcription factor PU.1. Therefore, as for other transcription factors downstream of extracellular signalling (51, 57), the GR binding landscape is shaped by enhancer elements already occupied by PU.1.
Evolutionary conservation has long been used to identify candidate functional regions of the genome (58). A very small subset of GC-induced genes and nearby GR-bound sites fit this pattern. These genes are enriched for a number of shared functional annotations, such as stimulus response and immune/wounding response (Fig. S1C), while the shared GR sites were found near to induced genes in both species (Fig. 3 and 4). The shared GR bound sites were subject to evolutionary constraint of the GR-binding motif (Fig. 5), suggesting they have been exposed to purifying selection to maintain their function in human and mouse.
The large majority of the GR-bound sites were not conserved between mouse and human. The level of divergence we found between GR binding sites and motifs between human and mouse macrophages is greater than that previously reported for a set of liver-specific transcription factors, where 20-30% of sites were conserved between primates and rodents (13). Although our dataset is limited to one factor and motif, it would appear that GC-responsive elements are even more divergent than the sites identified in liver, which would be consistent with previous observations for high divergence in long-range regulatory elements active in immune tissues (7).
The gain or loss of these GR bound sites is largely due to nucleotide substitutions, rather than insertion or deletion of sequence (Fig. 2) as previously reported for a set of liver enhancers (17). These substitutions cause gain or loss of the canonical GRE as well as partner motifs such as PU.1 (Fig. 5). These turnover events had a biological consequence: they were clearly associated with species differences in transcriptional regulation (Fig. 4C-F). The macrophage subtypes studied here are not directly comparable and there are known species differences in the response to CSF1, which produces a proatherogenic signal in human macrophages but not in mice (45). However, the association with sequence turnover strongly suggests that the divergent expression response to GC is unrelated to the differences in the regulatory network of different macrophage populations (59). Sequence variation also underlies the divergent response of macrophages to LPS, but in that case it was attributable to promoter sequence variation (8).
There was no clear evidence for selection at species-specific GR sites which show functional turnover between human and mouse (Fig. 5C and 5D). Some of these sites may be under lineage-specific selection, in the same way that LPS-responsive genes are shared by large animals (humans and pigs) but radically different in rodents (8, 60). Such variation would not be detectable with the available genomic sequence data. Alternatively, the associated species-specific inducible genes may not have any function in the feedback regulation of innate immunity. Some of the species-specific GR-bound peaks might have arisen by chance within regions that favor open chromatin in macrophages, and bind GR solely as a consequence of high transcription factor concentrations in the nucleus (61).
An interesting avenue for further research will be the physiological consequences of the functional genomic changes we have found. We cannot say from our study how the genomic changes are related to the substantial phenotypic differences between species, or how much effect change at any one locus might have. One way to begin to address this would be, at a locus that normally only responds to GC in humans, to insert the sequence of the human GRE at the equivalent position in the mouse genome and compare the response.
The general principles of the macrophage response to GC are strongly conserved between mouse and man, but the specific loci involved have diverged considerably. As previously reported (13, 62), we have shown large-scale turnover of candidate enhancers and have extended this work by demonstrating that these turnover events impact on inducible gene expression. Surprisingly, given the divergence at cis-regulatory sequence associated with immunity (7) and the potential drive of host-pathogen interactions on macrophages, we could not detect evidence of positive selection at species-specific GR bound sites. Conserved elements on the other hand showed clear evidence of selection to preserve their characteristic motifs. Despite much interest in the turnover of enhancers (63), the traditional approach of identifying regulatory elements through deep evolutionary conservation may still be most useful in identifying those sites associated with conserved gene regulation.
Supplementary Material
Acknowledgements
We thank R. Illingworth (MRC HGU) for assistance with optimising ChIP-seq assays and Edinburgh Genomics for running microarrays and sequencing samples.
Financial support1
Footnotes
AJ is supported by a Wellcome Trust Clinical PhD Fellowship (097481/Z/11/Z). WAB and RSY are supported by unit programme grants from the UK MRC. The Roslin Institute is supported by Institute Strategic Programme Grants from BBSRC.
References
- 1.Ponting CP. The functional repertoires of metazoan genomes. Nat. Rev. Genet. 2008;9:689–698. doi: 10.1038/nrg2413. [DOI] [PubMed] [Google Scholar]
- 2.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 3.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 4.McCarroll S. a, Murphy CT, Zou S, Pletcher SD, Chin C-S, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 5.Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 6.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, Knight JC. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams B. a., Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See L-H, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender M. a., Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu Y-C, Rasmussen MD, Bansal MS, Kellis M, Keller C. a., Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan C. a., Rosenbloom KR, de Sousa B. Lacerda, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent W. James, Santos M. Ramalho, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh T. a., Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Hansen R. Scott, Bruijn M. De, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang K-H, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel G. a., Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou X-Q, Pazin MJ, Feingold E. a., Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos J. a., Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer M. a., Ren B. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroder K, Irvine KM, Taylor MS, Bokil NJ, Le Cao K-A, Masterman K-A, Labzin LI, Semple C. a, Kapetanovic R, Fairbairn L, Akalin A, Faulkner GJ, Baillie JK, Gongora M, Daub CO, Kawaji H, McLachlan GJ, Goldman N, Grimmond SM, Carninci P, Suzuki H, Hayashizaki Y, Lenhard B, Hume D. a, Sweet MJ. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E944–53. doi: 10.1073/pnas.1110156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 12.Visel A, Bristow J, Pennacchio L. a. Enhancer identification through comparative genomics. Semin. Cell Dev. Biol. 2007;18:140–152. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballester B, Medina-Rivera A, Schmidt D, Gonzàlez-Porta M, Carlucci M, Chen X, Chessman K, Faure AJ, Funnell AP, Goncalves A, Kutter C, Lukk M, Menon S, McLaren WM, Stefflova K, Watt S, Weirauch MT, Crossley M, Marioni JC, Odom DT, Flicek P, Wilson MD. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife. 2014;3:1–29. doi: 10.7554/eLife.02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, Thurman RE, Johnson a. K., Vong S, Lee K, Bates D, Neri F, Diegel M, Giste E, Haugen E, Dunn D, Wilken MS, Josefowicz S, Samstein R, Chang K-H, Eichler EE, De Bruijn M, Reh T. a., Skoultchi a., Rudensky a., Orkin SH, Papayannopoulou T, Treuting PM, Selleri L, Kaul R, Groudine M, Bender M. a., Stamatoyannopoulos J. a. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science (80-. ) 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, Thurman RE, Vong S, Bates D, Neri F, Diegel M, Giste E, Dunn D, Vierstra J, Hansen RS, Johnson AK, Sabo PJ, Wilken MS, Reh T. a., Treuting PM, Kaul R, Groudine M, Bender M. a., Borenstein E, Stamatoyannopoulos J. a. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, Karagianni P, Brazma A, Adams DJ, Talianidis I, Marioni JC, Flicek P, Odom DT. Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell. 2013;154:530–40. doi: 10.1016/j.cell.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villar D, Berthelot C, Flicek P, Odom DT, Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M. Enhancer Evolution across 20 Mammalian Species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunarso G, Chia N-Y, Jeyakani J, Hwang C, Lu X, Chan Y-S, Ng H-H, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 19.Biggin MD. Animal Transcription Networks as Highly Connected, Quantitative Continua. Dev. Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC, Tonkin L. a., Biggin MD, Eisen MB. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related drosophila species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 22.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, López CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West M. a, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3507–12. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wells CA, Ravasi T, Faulkner GJ, Carninci P, Okazaki Y, Hayashizaki Y, Sweet M, Wainwright BJ, Hume D. a. Genetic control of the innate immune response. BMC Immunol. 2003;4:5. doi: 10.1186/1471-2172-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD. Initial sequencing and comparative analysis of the mouse genome [Review] Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 25.Nicolaides NC, Kyratzi E, Lamprokostopoulou A, Chrousos GP, Charmandari E. Stress, the stress system and the role of glucocorticoids. Neuroimmunomodulation. 2015;22:6–19. doi: 10.1159/000362736. [DOI] [PubMed] [Google Scholar]
- 26.Fardet L, Petersen I, Nazareth I. Prevalence of long-term oral glucocorticoid prescriptions in the UK over the past 20 years. Rheumatology (Oxford) 2011;50:1982–90. doi: 10.1093/rheumatology/ker017. [DOI] [PubMed] [Google Scholar]
- 27.Nixon M, Andrew R, Chapman KE. It takes two to tango: dimerisation of glucocorticoid receptor and its anti-inflammatory functions. Steroids. 2013;78:59–68. doi: 10.1016/j.steroids.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Ratman D, Vanden Berghe W, Dejager L, Libert C, Tavernier J, Beck IM, De Bosscher K. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol. Cell. Endocrinol. 2013;380:41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16701–6. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat. Struct. Mol. Biol. 2013;20:53–58. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrchen J, Steinmüller L, Barczyk K, Tenbrock K, Nacken W, Eisenacher M, Nordhues U, Sorg C, Sunderkötter C, Roth J. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood. 2007;109:1265–74. doi: 10.1182/blood-2006-02-001115. [DOI] [PubMed] [Google Scholar]
- 33.Varga G, Ehrchen J, Tsianakas A, Tenbrock K, Rattenholl A, Seeliger S, Mack M, Roth J, Sunderkoetter C. Glucocorticoids induce an activated, anti-inflammatory monocyte subset in mice that resembles myeloid-derived suppressor cells. J. Leukoc. Biol. 2008;84:644–50. doi: 10.1189/jlb.1107768. [DOI] [PubMed] [Google Scholar]
- 34.Galon J, Franchimont D, Hiroi N, Frey G, Boettner A, Ehrhart-Bornstein M, O’Shea JJ, Chrousos GP, Bornstein SR. Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J. 2002;16:61–71. doi: 10.1096/fj.01-0245com. [DOI] [PubMed] [Google Scholar]
- 35.Van de Garde MDB, Martinez FO, Melgert BN, Hylkema MN, Jonkers RE, Hamann J. Chronic exposure to glucocorticoids shapes gene expression and modulates innate and adaptive activation pathways in macrophages with distinct changes in leukocyte attraction. J. Immunol. 2014;192:1196–208. doi: 10.4049/jimmunol.1302138. [DOI] [PubMed] [Google Scholar]
- 36.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.John S, Sabo PJ, Johnson TA, Sung M-H, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos J. a, Hager GL. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008;29:611–24. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 38.John S, Sabo PJ, Thurman RE, Sung M-H, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trotter KW, Fan H-Y, Ivey ML, Kingston RE, Archer TK. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell. Biol. 2008;28:1413–26. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, Schwalie P, Hübner N, Evans RM. Insights into Negative Regulation by the Glucocorticoid Receptor from Genome-wide Profiling of Inflammatory Cistromes. Mol. Cell. 2013;49:158–171. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chinenov Y, Coppo M, Gupte R, Sacta M. a, Rogatsky I. Glucocorticoid receptor coordinates transcription factor-dominated regulatory network in macrophages. BMC Genomics. 2014;15:656. doi: 10.1186/1471-2164-15-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–88. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hume A, Stephens W, Warren HS, Curtin J. Preparation and Characterization of Human Bone Marrow-Derived Macrophages. J. Leukoc. Biol. 1985;38:541–552. doi: 10.1002/jlb.38.4.541. [DOI] [PubMed] [Google Scholar]
- 44.Ingersoll M. a, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, Lang R, Haniffa M, Collin M, Tacke F, Habenicht AJR, Ziegler-Heitbrock L, Randolph GJ. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood. 2010;115:e10–9. doi: 10.1182/blood-2009-07-235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irvine KM, Andrews MR, Fernandez-Rojo MA, Schroder K, Burns CJ, Su S, Wilks AF, Parton RG, Hume D. a, Sweet MJ. Colony-stimulating factor-1 (CSF-1) delivers a proatherogenic signal to human macrophages. J. Leukoc. Biol. 2009;85:278–88. doi: 10.1189/jlb.0808497. [DOI] [PubMed] [Google Scholar]
- 46.Hume D, Gordon S. The correlation between plasminogen activator activity and thymidine incorporation in mouse bone marrow-derived macrophages. Opposing actions of colony-stimulating factor, phorbol myristate acetate, dexamethasone and prostaglandin E. Exp. Cell Res. 1984;150:347–355. doi: 10.1016/0014-4827(84)90578-0. [DOI] [PubMed] [Google Scholar]
- 47.Stacey KJ, Fowles LF, Colman MS, Ostrowski MC, Hume DA. Regulation of urokinase-type plasminogen activator gene transcription by macrophage colony-stimulating factor. Mol. Cell. Biol. 1995;15:3430–3441. doi: 10.1128/mcb.15.6.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio T. a. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forrest ARR, Kawaji H, Rehli M, Baillie JK, de Hoon MJL, Lassmann T, Itoh M, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume D. a, Carninci P, Hayashizaki Y. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei C-L, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–28. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laurie S, Toll-Riera M, Radó-Trilla N, Albà MM. Sequence shortening in the rodent ancestor. Genome Res. 2012;22:478–85. doi: 10.1101/gr.121897.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 54.Tchen CR, Martins JRS, Paktiawal N, Perelli R, Saklatvala J, Clark AR. Glucocorticoid regulation of mouse and human dual specificity phosphatase 1 (DUSP1) genes: unusual cis-acting elements and unexpected evolutionary divergence. J. Biol. Chem. 2010;285:2642–52. doi: 10.1074/jbc.M109.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barczyk K, Ehrchen J, Tenbrock K, Ahlmann M, Kneidl J, Viemann D, Roth J. Glucocorticoids promote survival of anti-inflammatory macrophages via stimulation of adenosine receptor A3. Blood. 2010;116:446–55. doi: 10.1182/blood-2009-10-247106. [DOI] [PubMed] [Google Scholar]
- 56.So AY-L, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc. Natl. Acad. Sci. 2008;105:5745–5749. doi: 10.1073/pnas.0801551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent Enhancers Activated by Stimulation in Differentiated Cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 58.Haeussler M, Joly JS. When needles look like hay: How to find tissue-specific enhancers in model organism genomes. Dev. Biol. 2011;350:239–254. doi: 10.1016/j.ydbio.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 59.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapetanovic R, Fairbairn L, Beraldi D, Sester DP, Archibald AL, Tuggle CK, Hume D. a. Pig bone marrow-derived macrophages resemble human macrophages in their response to bacterial lipopolysaccharide. J. Immunol. 2012;188:3382–94. doi: 10.4049/jimmunol.1102649. [DOI] [PubMed] [Google Scholar]
- 61.MacArthur S, Li X-Y, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keränen SVE, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villar D, Flicek P, Odom DT. Evolution of transcription factor binding in metazoans - mechanisms and functional implications. Nat. Rev. Genet. 2014;15:221–33. doi: 10.1038/nrg3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using {R} and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 65.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy---analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 66.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat. Protoc. 2009;4:1535–50. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 68.Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford E. a. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–52. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.