Abstract
Integration of structural genomic data from a largely assembled rice genome sequence, with phylogenetic analysis of sequence samples for many other taxa, suggests that a polyploidization event occurred ≈70 million years ago, before the divergence of the major cereals from one another but after the divergence of the Poales from the Liliales and Zingiberales. Ancient polyploidization and subsequent “diploidization” (loss) of many duplicated gene copies has thus shaped the genomes of all Poaceae cereal, forage, and biomass crops. The Poaceae appear to have evolved as separate lineages for ≈50 million years, or two-thirds of the time since the duplication event. Chromosomes that are predicted to be homoeologs resulting from this ancient duplication event account for a disproportionate share of incongruent loci found by comparison of the rice sequence to a detailed sorghum sequence-tagged site-based genetic map. Differential gene loss during diploidization may have contributed many of these incongruities. Such predicted homoeologs also account for a disproportionate share of duplicated sorghum loci, further supporting the hypothesis that the polyploidization event was common to sorghum and rice. Comparative gene orders along paleo-homoeologous chromosomal segments provide a means to make phylogenetic inferences about chromosome structural rearrangements that differentiate among the grasses. Superimposition of the timing of major duplication events on taxonomic relationships leads to improved understanding of comparative gene orders, enhancing the value of data from botanical models for crop improvement and for further exploration of genomic biodiversity. Additional ancient duplication events probably remain to be discovered in other angiosperm lineages.
Keywords: colinearity, chromosome structural rearrangement, gene order, genome duplication, rice
The nearly completed sequences of Arabidopsis and Oryza shed light on the history of angiosperm genome evolution, and provide a foundation for advancing knowledge about many other flowering plants by using comparative approaches. Comparative genomics is especially important for the study of the large and highly repetitive genomes of many major crops. Detailed genetic maps, available for representatives of most major angiosperm groups, are of singular importance to comparative biology. Some genetic maps are near “saturation,” in that most available recombination events in the underlying populations have been detected (for example, see ref. 1). By integrating genetic maps with hybridization-based physical maps, resolution can be improved from centiMorgan scale to kilobase scale (for example, see ref. 2).
Comparisons of gene maps and/or other data, such as finished sequences of individual bacterial artificial chromosomes to one another and to the sequences of botanical models, reveal nonrandom patterns of similarity in gene order, but also much incongruence. Some incongruities may be caused by polyploidization followed by differential gene loss in different taxa. Widespread duplication is evident even in the small genome of Arabidopsis (3-6). The demonstration that one duplication event predates the divergence of Arabidopsis from most dicots, and an earlier event predates its divergence from the monocots, suggests that virtually all angiosperms are ancient polyploids, and that maximally informative genomic comparisons require mitigation of the effects of polyploidization/diploidization events that postdate divergence of relevant taxa (7). Using unfinished data emerging from an international effort to sequence the first cereal genome, two groups (8, 9) have reported duplication of rice chromatin, corroborating earlier suggestions (10-13) but reaching somewhat different conclusions about the extent of genome duplication.
Here, we refine a structural analysis of genomic duplication in rice, investigate the timing of the duplication event, and explore its impact on cereal comparative genomics. We show that the cereals evolved as independent lineages for about two-thirds of the period after the duplication event, and that differential gene loss after cereal divergence may explain many deviations from colinearity. The prevalence of ancient polyploidy suggests the need for a three-pronged approach to angiosperm comparative biology, integrating phylogenetic information about the relatedness among taxa with structural information about extant gene arrangements and “phylogenomic” approaches to determine the timing and mitigate the consequences of ancient duplications.
Materials and Methods
Duplication Analysis. A total of 56,055 Oryza gene sequences (www.tigr.org) encoded by their chromosomal order and transcriptional orientation were compared to each other by using blastp (14). The top five non-self protein matches that met a threshold of 1e-06 were considered in duplication analysis. Circumscription of individual duplicated segments was as described (7). A total of 10,882 sequences were removed because of blastn matches of <1e-10 with members of The Institute for Genomic Research (TIGR) rice repeat database.
Gene Tree Analysis. Each duplicated syntenic gene pair was compared to each taxon-specific sequence (using nucleotide databases created by batch NCBI download of taxon IDs indicated in Table 1 for Pinus, Allium, Sorghum, Zea, Hordeum, and Oryza minuta. For Arabidopsis, we used a recent (April 17th, 2003) set of predicted coding sequences from The Arabidopsis Information Resource, based on CDS sequences from the tigr 4.0 release (ftp://tairpub:tairpub@ftp.Arabidopsis.org/home/tair/Sequences/blast_datasets/OLD/ATH1_cds_20030417.Z). Musa ESTs were provided by ProMusa (www.promusa.org). Gene tree analysis is based on methods described in ref. 7, revised to incorporate recent improvements that are described elsewhere (15). Briefly, pairs of rice genes with similar sequences and in corresponding locations within duplicated blocks are compared to the sequences of best-matching genes from other organisms (identified as described in ref. 15). Inferences about the antiquity of genomic duplication are based on differences in the frequencies of “internal” trees, in which the foreign gene is more similar to one rice gene than the two rice genes are to each other, suggesting that taxon divergence is more recent than gene duplication. The fractions of “internal trees” associated with each duplication block (Table 1) were compared by using one-way ANOVA for correlated samples and Tukey's honestly significant difference analysis for post-ANOVA comparisons between organisms. A total of 100 bootstrapped Gaussian random samples were used for the analysis, calculated based on estimates of the population mean and variance from the rice duplication block data. Each duplicated segment pair was considered a treatment, and the indicated taxa were conditions, accounting for correlations that may result from comparing identical genes in different taxa. This is conservative, because in many cases different regions of an Oryza gene matched ESTs from different taxa, reducing the correlation problem. Data interpretation relies largely upon differences among taxa in the frequencies of internal trees. Incompleteness of EST data has the consequence that inferences about gene orthology will be imperfect, and paralogous associations usually form external trees, as discussed (15).
Table 1. Phylogenetic dating of a genomic duplication in the rice lineage.
| Pinus | Arabidopsis | Allium | Musa | Sorghum | Zea | Hordeum | O. minuta | |
|---|---|---|---|---|---|---|---|---|
| TaxID | 3,318 | Predicted | 4,678 | ProMusa | 4,557 | 4,577 | 4,512 | 63,629 |
| No. of reads | 114,628 | 28,581 | 20,272 | 33,922 | 220,670 | 257,255 | 376,301 | 5,286 |
| Block (chromosomes) | ||||||||
| 1 (1-5) | 0.02 (183) | 0.067 (240) | 0.045 (134) | 0.127 (110) | 0.303 (122) | 0.333 (129) | 0.358 (159) | 0.222 (63) |
| 2 (2-4) | 0.024 (84) | 0.031 (130) | 0.065 (77) | 0.086 (58) | 0.417 (72) | 0.391 (64) | 0.457 (81) | 0.375 (40) |
| 3 (2-6) | 0.050 (80) | 0.026 (117) | 0.072 (69) | 0.113 (62) | 0.400 (60) | 0.261 (69) | 0.361 (72) | 0.486 (35) |
| 4 (3-7) | 0.013 (80) | 0.086 (105) | 0.066 (61) | 0.100 (50) | 0.429 (56) | 0.357 (56) | 0.530 (66) | 0.375 (32) |
| 5 (3-10) | 0.041 (49) | 0.103 (68) | 0.056 (36) | 0.208 (24) | 0.370 (27) | 0.294 (34) | 0.359 (39) | 0.643 (14) |
| 6 (3-12) | 0.000 (10) | 0.067 (15) | 0.091 (11) | 0.125 (8) | 0.375 (8) | 0.111 (9) | 0.000 (8) | 0.500 (2) |
| 7 (4-8) | 0.111 (9) | 0.062 (16) | 0.100 (10) | 0.286 (7) | 0.375 (8) | 0.167 (6) | 0.500 (10) | 0.500 (4) |
| 8 (8-9) | 0.021 (48) | 0.101 (89) | 0.024 (42) | 0.138 (29) | 0.480 (50) | 0.425 (40) | 0.364 (55) | 0.231 (13) |
| 9 (11-12) | 0.000 (43) | 0.026 (77) | 0.024 (42) | 0.098 (41) | 0.113 (53) | 0.098 (41) | 0.118 (51) | 0.111 (18) |
| Total | 0.026 (586) | 0.061 (857) | 0.054 (482) | 0.121 (389) | 0.353 (456) | 0.310 (448) | 0.370 (541) | 0.339 (221) |
| Significance | A | A | A | A | B | B | B | B |
Primary data represent (decimal) fraction of gene trees that are internal, i.e., for which the gene from the taxon indicated by the column heading is more closely related to one rice homolog than to the other rice homolog. Values in parentheses indicate the number of genes that were informative (i.e., for which homologs could be identified that met the criteria required for building trees, and thus could be used in analyses) for each duplicated block × taxon combination.
Rice-Sorghum Synteny. The published order of 2,509 sequence-tagged sites along the sorghum chromosomes (1) was compared to the Oryza sequence assembly by using blastn, with a match of e < 10-6 considered significant.
Third-Nucleotide Substitution (Ks) Values. Ks values were calculated by using the yn00 method of the paml package (16) according to the method of Yang et al. (17).
Results
Fine-Scale Analysis of Ancient Duplication in the Rice Genome. We show (Fig. 1) the structure of an ancient duplication event based on analysis of 45,174 genes in a rice pseudomolecule, improving significantly on a prior study using a coarse assembly at whole bacterial artificial chromosome-level resolution from a less-complete physical map (9). Nine nonoverlapping “duplicated blocks” account for 61.9% of the rice transcriptome (excluding repetitive genes), with individual blocks ranging from 1.8 to 13.8% of the transcriptome. Individual blocks have retained syntenic duplicate copies for an average of 21.1% of genes, ranging from 16.2 to 24.6%, and excluding duplication 9 (chromosomes 11-12, 5.8% of transcriptome) that shows an unusually high 33.2% of duplicated genes but may be affected over part of its length by misassigned bacterial artificial chromosome (J.E.B. and A.H.P., unpublished data). Many nonduplicated regions correspond to the rice centromeres, as has been found for Arabidopsis (3-6). Gene lists and associated statistics comprising each duplicated segment are provided (Tables 3-5, which are published as supporting information on the PNAS web site).
Fig. 1.
Arrangement of duplicated protein-encoding genes in Oryza. Both X and Y axes represent 45,174 genes in their chromosomal order (based on a 7/23/2003 assembly from www.tigr.org). The single best-matching gene pairs (identified as described in Materials and Methods) are plotted and color-coded to indicate same (red) or opposite (green) transcriptional orientations. The lower left area shows only the subset of best-matching gene pairs for which synonymous substitution rates range from 0.75 to 0.95. Duplicated regions are highlighted in blue along the y axis. Red dots indicate locations of centromeres.
The distribution of DNA sequence divergence levels among syntenic duplicated rice genes based on synonymous substitution (Ks) rates has a modal value of 0.85, with a long tail extending beyond 3. The mode is a more representative statistic than the average Ks (1.05), because the latter is disproportionately affected by extreme values in the long tail of the distribution. Pairs of loci with Ks values within 0.1 units of the peak (0.75-0.95: Fig. 1 Lower) show much less “noise” obscuring chromosomal-level duplications, suggesting that such noise may be caused by smaller-scale duplications (such as individual genes) that are more recent or more ancient than genome-wide duplication.
Phylogenetic Dating of the Rice Genomic Duplication. To relate the duplication event to the divergence time of rice from other angiosperms, we analyzed gene trees including one pair of syntenic duplicated rice genes, the best-matching sequence from a test organism, and Physcomitrella as an outgroup, as described (15). Sorghum (representing the Panicoideae) and Hordeum (Pooideae) ESTs each showed similarly high frequencies (31-37%) of internal trees with the rice gene pairs, i.e., the test sequence is more similar to one rice duplicate than is the other rice duplicate. By contrast, ESTs for divergent monocot lineages (Musa, banana, and Allium, onion), a dicot (Arabidopsis), and a gymnosperm (Pinus) each show similarly low frequencies (2.6-12.1%) of internal trees with rice duplicates, suggesting that rice duplication is more recent than its divergence from its closest relative among these lineages (banana), but more ancient than its divergence from Sorghum and Hordeum. Sufficient EST resources to study the other major Poaceae clades, Chloridoideae and Arundinoideae, are presently lacking.
Duration of Independent Evolution of Cereal Lineages. To estimate the duration of independent evolution of the cereal lineages, we calculated Ks values between the duplicated rice genes and best-matching ESTs from other taxa that were used for gene tree analysis. Like rice, Sorghum and Hordeum each showed Ks distributions that were skewed right, but with sharp peaks near 0.51 and 0.57, respectively. Based on an average synonymous substitution rate of 6.1-6.5 per 109 years (18, 19), these values suggest divergence times of ≈42 and ≈47 million years ago (MYA), respectively, close to the 50 MYA estimated elsewhere to approximate the time of divergence of the cereals (20). The Ks peak for the rice duplicates corresponds to an age of ≈70 million years, suggesting that the cereals evolved as independent lineages for roughly two-thirds of the time since the duplication event.
Genomic Distributions of Comparative Loci That Are Incongruent with Patterns of Rice-Sorghum Colinearity. A duplication/diploidization event that predates divergence of taxa from a common ancestor may account for some incongruence in “comparative maps.” Specifically, if gene loss were still continuing at an appreciable rate after taxon divergence occurred, then differential gene loss in independent lineages would cause incongruities in their comparative maps. To test this possibility, we examined a sorghum-rice comparative map (Fig. 2) developed by blasting sequences from 2,509 genetically mapped sorghum loci (1) against the rice genome assembly. The positions of 1,626 corresponding loci could be plotted based on the rice physical location and sorghum genetic location. This revealed much colinearity, with eight sorghum linkage groups (A, D, E, F, G, H, I, and J) corresponding to single rice chromosomes (1, 4, 12, 2, 5, 11, 6, and 8), and two sorghum linkage groups (B and C) differing from rice by translocations (between chromosomes 7/9 and 3/10, respectively). However, many loci deviate from these syntenic/colinear relationships (Table 2). Across the entire genome, incongruent loci occurred on homoeologous rice chromosomes (identified based on Fig. 1) at more than twice the random frequency, a highly significant enrichment (based on a contingency test, χ2 = 101.54, 1 df; P << 0.001). This suggests that many cases of incongruent loci may be explained by loss of the true ortholog in either rice or sorghum, leaving the homoeolog as the best match.
Fig. 2.
Patterns of colinearity between sorghum and rice. The x axis represents 45,174 rice genes in their chromosomal order, and the y axis represents 2,509 loci in their recombinational arrangement along a sorghum STS-based genetic map (1). Rice chromosomes (1-12) and sorghum linkage groups (A-J) are arranged consecutively, and labeled at top and left, respectively. Red circles represent inferred locations of sorghum centromeres (1). Each dot represents a best match (≤1e-06) between a sorghum STS and a rice gene. The total number of probes mapping to each intersection of sorghum and rice chromosomes is shown at upper left in each cell. Horizontal subdivisions of sorghum linkage groups B, C, and F delineate locations at which sorghum-rice syntenic relationships change, consistent with subdivisions of the counts of incongruent loci shown in Table 2.
Table 2. Genomic distribution of comparative loci that are incongruent with patterns of colinearity between sorghum and rice.
| Sorghum linkage gp. | Rice chromosome | Rice homoeolog | No. of incongruent loci on rice homoeolog | No. of incongruent loci on other rice chromosomes | Inferred sorghum homoeolog* | No. of incongruent loci on sorghum homoeolog | No. of incongruent loci on other sorghum groups |
|---|---|---|---|---|---|---|---|
| A | 1 | 5 | 10 | 46 | G | 24 | 99 |
| B | 7 | 3 | 18 | 47 | C | 22 | 83 |
| B | 9 | 8 | 5 | 19 | J | 7 | 33 |
| C | 3 | 7 | 12 | 70 | B | 18 | 135 |
| C | 10 | 3 | 13 | 43 | ? | - | - |
| D | 4 | 2 | 9 | 40 | F | 11 | 75 |
| E | 12 | 11 | 16 | 40 | H | 25 | 70 |
| F | 2 | 6 | 10 | 27 | I | 19 | 57 |
| F | 2 | 4 | 2 | 12 | D | 6 | 34 |
| G | 5 | 1 | 17 | 34 | A | 24 | 73 |
| H | 11 | 12 | 8 | 44 | E | 25 | 105 |
| I | 6 | 2 | 15 | 42 | F | 21 | 97 |
| J | 8 | 9 | 5 | 33 | B | 11 | 66 |
| Total | 140 | 497 | 213 | 927 |
If duplication occurred in a common ancestor of rice and sorghum, then a disproportionate share of duplicated loci in sorghum should fall on chromosome pairs that are predicted to be homoeologs based on their correspondence to duplicated rice chromosomes. We have previously shown that the genomic distribution of duplicated loci in sorghum is not random (1, 21). Based on the observed duplication patterns in rice (Fig. 1) and syntenic/colinear relationships between rice and sorghum (Fig. 2), we identified pairs of sorghum chromosomes (or segments) that correspond to duplicated rice chromosomes/segments (Table 2 and Fig. 4, which is published as supporting information on the PNAS web site). These pairs of sorghum chromosomes/segments account for 22 (50%) of 44 regions that showed enrichment for duplicated DNA markers (highlighted in Fig. 4). Closely following the findings for rice-sorghum incongruence, duplicated loci occurred on putatively homoeologous regions of sorghum at about twice the frequency expected by chance (Table 2), a highly significant (χ2 = 95.5, 1 df, P < 0.001) enrichment. This supports the hypothesis that the most recent genome-wide duplication of rice and sorghum occurred in a common ancestor.
Estimates of the extent of incongruent markers between sorghum and rice and nonrandom distribution of duplicated markers in sorghum are both conservative in that many areas of the rice genome remain incompletely sequenced and/or annotated. Additional data may extend patterns of duplication beyond the 62% of the genome for which we can presently infer them.
Discussion
Divergence of the Cereals Closely Followed Genome-Wide Duplication in a Common Ancestor. Two lines of evidence show that a large-scale, perhaps genome-wide, duplication occurred ≈20 million years before the divergence of Oryza, Sorghum, and Hordeum from common ancestors that existed ≈41-47 MYA. In other words, the cereals have evolved independently for about two-thirds of the period since their most recent genome-wide duplication.
Our findings suggest that the rice duplication event involved most, if not all, of the genome, somewhat different from a prior interpretation that suggests only partial duplication (8) based on an earlier assembly. We find nine nonoverlapping “duplicated blocks” to account for 62% of the rice transcriptome, including a chromosome 1-5 duplication suggested by early studies (10) but not found in the prior whole-genome interpretation (8). Many nonduplicated regions are near the rice centromeres, as has been found for Arabidopsis (3-6). Similar percentages of duplicated genes (Table 3), frequencies of internal gene trees (Table 1), and Ks values (not shown) for most of the duplicated segments suggest that a single duplication event may account for most of them, except the chromosome 11-12 duplication, which appears to be more recent. Although the lengths of the duplicated segments vary widely, a completed rice sequence will be necessary for this criterion to be reliable enough to consider in assessing their age.
Diploidization appears somewhat more extensive in rice than Arabidopsis (7), with only 21.4% of rice genes retaining a syntenic homoeolog, corresponding to levels that are between those of the α (29.7%) and much older β (16.3%) duplication event in Arabidopsis. Improved annotation of both Arabidopsis and Oryza may necessitate revisions of these estimates. Nonetheless, in each case retention of duplicated gene copies is far greater than would be predicted based on theoretical considerations (18), suggesting that gene loss is not random.
Differential Gene Loss Contributes to Apparent Incongruities in Comparative Grass Genomics. Ancient duplication and subsequent diploidization has shaped the genomes of all Poaceae crops, including the major cereals, many forages, and leading biomass crops such as sugarcane. This is exemplified by our findings that a preponderance of loci that are incongruent with the most parsimonious syntenic/colinear relationships among rice and sorghum (for example), are located on the homoeologous chromosomal regions that resulted from ancient duplication. The finding that duplication patterns within sorghum parallel those within rice suggests that the same duplication event affected both genomes. Although loss of some DNA sequences after polyploid formation is rapid (22), the progressively lower fraction of genes remaining duplicated since progressively more ancient events in Arabidopsis, dropping from 29.7% for the α event of 20-80 MYA to 13.1% for the γ event of 300 MYA or more (7), suggest that diploidization is an ongoing process.
The extent to which differential gene loss accounts for incongruity in comparative maps should be related to the duration of the period between the duplication event and the divergence of the respective lineages. Rapid diploidization events that occurred shortly after polyploidization would be expected to affect all Poaceae (thus representing common features of their respective genomes), whereas gene loss after taxon divergence would contribute to incongruities among Poaceae comparative maps. The finding that the cereals have evolved independently for two-thirds of the postduplication period suggests that there has been appreciable opportunity for differential gene loss to occur.
In other taxa, a good case can be made for the possibility that fixation of differential gene losses (or “nonfunctionalization” by mutation) in small populations may contribute to reproductive isolation (23). However, the >20-million-year lag between duplication and divergence raises questions about the contribution of this mechanism to cereal divergence.
Implications for the Relationship of Sorghum and Maize. The finding that the most recent genome-wide duplication event in sorghum occurred ≈70 MYA raises perplexing questions about cereal karyotypic evolution. It is well established that maize, which has 10 chromosomes in its gametes (n = 10), underwent genome-wide duplication ≈11 MYA, most probably involving fusion of nuclei from ancestors that had five chromosomes in their gametes (24). Traditionally, the fact that there exist Sorghum species with n = 5 has been viewed as generally supportive of this model. However, if divergence of maize and sorghum (variously estimated at 11-28 MYA) was from a common n = 5 ancestor, then perplexing questions arise about how and when sorghum reached n = 10, which it had presumably done by 5 MYA when the sorghum and sugarcane (n = 8, 10) lineages diverged (25). The fact that modern sorghum chromosomes show only vestiges of duplication, apparently dating to the 70-MYA event shared with rice, suggests that evolution of the n = 10 nucleus of sorghum by polyploidization would need to have involved hybridization between n = 5 genotypes that diverged from a common ancestor 70 MYA. Although we cannot rule this out, it seems farfetched. Key to resolving this question is future, detailed, structural analysis of the genomes of n = 5 members of the Sorghum genus.
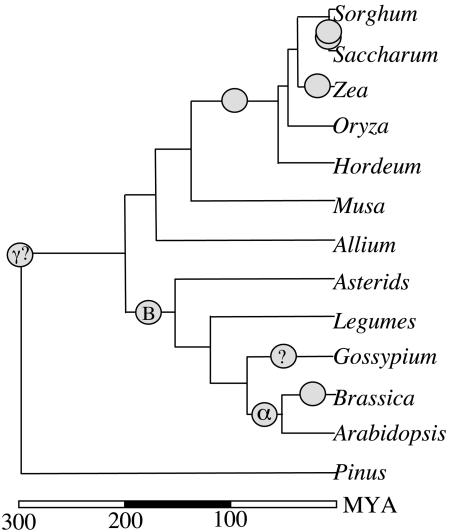
Implications for Use of Botanical Models in Angiosperm Comparative Genomics. Understanding the relative order of genome-wide duplication and taxonomic divergence is central to comparative genomic biology (26). The application of completed sequences and associated gene functional annotations from botanical models, to the improvement of the world's leading crops and more generally to dissecting the molecular basis of plant biodiversity, will benefit greatly from superimposition of the timing of major duplication events on taxonomic relationships. In addition to the ancient monocot event described herein and previously reported dicot events (7), additional duplication events in the past few million years have long been known to influence the comparative genomics of individual lineages such as maize (27), sugarcane (25), and Brassica (28). By integrating phylogenetic information about the relatedness among taxa with structural information about extant gene arrangements and “phylogenomic” approaches to describe the timing and mitigate the consequences of ancient duplications, a more detailed picture of angiosperm genome evolution is beginning to unfold (Fig. 3).
Fig. 3.
An early phylogenetic tree of genomic duplications for the angiosperms. By integrating data described herein and elsewhere (7), ancient duplications in the monocots and dicots, respectively, are superimposed on a partial angiosperm phylogenetic tree that also represents well established recent duplications in several lineages. Open circles indicate possible chromosomal duplication or polyploid formation events. Question marks indicate (i) the need for additional data to support tentative indications of a polyploidization event in the Gossypium lineage (J. Rong, J.E.B., and A.H.P., unpublished data) and (ii) uncertainty about the dating of the γ event (7). Gene tree analyses (see text and Table 1) support largely “one-to-one” correspondence of the rice chromosomes to those of other diploid cereals, but suggest the need for “one-to-two (or more)” comparisons to more distant lineages. More recent duplications and/or polyploid formation within many lineages further complicate comparative genomics. Branch lengths along the y axis approximate divergence times cited (7) or Ks data reported herein, converted to MY (millions of years) by using the average of current molecular clocks (18, 19).
Comparative gene orders along homoeologous chromosomal segments provide a means to make phylogenetic inferences about chromosome structural rearrangements that differentiate among the grasses. For example, an apparent inversion distinguishes rice chromosome 2 from sorghum LG F (Fig. 2), but additional information is needed to infer which arrangement is ancestral. The finding (Fig. 1) that rice chromosome 2 shows no difference from its ancient homoeolog (chromosome 4) in this region suggests that the inversion occurred in the Sorghum lineage after its divergence from a common ancestor shared with rice. This approach may partly mitigate the present lack of information about gene order in monocot groups that would be suitable as outgroups for the Poaceae.
It seems likely that completed sequences for representatives of other branches of the angiosperms may reveal additional, ancient, events that occurred in these lineages since their divergence from Arabidopsis and the monocots. Particularly important gaps in information exist for the asterids and rosids that are distant from Arabidopsis, such as the legumes. Within the monocots, additional sequences are clearly needed to characterize, for example, the tremendous diversity that is reflected by Ks statistics of 1.73 for Musa and 1.94 for Allium (calculated as described above) in comparison to rice genes, suggesting divergence times of 142 and 159 MYA, respectively. Characterization of basal angiosperms may help to shed light on the provenance of events near the monocot-dicot divergence. The sequences of additional Poales and Brassicales taxa will provide for phylogenetic “triangulation” of events that contribute to the diversity within each of these groups.
Supplementary Material
Acknowledgments
We thank our labmates for technical support. This work was supported by the U.S. Department of Agriculture National Research Initiative, U.S. National Science Foundation Plant Genome Research Program, Howard Hughes Medical Institute Graduate Fellowship Program, International Consortium for Sugarcane Biotechnology, and the Georgia Agricultural Experiment Station.
This report was presented at the international Congress, “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution,” held May 27-31, 2003, at the University of Bologna, Bologna, Italy. The scientific organizers were Roberto Tuberosa, University of Bologna, Bologna, Italy; Ronald L. Phillips, University of Minnesota, St. Paul, MN; and Mike Gale, John Innes Center, Norwich, United Kingdom. The Congress web site (www.doublehelix.too.it) reports the list of sponsors and the abstracts.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: MYA, million years ago.
References
- 1.Bowers, J. E., Abbey, C., Anderson, S., Chang, C., Draye, X., Hoppe, A. H., Jessup, R., Lemke, C., Lennington, J., Li, Z., et al. (2003) Genetics 165, 367-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Draye, X., Lin Y.-R., Qian, X. Y., Bowers, J. E., Burow, G. B., Morrell, P. L., Peterson, D. G., Presting, G. G., Ren, S. X., Wing, R. A. & Paterson, A. H. (2001) Plant Physiol. 125, 1325-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterson, A. H., Bowers, J. E., Burow, M. D., Draye, X., Elsik, C. G., Jiang, C. X., Katsar, C. S., Lan, T. H., Lin, Y. R., Ming, R. G. & Wright, R. J. (2000) Plant Cell 12, 1523-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc, G., Barakat, A., Guyot, R., Cooke, R. & Delseny, I. (2000) Plant Cell 12, 1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vision, T., Brown, D. & Tanksley, S. (2000) Science 290, 2114-2117. [DOI] [PubMed] [Google Scholar]
- 6.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 7.Bowers, J. E., Chapman, B. A., Rong, J. K. & Paterson, A. H. (2003) Nature 422, 433-438. [DOI] [PubMed] [Google Scholar]
- 8.Vandepoele, K., Simillion, C. & Van de Peer, Y. (2003) Plant Cell 15, 2192-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paterson, A., Bowers, J., Peterson, D., Estill, J. & Chapman, B. (2003) Curr. Opin. Genet. Dev. 13, in press. [DOI] [PubMed]
- 10.Kishimoto, N., Higo, H., Abe, K., Arai, S., Saito, A. & Higo, K. (1994) Theor. Appl. Genet. 88, 722-726. [DOI] [PubMed] [Google Scholar]
- 11.Nagamura, Y., Inoue, T., Antonio, B., Shimano, T., Kajiya, H., Shomura, A., Lin, S., Kuboki, Y., Harushima, Y., Kurata, N., Minobe, Y., Yano, M. & Sasaki, T. (1995) Breeding Sci. 45, 373-376. [Google Scholar]
- 12.Wang, S. P., Liu, K. D. & Zhang, Q. F. (2000) Acta Bot. Sinica 42, 1150-1155. [Google Scholar]
- 13.Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R. L., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002) Science 296, 92-100. [DOI] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J (1990) J. Mol. Biol. 215, 403-10. [DOI] [PubMed] [Google Scholar]
- 15.Chapman, B. A., Bowers, J. E. & Paterson, A. (2004) Bioinformatics 20, 180-185. [DOI] [PubMed] [Google Scholar]
- 16.Yang, Z. H. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 17.Yang, Z. H., Nielsen, R., Goldman, N. & Pedersen, A. M. K. (2000) Genetics 155, 431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch, M. & Conery, J. S. (2000) Science 290, 1151-1155. [DOI] [PubMed] [Google Scholar]
- 19.Gaut, B. S., Morton, B. R., McCaig, B. C. & Clegg, M. T. (1996) Proc. Natl. Acad. Sci. USA 93, 10274-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kellogg, E. A. (1998) Proc. Natl. Acad. Sci. USA 95, 2005-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chittenden, L. M., Schertz, K. F., Lin, Y. R., Wing, R. A. & Paterson, A. H. (1994) Theor. Appl. Genet. 87, 925-933. [DOI] [PubMed] [Google Scholar]
- 22.Eckhardt, N. (2001) Plant Cell 13, 1699-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch, M. & Force, A. G. (2000) Am. Nat. 156, 590-605. [DOI] [PubMed] [Google Scholar]
- 24.Gaut, B. S. & Doebley, J. F. (1997) Proc. Natl. Acad. Sci. USA 94, 6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ming, R., Liu, S. C., Lin, Y. R., da Silva, J., Wilson, W., Braga, D., van Deynze, A., Wenslaff, T. F., Wu, K. K., Moore, P. H., et al. (1998) Genetics 150, 1663-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellogg, E. A. (2003) Nature 422, 383-384. [DOI] [PubMed] [Google Scholar]
- 27.Wendel, J. F., Stuber, C. W., Edwards, M. D. & Goodman, M. M. (1986) Theor. Appl. Genet. 72, 178-185. [DOI] [PubMed] [Google Scholar]
- 28.Lan, T. H., DelMonte, T. A., Reischmann, K. P., Hyman, J., Kowalski, S. P., McFerson, J., Kresovich, S. & Paterson, A. H. (2000) Genome Res. 10, 776-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.