Abstract
In this paper, we propose a new approach for computing a distance between two shapes embedded in three-dimensional space. We take as input a pair of triangulated genus zero surfaces that are topologically equivalent to spheres with no holes or handles, and construct a discrete conformal map f between the surfaces. The conformal map is chosen to minimize a symmetric deformation energy Esd(f) which we introduce. This measures the distance of f from an isometry, i.e. a non-distorting correspondence. We show that the energy of the minimizing map gives a well-behaved metric on the space of genus zero surfaces. In contrast to most methods in this field, our approach does not rely on any assignment of landmarks on the two surfaces. We illustrate applications of our approach to geometric morphometrics using three datasets representing the bones and teeth of primates. Experiments on these datasets show that our approach performs remarkably well both in shape recognition and in identifying evolutionary patterns, with success rates similar to, and in some cases better than, those obtained by expert observers.
Keywords: conformal mapping, geometric morphometrics, primates
1. Introduction
Geometry and topology have had increasing impact in biology over the last two decades. The application of mathematical methods in biological studies has not only become viable but in fact is assuming a central role, owing to the increasing performance of computers and the improvement of the devices used for imaging biological systems. In medicine, for example, diagnosis and treatment planning previously relied on conventional X-ray images, recorded on an analogue film. Today, however, more and more digital three-dimensional images, such as those generated with computed tomography, magnetic resonance imaging, and functional, marker-based images are acquired from patients to detect and monitor putative pathologies. This has driven the need for software development to analyse those images, which in turn has provided a major impetus for the development of new mathematical methods and algorithms for image processing. With these new software tools, the three-dimensional images can be quantitatively analysed and visualized, making medical diagnosis, the assessment of therapeutic strategies, and even surgery more reliable and reproducible [1].
The comparison of images and of the shapes they represent is by no means limited to medicine. Advances in geometry and topology, and the parallel transformation of biology into a quantitative science, have led to a renewed interest in applying geometric methods to representing, searching, simulating, analysing and comparing biological systems [2]. These methods are developed and applied in a wide range of fields, including computer vision, biological imaging, brain mapping, target recognition and for satellite image analysis. In molecular biology, the notion that the structure (or shape) of a protein is a major determinant of its function has led to the development of methods for representing, measuring and comparing protein structures [3–5]. Brain morphometry, concerned with the measurement of the brain geometric structures and the changes they undergo during development, ageing, learning, disease and evolution, has become central in neurobiology [6–10]. Morphometrics, the quantitative analysis of forms, has equal potential in evolutionary biology [11–14], though its impact in this field has been somewhat obscured by the phenomenal success of molecular phylogenetics [15]. There is hope however that information from both approaches can be combined to reach a synthetic, comprehensive and quantitative view of phylogeny (see, for example, [16,17] for successful integration in the field of palaeoanthropology). To reach this goal, geometric morphological studies need to be automated and standardized, starting with the determination of geometric correspondence between shapes [14,18]. This paper concerns mathematical and computational aspects of this problem.
While three-dimensional data representing a shape come in many forms, we concentrate on the important and commonly occurring case where the surface of the shape is available and described with a discrete triangular mesh. Thus, we are interested in understanding the structure of surfaces situated in our three-dimensional world. Mathematically, these objects are two-dimensional Riemannian manifolds in the smooth case, and piecewise-flat surfaces in the discrete setting. We work with both descriptions. We restrict ourselves to surfaces of genus zero. These are the surfaces that can be continuously deformed to a sphere, or, alternatively, surfaces that have no holes or handles.
In a chapter titled ‘The comparison of related forms’, Thompson [19] explored how differences in the forms of related animals can be described by means of simple mathematical transformations. This inspired the development of several shape comparison techniques whose aim is to define a map between two shapes that can be used to measure their similarity. This is a challenging problem, as the space of possible maps is extremely large and difficult to characterize mathematically. In this paper, we develop methods that generate maps between two shapes corresponding to surfaces of genus zero.
The dimension of the space of maps between two shapes can be reduced by enforcing correspondence between specific landmarks. Ideally, these landmark points should identify homologous structures on the surfaces of the two shapes, should conserve their relative positions, should provide adequate coverage and should be found reliably and consistently [2]. The task of finding such landmark points is usually performed manually by skilled morphometricians with extensive training. The resulting human choices can lead to error due to the variability and inconsistency of human input [20]. Many methods have been developed to circumvent this inherent limitation, either through automation of the landmark selection process or by eliminating the need to use specific point correspondence in the process of aligning the surface altogether. Automatically selected points may relate to distinctive geometric features such as local curvature maxima [21], be inferred from an atlas for the shape of interest [22], or be optimally distributed on the surface of interest based on some statistical criteria [23,24]. Spectral techniques, for example, assign a signature to each vertex in the mesh, under the premise that points with similar signatures are more likely to correspond [25,26]. There is a growing interest in the concept of semi-landmarks. These are points that characterize the outlines of the shapes of interests. The positions of these points are optimized to match the positions of corresponding points along an outline in a reference conformation [12,27–31]. Despite the optimization procedures, there is no guarantee that these points are placed accurately and consistently across collections of surfaces, unless those surfaces are highly homologous. In addition, most of the techniques based on semi-landmarks for three-dimensional shapes still rely on a few user-defined landmarks [29,32]; as such, they do not fully remove the inherent limitation of the variability of human input. Landmark-based methods that find maps between two shapes work on the premise that knowledge of a mapping on a small number of correspondences can be extended to give the full map between the two surfaces of interest [33–35]. By contrast, landmark-free methods skip the search for landmarks altogether. For example, Valliant & Glaunès [36] introduced a representation of surface in the form of currents and then imposed a Hilbert space structure on it, whose norm is used to quantify the similarity between two surfaces. McCane [37] developed a variational method for matching curves in two or three dimensions by optimizing their parametrizations. In parallel, Laga et al. [38] developed statistical models of shapes based on the squared root velocity function that allow for the modelling of shape variability without considering landmarks. They recently implemented this procedure to study the shapes of plant leaves [38].
In a groundbreaking recent paper, Boyer et al. [13] introduced several distance measures that can be used to generate fully automated correspondences between surfaces. They tested their approaches on three datasets representing the skeletal anatomy of a collection of primates, showing success in taxonomic classifications [13]. These approaches were tailored to topological discs, as all the datasets represent surfaces having the topology of a disc. That is, the surfaces can be obtained from a flat disc in the plane by stretching and bending, but without tearing or gluing. We present here a new algorithm that has a similar philosophy to their work, but uses a very different geometric distance measure. As with their method, our approach fully eliminates the use of landmarks. While they measured distortion based on area-preserving maps with a continuous Procrustes distance and general maps with a Wasserstein distance, we work entirely within the framework of conformal (angle-preserving) maps and focus on finding a globally optimal conformal mapping between two genus zero closed surfaces. We associate an energy to any conformal map between two such surfaces. We show that the energy of the optimal map defines a metric on the space of surfaces of genus zero. Experiments on the same datasets as those used by Boyer et al. [13] show that our method outperforms their distance measures in identifying evolutionary patterns between the specimens whose bones are included in the datasets and, indeed, performs as well as or better than trained observers. We note that while our algorithm was developed to compare spherical or genus zero surfaces, it can also be used to compare disc-like surfaces. To do so, we introduce a preliminary step where the holes of a surface are filled in (coned) to create a closed surface. We expect that our method would perform even better in studies where the compared surfaces were already of genus zero.
This paper develops previous preliminary studies [39,40]. We have modified the elastic energy used to measure the distance of an optimal conformal mapping from an isometry so that it now defines a mathematical metric on the space of shapes. The paper is organized as follows. Section 2 provides the mathematical background for our method: conformal geometry and a metric to measure the similarity between surfaces of genus zero. The details of its implementation on discrete surfaces are provided in the electronic supplementary material. Section 3 presents and discusses the results obtained by our algorithm on three test cases introduced by Boyer et al. [13]. We conclude the paper with a brief discussion on the implications of this work on using phenetics to reconstruct phylogeny, and on future developments of the method itself.
2. A new distance between shapes
2.1. Optimal conformal map between genus zero surfaces
Let F1 and F2 be two surfaces of genus zero and equal area. By rescaling each surface, it is straightforward to arrange for all surface areas to be equal to 1. While our method allows us to compare surfaces of different areas, we focus here on scale invariant shape properties. A map f from F1 to F2 defines for each point 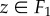 a corresponding point
a corresponding point 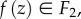 called the image of z under f. For smooth surfaces, one can specify the angles between two curves. Maps that preserve these angles are called conformal. Such maps do not need to preserve length. Examples of conformal maps include the Mercator projection used in cartography and the stereographic projection that maps a sphere (minus its North Pole) onto the plane.
called the image of z under f. For smooth surfaces, one can specify the angles between two curves. Maps that preserve these angles are called conformal. Such maps do not need to preserve length. Examples of conformal maps include the Mercator projection used in cartography and the stereographic projection that maps a sphere (minus its North Pole) onto the plane.
A conformal map preserves angles but usually distorts distances, with isometries being the exception. This distortion is characterized by a dilation factor, 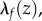 that measures the stretching of vectors by f at each point z in F1. This stretching is the same in all directions.
that measures the stretching of vectors by f at each point z in F1. This stretching is the same in all directions.
Our objective is to find a conformal map between the two genus zero surfaces F1 and F2 that is as close to an isometry as possible. An isometry has two distinct local properties. It preserves angles at each point (conformality) and it preserves area. There is a natural choice for picking a map that is as close to an isometry as possible. One first restricts to finding a conformal map between the two surfaces. The uniformization theorem ensures the existence of such a map, and indeed of many such maps between any two surfaces of genus zero [41]. To pick the best conformal map, it is natural to use the second criterion of an isometry, area preservation, and choose a conformal map that minimizes the local area distortion. This is the underlying idea of our method, as described in [39,40,42].
Following this idea, the conformal map f between F1 and F2 can be described as the composition of three conformal maps, 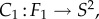
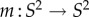 and
and 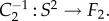 The map m is an element of the six-dimensional group PSL
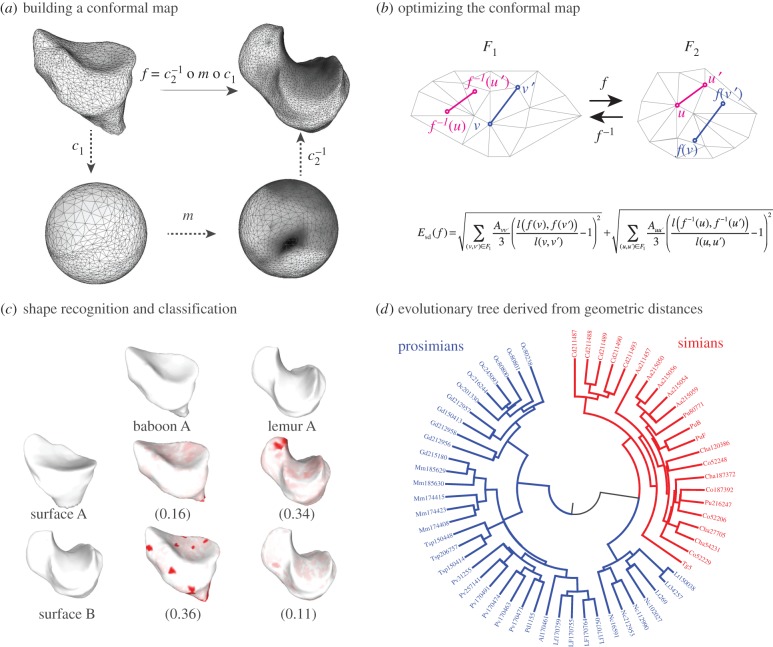
The map m is an element of the six-dimensional group PSL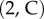 that describes all conformal maps from the round sphere S2 to itself. Figure 1a illustrates this process for two proximal metatarsal bones of primates.
that describes all conformal maps from the round sphere S2 to itself. Figure 1a illustrates this process for two proximal metatarsal bones of primates.
Figure 1.
An optimal conformal map and its applications. (a) The comparison of two surfaces F1 and F2 relies on the existence of a map f between these surfaces. When the two surfaces are of genus zero, it is possible to construct f as a composition of three maps C1, m and C2, where C1 and C2 are conformal maps from the surfaces F1 and F2 to the sphere and m is a bijective conformal map of the sphere to itself. The key to our approach is that the group of conformal self-maps of the sphere is the well-understood group of Möbius transformations. As such, m is defined by six parameters that can be optimized to yield minimal distortion. (b) Distortion is computed as the sum of the stretching induced by f on all edges 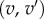 in the mesh describing F1, and of the stretching induced by
in the mesh describing F1, and of the stretching induced by 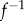 on all edges
on all edges 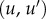 in the mesh describing F2. In the corresponding symmetric distortion energy Esd(f), each edge is weighted by the areas of its adjacent triangles. The energy of the minimizing map defines a distance dsd on the space of surfaces of genus 0. (c) Shape recognition is a natural application for the method described here. All surfaces shown here correspond to proximal metatarsal bones from baboons and lemurs. When the surface A is mapped onto the surfaces for baboon A and lemur A, the corresponding symmetric distortion distances dsd are 0.16 and 0.34, respectively, identifying surface A as corresponding to a baboon. By contrast, surface B is closer to lemur A, and therefore corresponds to a lemur. Regions on the images with large stretching are coloured. (d) The unweighted pair group method with arithmetic mean (UPGMA) tree built from the distance matrix computed from a comprehensive comparison of all bones in database A clearly separates those bones into two categories, those from prosimians (left) and those from simians (right). This geometry-based categorization of the primates obtained from the metatarsal shapes can then be combined with other classifications to help understand their phylogeny. (Online version in colour.)
in the mesh describing F2. In the corresponding symmetric distortion energy Esd(f), each edge is weighted by the areas of its adjacent triangles. The energy of the minimizing map defines a distance dsd on the space of surfaces of genus 0. (c) Shape recognition is a natural application for the method described here. All surfaces shown here correspond to proximal metatarsal bones from baboons and lemurs. When the surface A is mapped onto the surfaces for baboon A and lemur A, the corresponding symmetric distortion distances dsd are 0.16 and 0.34, respectively, identifying surface A as corresponding to a baboon. By contrast, surface B is closer to lemur A, and therefore corresponds to a lemur. Regions on the images with large stretching are coloured. (d) The unweighted pair group method with arithmetic mean (UPGMA) tree built from the distance matrix computed from a comprehensive comparison of all bones in database A clearly separates those bones into two categories, those from prosimians (left) and those from simians (right). This geometry-based categorization of the primates obtained from the metatarsal shapes can then be combined with other classifications to help understand their phylogeny. (Online version in colour.)
Varying m gives all conformal maps from F1 to F2. We specify the optimal m to be the one that leads to a minimum of the following symmetric distortion energy integral energy Esd:
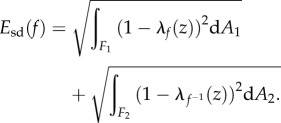 |
2.1 |
A conformal map f is an isometry if and only if its dilation 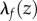 at every point z is equal to 1.
at every point z is equal to 1. 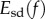 is the natural measure of how
is the natural measure of how 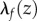 and
and 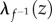 differ from 1, and therefore of how f and
differ from 1, and therefore of how f and 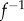 deviate from an isometry. Note that
deviate from an isometry. Note that 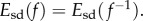 The infimum of the magnitude of
The infimum of the magnitude of 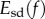 as f varies over all conformal diffeomorphisms from F1 to F2 exists [42] and is used to define the distance between the two surfaces.
as f varies over all conformal diffeomorphisms from F1 to F2 exists [42] and is used to define the distance between the two surfaces.
The symmetric distortion energy defined in equation (2.1) has the following properties [42]:
(1) for any pair of genus zero surfaces, there exists a smooth conformal diffeomorphism fmin between them that minimizes the symmetric distortion energy,
(2) the symmetric distortion energy of a map is zero if and only if the map is an isometry,
(3) the symmetric distortion energy of fmin defines a metric dsd on the space of genus zero surfaces, so that dsd satisfies the following three properties, (i)
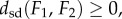 with equality if and only if F1 and F2 are isometric, (ii)
with equality if and only if F1 and F2 are isometric, (ii) 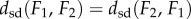 , and (iii)
, and (iii) 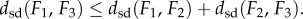
Property (iii) of the metric dsd, the triangle inequality, is important for applications, as it implies robustness. Namely, if the distances 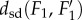 and
and 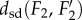 are small, this property guarantees that
are small, this property guarantees that 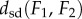 is close to
is close to 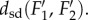 Thus, the distance measure is stable under noise and measurement errors.
Thus, the distance measure is stable under noise and measurement errors.
2.2. Comparing discrete genus zero surfaces
In practice, the two surfaces F1 and F2 are discrete and represented by meshes 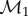 and
and 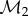 , respectively. Meshes are taken to be triangular, so that
, respectively. Meshes are taken to be triangular, so that 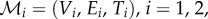 where
where 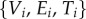 denote the vertices, edges and faces, respectively. We do not restrict the topologies of the meshes to be the same. The number of vertices, edges and faces of
denote the vertices, edges and faces, respectively. We do not restrict the topologies of the meshes to be the same. The number of vertices, edges and faces of  and
and  can be different. The method described above for computing an optimal conformal map between two smooth surfaces needs to be adapted for its applications to their discrete counterparts. While we refer the reader to our previous papers [39,40] and to the electronic supplementary material for a full description of this adaptation, we summarize here the changes that are relevant to this paper.
can be different. The method described above for computing an optimal conformal map between two smooth surfaces needs to be adapted for its applications to their discrete counterparts. While we refer the reader to our previous papers [39,40] and to the electronic supplementary material for a full description of this adaptation, we summarize here the changes that are relevant to this paper.
The total distortion for a mesh is a discrete version of the symmetric distortion energy given by equation (2.1) and is computed as a sum over all edges of the two surface meshes
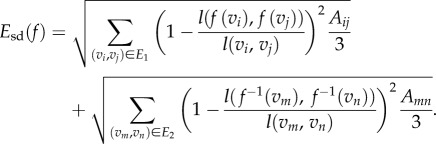 |
2.2 |
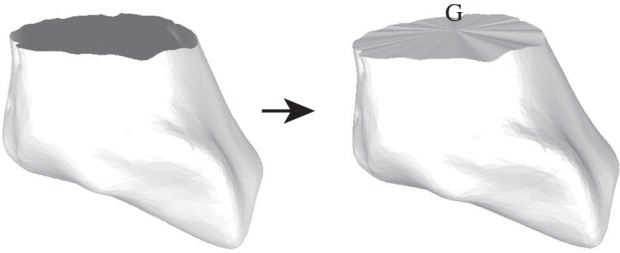
Here E1 and E2 denote the sets of edges in the meshes on F1 and F2, respectively. Aij is the sum of the areas of the two triangles adjacent to the edge 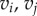 and
and 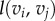 is the distance between the two vertices
is the distance between the two vertices 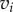 and
and 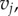 namely the length of the edge
namely the length of the edge 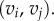
The image 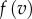 of a vertex v of the mesh
of a vertex v of the mesh 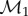 is not artificially restricted to correspond to a vertex of the mesh
is not artificially restricted to correspond to a vertex of the mesh 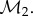 Instead, v is mapped to an arbitrary point belonging to one face of
Instead, v is mapped to an arbitrary point belonging to one face of 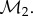 Special care is then needed for computing distances. The distance between two vertices
Special care is then needed for computing distances. The distance between two vertices 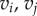 forming an edge
forming an edge 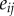 of F1 is simply the length of this edge. The images
of F1 is simply the length of this edge. The images 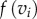 and
and 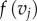 most likely do not form an edge of F2. The distance
most likely do not form an edge of F2. The distance 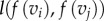 is computed using a flat Euclidean metric on each face of the triangulation. Figure 1b illustrates the calculation of
is computed using a flat Euclidean metric on each face of the triangulation. Figure 1b illustrates the calculation of 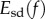 on two edges.
on two edges.
We have implemented this procedure into the program MatchSurface. A complete description of its algorithms is given in the electronic supplementary material. For a pair of surfaces F1 and F2 represented by meshes 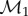 and
and 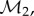 MatchSurface produces an approximation for the map fmin that minimizes the symmetric distortion energy Esd over all conformal maps between F1 and F2. Its output consists of the image
MatchSurface produces an approximation for the map fmin that minimizes the symmetric distortion energy Esd over all conformal maps between F1 and F2. Its output consists of the image 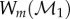 of
of 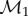 warped by fmin onto F2, the image
warped by fmin onto F2, the image 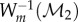 of
of 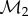 warped by
warped by 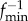 onto F1, and the distance
onto F1, and the distance 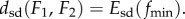 Applications of MatchSurface include, but are not limited to, (i) comparing a surface
Applications of MatchSurface include, but are not limited to, (i) comparing a surface 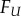 of unknown origin with a library of surfaces that are well characterized, using the best matching surface as a template to infer properties for
of unknown origin with a library of surfaces that are well characterized, using the best matching surface as a template to infer properties for 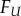 (figure 1c) and (ii) construction of a neighbour joining tree that captures the hierarchical geometric similarities between a set of surfaces. This tree can then be related to properties of the objects it represents, such as phylogeny, as illustrated in figure 1d.
(figure 1c) and (ii) construction of a neighbour joining tree that captures the hierarchical geometric similarities between a set of surfaces. This tree can then be related to properties of the objects it represents, such as phylogeny, as illustrated in figure 1d.
3. Results
To test the effectiveness of MatchSurface, we applied it on three independent datasets, representing three regions of the skeletal anatomy of a collection of primates. The first dataset contains 61 proximal first metatarsals of prosimian primates, New and Old World monkeys, the second dataset contains 45 distal radii of apes and humans and the third dataset includes 116 second mandibular molars of prosimian primates and non-primate close relatives. They were originally assembled by Boyer et al. [13], who used them in a similar study with different shape comparison measures based on an ‘earth mover’ metric, and on a ‘continuous Procrustes' (cP) distance. We note that all meshes included in the three datasets represent genus zero surfaces with one boundary. The position of this boundary is somewhat arbitrary. Its impact on the study by Boyer et al. was not discussed. Similarly, it will not be considered here. We did however detect and clean up those boundaries by removing ‘dangling’ triangles, i.e. triangles with two boundary edges (see the electronic supplementary material for details). Our method is designed for genus zero closed surfaces. It easily extends however to genus zero surfaces with boundaries by coning off the boundary curves (see figure 2 for an illustration of this process). Using these datasets allows us to evaluate the performances of the algorithm implemented in MatchSurface. The evaluation is done by comparing the distances dsd between the surfaces included in the three datasets with the continuous Procrustes distances provided by Boyer et al., and with the distances based on landmarks identified by trained morphologists. The datasets include two sets of the latter for the metatarsal dataset, referred to as ‘observer1’ (obs1) and ‘observer2’ (obs2), and one set for the radii dataset and the teeth dataset, ‘observer1’. We note that the cP distance, in common with dsd, does not require preliminary selection of landmarks on the surfaces. Geometric morphometricians on the other hand have determined landmarks on each surface, choosing them to reflect correspondences considered biologically and evolutionarily meaningful (see SI Appendix, Materials of [13]). These landmarks determine a ‘discrete’ Procrustes distance between any two surfaces, which we refer to as dobs. Each of the three distances (dsd, dcP and dobs) defines a matrix for each dataset, containing all pairwise distances between the surfaces included in that dataset. To measure the effectiveness of each distance, we compare those matrices in three different ways.
Figure 2.
Coning off a surface with boundary. The method presented in this paper is currently optimized for surfaces of genus zero. These surfaces do not have a boundary. The meshes included in our test sets however have one boundary, whose limits are somewhat arbitrary. To fill in the corresponding hole, we detect all vertices and edges on the boundary, compute the centre of gravity G of these vertices, and add to the mesh all the triangles formed by connecting G to the edges of the boundary. The result is a genus zero surface with no boundary.
All three distances rank pairs of surfaces according to their similarity. The relative performance of dsd and dcP with respect to the observer distance dobs can then be computed using a correlation analysis. Table 1 gives the corresponding coefficients.
Table 1.
Correlations between observer distances, cP and sd distances.
| dataset | no. pairs | obs1/cP | obs2/cP | obs1/sd | obs2/sd | obs1/obs2 |
|---|---|---|---|---|---|---|
| metatarsal (all) | 1830 | 0.62 | 0.63 | 0.82 | 0.81 | 0.87 |
| metatarsal (same species) | 112 | 0.31 | 0.17 | 0.55 | 0.31 | 0.46 |
| metatarsal (different species) | 1718 | 0.57 | 0.58 | 0.79 | 0.78 | 0.85 |
| radius (all) | 990 | 0.28 | n.a. | 0.59 | n.a. | n.a. |
| radius (same species) | 198 | 0.29 | n.a. | 0.40 | n.a. | n.a. |
| radius (different species) | 792 | 0.13 | n.a. | 0.46 | n.a. | n.a. |
| teeth (all) | 4851 | 0.55 | n.a. | 0.58 | n.a. | n.a. |
| teeth (same genus) | 180 | 0.01 | n.a. | 0.63 | n.a. | n.a. |
| teeth (different genus) | 4671 | 0.51 | n.a. | 0.51 | n.a. | n.a. |
The distances based on manual assignments of landmarks by morphometricians may be considered as a reference, since they are based on extensive expert knowledge, though they are not deemed perfect. Note that there is variability between morphometricians, though the correlations between the results of two such observers is high. On all three datasets, the distances based on sd match observer distances better than the distances based on cP.
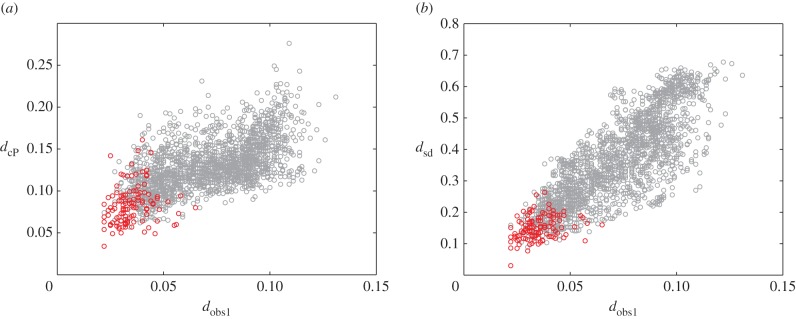
This advantage is further illustrated in figure 3.
Figure 3.
Comparison of the observer distance with the cP distance (a), and the sd distance (b). Results are shown for the metatarsal database. Red circles correspond to pairs of surfaces from specimens that belong to the same species. The distances between same species pairs are expected to be small for any of the three distance measures. This is indeed the case, as the corresponding points are found to cluster in the lower left corner of the plots. The correlation coefficients between dcP and 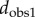 and between dsd and
and between dsd and 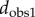 over all pairs of surfaces are 0.62 and 0.82, indicating that dsd matches better with the observer1 distance, over a broader range or values. We also compute the ratio of the range of distance values for the pairs corresponding to specimens from the same species over the range of values for all pairs of surfaces in the dataset. A lower ratio indicates better performance, as a large ratio would indicate lower discrimination of the significant pairs. We find ratios of 50%, 39% and 36% for the cP distance, the observer1 distance, and the sd distance, respectively.
over all pairs of surfaces are 0.62 and 0.82, indicating that dsd matches better with the observer1 distance, over a broader range or values. We also compute the ratio of the range of distance values for the pairs corresponding to specimens from the same species over the range of values for all pairs of surfaces in the dataset. A lower ratio indicates better performance, as a large ratio would indicate lower discrimination of the significant pairs. We find ratios of 50%, 39% and 36% for the cP distance, the observer1 distance, and the sd distance, respectively.
The match between dsd and the observer distance dobs is more consistent over the whole range of values. We note that all three distances identify the pairs of surfaces corresponding to specimens from the same species as being similar (red circles in figure 3). Even on this subset of all pairs, however, dsd is better correlated to the observer distances (table 1). The same behaviour is observed for pairs of surfaces from specimens that belong to the same family or to the same superfamily (results not shown). Note that the similarities found between the dsd distances and the observer distances are comparable to those between the two observers.
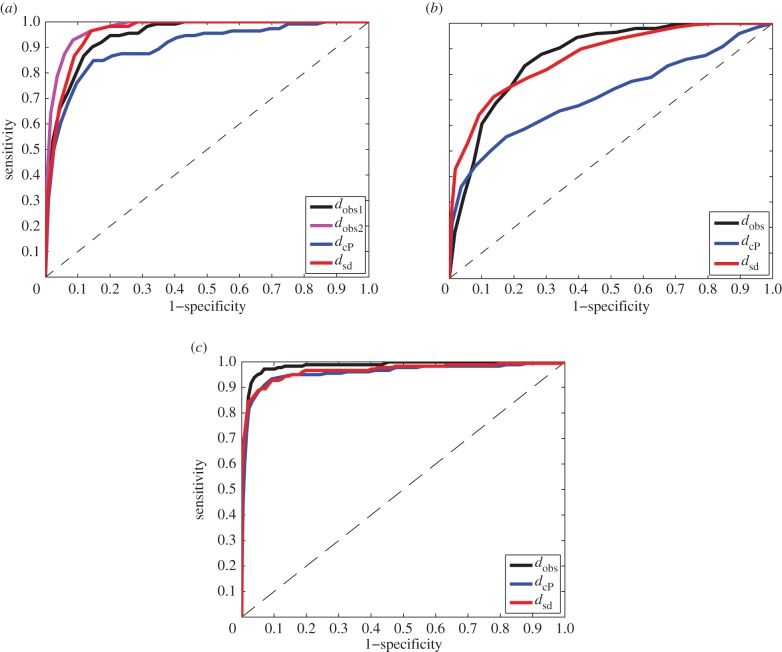
For a second comparison of the three distance measures, we evaluated their performance using a receiver operating characteristic (ROC) analysis [43]. In this approach, a ‘gold standard’ is defined, based on a choice of level in the phylogeny of the specimens, either species, genus, family or superfamily. A pair of surfaces is then defined as similar, or ‘positive’, if the corresponding specimens belong to the same taxonomic group considered, and ‘negative’ otherwise. For varying thresholds of the distance measure under study, all pairs of surfaces whose distances fall below the threshold are then assumed positive, while those above it are deemed negative. The pairs that agree with the standard are called true positives (TPs), while those that do not are false positives (FPs). An ROC analysis compares the rate of TPs (also called sensitivity) to the rate of FPs (which corresponds to 1 minus the sensitivity). It is scored with the proportion of area under the corresponding curve (AUC). An AUC of 1 indicates that all TPs are detected first. This corresponds to the ideal distance measure. On the other hand, the diagonal curve leads to an AUC of 0.5. In this case, TP and FP appear at the same rate, and the distance measure contains no information.
The results of ROC analyses based on the three distances dsd, dcP and dobs are given in table 2 and illustrated in figure 4.
Table 2.
Area under the curve (AUC) of ROC analyses on the observer, cP and sd distances.
| dataset | first metatarsal |
radius |
teeth |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| classification | N | obs1 | obs2 | cP | sd | N | obs | cP | sd | N | obs | cP | sd |
| species | 13 | 0.94 | 0.97 | 0.90 | 0.96 | 5 | 0.87 | 0.72 | 0.87 | n.a. | n.a. | n.a. | n.a. |
| genera | 13 | 0.94 | 0.97 | 0.90 | 0.96 | 4 | 0.86 | 0.71 | 0.80 | 24 | 0.98 | 0.96 | 0.97 |
| families | 9 | 0.91 | 0.95 | 0.84 | 0.95 | n.a. | n.a. | n.a. | n.a. | 17 | 0.87 | 0.79 | 0.86 |
| superfamilies | 2 | 0.96 | 0.97 | 0.73 | 0.86 | n.a. | n.a. | n.a. | n.a. | 5 | 0.64 | 0.62 | 0.77 |
Figure 4.
ROC analyses of three distance measures of surface similarities. We compare the effectiveness of an observer-based distance, 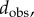 a continuous Procrustes distance,
a continuous Procrustes distance, 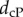 , and a symmetric distortion distance, dsd, to detect similarities that identify with membership of the same species in a set of 61 metatarsal surfaces (a), in a set of 45 radius surfaces (b), and with genera in a set of 99 teeth surfaces (c). ‘True’ relationships are defined by the phylogeny of the specimens included in those datasets. An ROC curve that falls close to the first diagonal (shown as a dashed line) reveals poor performance, while an ROC curve that first follows the y-axis, i.e. whose area under the curve is the largest, indicates good performance. The results indicate that dsd outperforms
, and a symmetric distortion distance, dsd, to detect similarities that identify with membership of the same species in a set of 61 metatarsal surfaces (a), in a set of 45 radius surfaces (b), and with genera in a set of 99 teeth surfaces (c). ‘True’ relationships are defined by the phylogeny of the specimens included in those datasets. An ROC curve that falls close to the first diagonal (shown as a dashed line) reveals poor performance, while an ROC curve that first follows the y-axis, i.e. whose area under the curve is the largest, indicates good performance. The results indicate that dsd outperforms 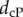 and is comparable to an observer-based distance.
and is comparable to an observer-based distance.
Species or genera identification based on landmarks manually defined by experts is expected to perform best. Indeed, the ROC curves derived from the observer distances illustrate excellent classification results, with AUC values above 0.85 in all cases, above 0.9 for the metatarsal dataset, and above 0.95 for the teeth dataset. Note that even with human expertise included, the classification is not perfect. In addition, we observe differences between the results obtained by two distinct experts. The two distance measures dcP and dsd alleviate the difficulty of defining landmarks on the two surfaces to be compared. Instead, both methods construct a map between the two surfaces using only their geometric properties. The distances they compute reflect different geometric properties. We find that the distance dsd introduced in this study outperforms dcP on all three datasets, at all phylogenetic classification levels. In fact, dsd performs as well as the observer, landmark-based distances, with differences that are of similar magnitude to the differences measured between distinct observers.
In such an ROC analysis, it is worth focusing on the small distances, as those are often the most reliable ones and the most relevant for applications. For each distance measure, we defined S200 to be the set of 200 pairs of surfaces with the lowest distances. Each pair was deemed to be ‘true’ or ‘false’ if the corresponding pair of specimens belonged to the same species or not, respectively. Table 3 reports the repartition of true and false pairs within S200 for the three distances considered here, for the three datasets. The higher the number of true pairs, the better. The rankings provided by dsd are similar to those provided by the expert observers, outperforming the cP distance on all three datasets.
Table 3.
Repartition of the 200 smallest distances for the observer, cP and sd distances.
| dataset | first metatarsal |
radius |
teeth |
|||
|---|---|---|---|---|---|---|
| distance | true (112)a | false (1718) | true (198) | false (792) | true (180) | false (4671) |
| observer1 | 81 (72%) | 119 (7%) | 120 (60%) | 80 (10.1%) | 134 (74.4%) | 66 (1.4%) |
| observer2 | 98 (87.5%) | 102 (5.9%) | n.a. | n.a. | n.a. | n.a. |
| dcP | 77 (68.7%) | 123 (7.2%) | 99 (50%) | 101 (12.8%) | 130 (72.2%) | 70 (1.5%) |
| dsd | 86 (76.8%) | 114 (6.6%) | 127 (64%) | 73 (9.2%) | 138 (76.7%) | 62 (1.3%) |
aThe distance between two surfaces is said to be true if the corresponding specimens belong to the same species, and false otherwise. The total number of such pairs is given in parenthesis. We chose the first 200 distances in both sets to guarantee that we would include both types of distances. We note that the best possible performance for a distance measure would find 112 true and 83 false distances for the metatarsal dataset, 198 true and 2 false for the radius dataset, and 180 true and 20 false for the teeth dataset.
The ROC analysis ranks distances between specimens and assesses if this ranking is compatible with an existing classification; it does not perform the classification itself. We extend the ROC analysis to the actual problem of classification by performing a third set of computational experiments. Each experiment involves a dataset of surfaces/specimens, D, a taxon, T, and a distance measure, d. We begin by randomly dividing the sets of surfaces in D into two groups of approximately equal size. The first group serves as a training set to define the taxa, while the second group serves as a test set. A test surface is classified by assigning it to the taxon of its nearest neighbour in the training set. This is much akin to the ‘threading’ method illustrated in figure 1c and used in the protein structure prediction community [44]. The results are stored in a confusion matrix, C, whose element 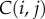 reports the number of test surfaces corresponding to specimens that belong to taxon i that have been classified as belonging to taxon j. The accuracy of the classifier d is then defined to be the ratio of the trace of the confusion matrix over the sum of all its elements (i.e. the percentage of correctly classified specimens). To remove the influence of the initial division of the dataset into test and training sets, the procedure is repeated 5000 times. We performed these experiments for the three datasets, for the three distance measures and for different levels in the taxonomy of the specimens. The results are reported in table 4.
reports the number of test surfaces corresponding to specimens that belong to taxon i that have been classified as belonging to taxon j. The accuracy of the classifier d is then defined to be the ratio of the trace of the confusion matrix over the sum of all its elements (i.e. the percentage of correctly classified specimens). To remove the influence of the initial division of the dataset into test and training sets, the procedure is repeated 5000 times. We performed these experiments for the three datasets, for the three distance measures and for different levels in the taxonomy of the specimens. The results are reported in table 4.
Table 4.
Success rates of classification experiments, based on the observer, cP and sd distances.
| dataset | first metatarsal |
radius |
teeth |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| classification | N | obs1 | obs2 | cP | sd | N | obs | cP | sd | N | obs | cP | sd |
| species | 13 | 74a | 86 | 71 | 81 | 5 | 76 | 78 | 85 | n.a. | n.a. | n.a. | n.a. |
| genera | 13 | 74 | 86 | 71 | 81 | 4 | 85 | 85 | 90 | 24 | 87 | 87 | 94 |
| families | 9 | 83 | 94 | 82 | 93 | n.a. | n.a. | n.a. | n.a. | 17 | 91 | 89 | 93 |
| superfamilies | 2 | 100 | 100 | 98 | 100 | n.a. | n.a. | n.a. | n.a. | 5 | 94 | 95 | 95 |
aPercentage of correctly classified specimens, computed over 5000 experiments.
Classifications based on the sd distances outperform those based on the cP distances for the three anatomical datasets, and are similar in accuracy to those based on the observer distances, even outperforming them for the radius and teeth datasets. We note the significant differences between the two observers on the first metatarsal dataset. These differences indicate the difficulties in defining consistent landmarks on anatomical surfaces even for experienced morphometricians.
A distance matrix that contains the results of an all-against-all comparison of all specimens included in a dataset can be turned into a tree using a variety of clustering techniques. We used the unweighted pair group method with arithmetic mean (UPGMA) to build trees based on the four distance measures (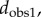
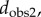 and
and 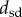 ) for the first metatarsal dataset, as implemented in the software package PHYLIP [45]. UPGMA constructs a tree by minimizing the net disagreement between the matrix pairwise distances and the distances measured on the tree. Results are shown in figure 5. We note that those are phenetic trees, in opposition to phylogenetic trees that are built from molecular sequencing data.
) for the first metatarsal dataset, as implemented in the software package PHYLIP [45]. UPGMA constructs a tree by minimizing the net disagreement between the matrix pairwise distances and the distances measured on the tree. Results are shown in figure 5. We note that those are phenetic trees, in opposition to phylogenetic trees that are built from molecular sequencing data.
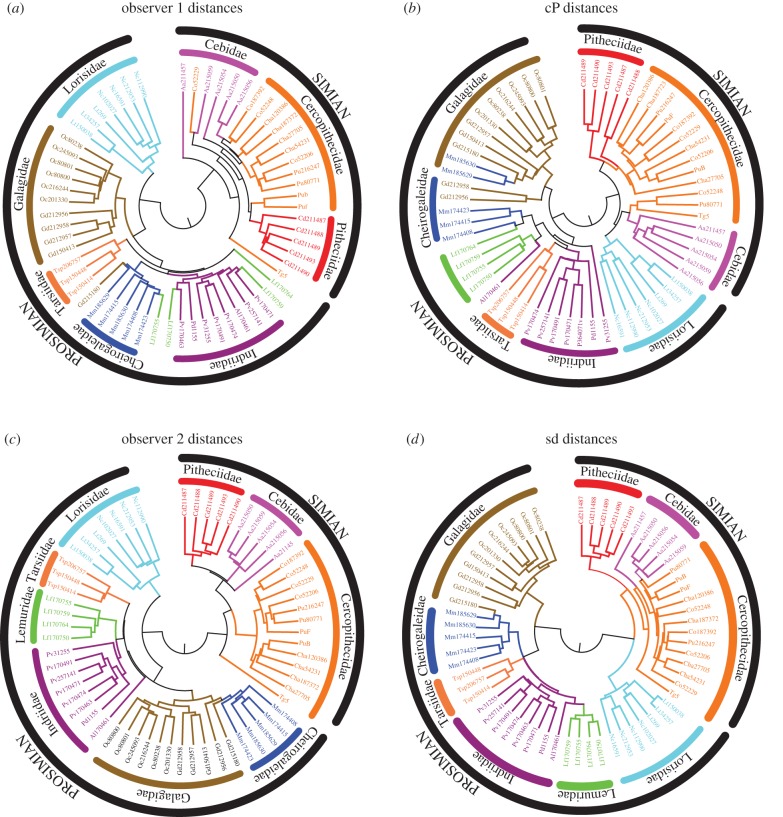
Figure 5.
Morphology-based UPGMA trees for the specimens included in the first metatarsal database: (a,c) are derived from distances computed using landmarks placed on each surface by experienced morphometricians, while (b,d) are computed based on landmark-free shape comparison methods, namely cP [13] and sd (this work). Branches of the trees are coloured according to the actual species of the specimens. The trees were computed using the program Neighbor from the software package PHYLIP [45], and subsequently drawn using the package MEGA6 [46].
The UPGMA tree based on dsd shows a high level of agreement with the actual phylogeny of the specimens considered, both at the superfamily and at the family level. The two superfamilies, simians and prosimians, are clearly separated on the tree. In addition, nine clades with three or more branches only include specimens from the same family, with four of those exactly corresponding to one family, namely the Tarsiidae, Lemuridae, Lorisidae and Pitheciidae. We only observe one unusual association, namely the first metatarsal of one Galago is found to be very similar to the first metatarsal of a Cheirogaleidae, while other members of these two families are clearly distinguished. We note that similar, and sometimes larger, overlaps between these two families are observed in the phylogeny trees built from the observers' distances or from the cP distances. The tree based on observer1 distances shows even more misassociations, with members of the Lemuridae families (in green) being spread out over the Indridae and Cheirogaleidae families.
To quantify the differences between the trees generated from the observers, cP and sd distance matrices computed for the metatarsal dataset, we first rescaled those distance matrices so that all distances ranged between 0 and 1, and regenerated the UPGMA trees. The four trees are then compared using the TreeDist program from the software package PHYLIP. TreeDist computes the symmetric distance of Robinson & Foulds [47] to evaluate the smilarity between two trees. We find that the distances between the observers' trees and the cP and sd trees are 96 and 84, and 86 and 82, respectively. While we cannot assess the meaning of the absolute values of these distances, and the significance of the differences between those values, we do notice that the observers' trees resemble most the tree computed with the sd method introduced here.
4. Discussion
Finding efficient algorithms to describe, measure and compare shapes is a central problem in numerous disciplines that generate extensive quantitative and visual information. Among these, biology occupies a central place. Structural biologists studying bio-molecular structures, neurobiologists studying the shapes of brain structures and their variations during ageing or in diseases, as well as morphometrists who use three-dimensional geometric morphometry are all concerned with characterizing three-dimensional shapes and computing distances between those shapes. In this context, we have developed a new method for automatically generating a conformal map between two surfaces of genus zero. This new approach leads to flexible registration of the two surfaces and accurate measurements of their geometric dissimilarities based on an actual metric on the space of surfaces of genus zero, without the need for the selection of landmark points. Our use of conformal maps is taking advantage of a tool to reduce the dimension of the space of correspondences down to a six-dimensional subspace which retains geometric information and is mathematically natural. Its implementation within the program MatchSurface is based on fast and robust numerical methods, making surface comparisons feasible for a wide range of datasets. We have illustrated its use in the field of geometric morphometry, using three datasets representing bones and teeth of primates. Experiments on these datasets show that it performs remarkably well both in shape recognition and in identifying evolutionary patterns.
While we show successful taxon recognition based on geometric information for one of the datasets considered (the Metatarsal dataset, see figure 5), taxonomy is not the intended purpose of this method. Instead, we restrict its current applications to providing robust estimates of the distances between three-dimensional shapes, with one finality being to support phylogeny reconstruction. Since the advent of computers and the developments of robust and fast scientific computing techniques to quantify phenotypes, there has been a wealth of studies attempting to provide a quantitative view of evolution in biology, starting with classification and taxonomy. The concept of numerical taxonomy [48] and the development of three-dimensional geometric morphometrics are both part of this movement (for an excellent review on this topic, see the recent paper by Boyer et al. [14]). The development of genomics, however, in the last three decades has led many to question the relevance of geometric morphometrics for phylogenetic studies. Indeed, the assessments of genetic variations from automated genomic analyses have greatly improved our understanding of the relationships between the genotype and phenotype of individuals of many species (for a review, see [49]). Those relationships serve as the basis for building the phylogeny of organisms, i.e. the history of their lineages as they change through time. As a consequence, phylogenetics is playing a central role in the study of evolution [50]. In comparison, it is much harder to develop good models of the genetics that underlie morphological changes [51]. It should be noted also that genomics studies are comparatively easier than morphometric studies, with the cost of sequencing whole genomes being so low today, and with the analyses of the one-dimensional information contained in a genome being significantly simpler than the analyses of the three-dimensional information contained in a shape. As a consequence, the utility of morphological data in phylogenetic research has become increasingly questioned (see, for example, Wiens [11], a comment to a paper by Scotland et al. [15]). As a response to the criticisms expressed against geometric morphometry, MacLeod et al. [18] and more recently Boyer et al. [14] have emphasized the need to automate and standardize morphological studies, starting with the determination of geometric correspondence between shapes. The method developed in this paper is one contribution towards this goal. We have shown that it is robust and versatile, albeit currently limited to studying shapes with surfaces of genus zero.
The method described here extends earlier work presented in [39,40]. The earlier work used energies that did not lead to a metric on the space of shapes. We note that these approaches are most accurate on surfaces that have uniform geometry, without long protrusions or spikes [39]. The method is constrained to finding a conformal map between two surfaces, and, while always possible for genus zero, this cannot in general be done for surfaces of positive genus. The basic idea behind using conformal parametrization for surface mapping is deceptively simple and ultimately very powerful. As genus zero surfaces can always be mapped conformally onto the sphere, the search for (near) isometries between them can be made more tractable by restricting to a search within the Möbius group, which is parametrized with six degrees of freedom only. Spheres can be formed from topological discs by coning their boundaries to their centre of mass. Thus, a method for comparing genus zero surfaces, such as MatchSurface, also gives a method to compare surfaces having the topology of a disc.
Finally, we note that the symmetric deformation energy of a conformal map between two surfaces F1 and F2 defined in equation (2.1) establishes a metric on the space of genus zero surfaces. This property is highly desirable for surface comparison, as such a metric is robust and not overly sensitive to noise and measurement errors. The applications of our method extend beyond comparing anatomical surfaces with fields as varied as motion capture, medical imaging and computational biology.
Supplementary Material
Acknowledgements
We thank Yaron Lipman for making the data from [13] freely available on his website (http://www.wisdom.weizmann.ac.il/~ylipman/CPsurfcomp/).
Authors' contributions
Both authors contributed to the conception, design and interpretation of the methods and experiments presented in the paper. They both participated in the drafting and revision of the paper, and approve its final version.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by grant no. MOE2012-T3-1-008 from the Ministry of Education of Singapore (to P.K.) and by grant no. IIS-1117663 from the National Science Foundation (to J.H.).
References
- 1.Angenent S, Pichon E, Tannenbaum A. 2006. Mathematical methods in medical image processing. Bull. Am. Math. Soc. 43, 365–396. ( 10.1090/S0273-0979-06-01104-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zelditch ML, Swiderski DL, Sheets HD. 2012. Geometric morphometrics for biologists: a primer. London, UK: Elsevier. [Google Scholar]
- 3.Max N, Getzoff E. 1988. Spherical harmonic molecular surfaces. IEEE Comput. Graph. Appl. 8, 42–50. ( 10.1109/38.7748) [DOI] [Google Scholar]
- 4.Koehl P. 2006. Protein structure classification. In Reviews in computational chemistry, vol. 22 (eds Lipkowitz KB, Cundari TR, Gillet VJ, Boyd B), pp. 1–56. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 5.Kolodny R, Petrey D, Honig B. 2006. Protein structure comparison: implications for the nature of fold space, and structure and function prediction. Curr. Opin. Struct. Biol. 16, 393–398. ( 10.1016/j.sbi.2006.04.007) [DOI] [PubMed] [Google Scholar]
- 6.Kötter R, Wanke E. 2005. Mapping brains without coordinates. Phil. Trans. R. Soc. B 360, 751–766. ( 10.1098/rstb.2005.1625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otte A, Halsband U. 2006. Brain imaging tools in neurosciences. J. Physiol. Paris 99, 281–292. ( 10.1016/j.jphysparis.2006.03.011) [DOI] [PubMed] [Google Scholar]
- 8.Gholipour A, Kehtarnavaz N, Briggs R, Devous M, Gonipath K. 2007. Brain functional localization: a survey of image registration techniques. IEEE Trans. Med. Imaging 26, 427–451. ( 10.1109/TMI.2007.892508) [DOI] [PubMed] [Google Scholar]
- 9.MacKenzie-Graham A, Boline J, Toga AW. 2007. Brain atlases and neuroanatomic imaging. Methods Mol. Biol. 401, 183–194. ( 10.1007/978-1-59745-520-6_11) [DOI] [PubMed] [Google Scholar]
- 10.Pantazis D, Joshi A, Jiang J, Shattuck D, Bernstein LE, Damasio H, Leahy RM. 2010. Comparison of landmark based and automatic methods for cortical surface registration. Neuroimage 49, 2479–2493. ( 10.1016/j.neuroimage.2009.09.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiens JJ. 2004. The role of morphological data in phylogeny reconstruction. Syst. Biol. 53, 653–661. ( 10.1080/10635150490472959) [DOI] [PubMed] [Google Scholar]
- 12.Adams DC, Rohlf FJ, Slice DE. 2004. Geometric morphometrics: ten years of progress following the revolution. Ital. J. Zool. 71, 5–16. ( 10.1080/11250000409356545) [DOI] [Google Scholar]
- 13.Boyer DM, Lipman Y, StClair E, Puente J, Patel BA, Funkhouser T, Jernvall J, Daubechies I. 2011. Algorithms to automatically quantify the geometric similarity of anatomical surface. Proc. Natl Acad. Sci. USA 108, 18 221–18 226. ( 10.1073/pnas.1112822108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyer DM, Puente J, Gladman JT, Glynn C, Mukherjee S, Yapuncich GS, Daubechies I. 2015. A new fully automated approach for aligning and comparing shapes. Anat. Rec. 298, 249–276. ( 10.1002/ar.23084) [DOI] [PubMed] [Google Scholar]
- 15.Scotland RW, Olmstead RG, Bennett JR. 2003. Phylogeny reconstruction: the role of morphological data. Syst. Biol. 52, 539–548. [DOI] [PubMed] [Google Scholar]
- 16.Weaver T. 2014. Tracing the paths of modern humans from Africa. Proc. Natl Acad. Sci. USA 111, 7170–7171. ( 10.1073/pnas.1405852111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reyes-Centero H, Ghirotto S, Détroit F, Grimaud-Hervé D, Barbujani G, Harvati K. 2014. Genomic and cranial phenotype data support multiple modern human dispersals from Africa and a southern route into Asia. Proc. Natl Acad. Sci. USA 111, 7248–7253. ( 10.1073/pnas.1323666111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLeod N, Benfield M, Culverhouse P. 2010. Time to automate identification. Nature 467, 154–155. ( 10.1038/467154a) [DOI] [PubMed] [Google Scholar]
- 19.Thompson DW. 1917. On growth and form. Cambridge, UK: University Press. [Google Scholar]
- 20.Gartus A, Geissler A, Foki T, Tahamtan AR, Pahs G, Barth M, Pinker K, Trattnig S, Beisteiner R. 2007. Comparison of fMRI coregistration results between human experts and software solutions in patients and healthy subjects. Eur. Radiol. 17, 1634–1643. ( 10.1007/s00330-006-0459-z) [DOI] [PubMed] [Google Scholar]
- 21.Turner WD, Brown RE, Kelliher TP, Tu PH, Taister MA, Miller KW. 2005. A novel method of automated skull registration for forensic facial approximation. Forensic Sci. Int. 159, 149–158. ( 10.1016/j.forsciint.2004.10.003) [DOI] [PubMed] [Google Scholar]
- 22.Lu H, Nolte L-P, Reyes M. 2012. Interest points location for brain image using landmark-annotated atlas. Int. J. Imaging Syst. Technol. 22, 145–152. ( 10.1002/ima.22015) [DOI] [Google Scholar]
- 23.Cates J, Meyer M, Fletcher PT, Whiteker R. 2006. Entropy-based particle systems for shape correspondence. In Proc. 1st MICCAI workshop on Mathematical Foundations of Computational Anatomy: Copenhagen, Denmark, 1 October 2006, pp. 90–99. Lecture Notes in Computer Science. New York, NY: Springer.
- 24.Cates J, Fletcher PT, Styner M, Shenton M, Whitaker R. 2007. Shape modeling and analysis with entropy-based particle systems. Inf. Process. Med. Imaging 20, 333–345. ( 10.1007/978-3-540-73273-0_28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rustamov RM.2007. Laplace–Beltrami eigenfunctions for deformation invariant shape representation. In SGP'07, Proc. of the Symposium on Geometry Processing, Eurographics Association, Barcelona, Spain, 46 July 2007, pp. 225–233. Aire-la-Ville, Switzerland: Eurographics Association.
- 26.Sun J, Ovsjanikov M, Guibas L.2009. A concise and provably informative multi-scale signature based on heat diffusion. In SGP'09, Proc. of the Symposium on Geometry Processing, Eurographics Association, Berlin, Germany, 15–17 July 2009, pp. 1383–1392. Aire-la-Ville, Switzerland: Eurographics Association.
- 27.Sampson PD, Bookstein FL, Sheenan FH, Bolson EL. 1996. Eigenshape analysis of left ventricular outlines from contrast ventriculograms. In Advances in morphometrics, vol. 22 (eds Marcus LF, Corti M, Loy A, Naylor GJP, Slice DE), pp. 211–233. New York, NY: Plenum Press. [Google Scholar]
- 28.Bookstein FL. 1977. Landmark methods for forms without landmarks: localizing group differences in outline shape. Med. Image Anal. 1, 225–243. ( 10.1016/S1361-8415(97)85012-8) [DOI] [PubMed] [Google Scholar]
- 29.Gunz P, Mitteroecker P, Bookstein FL. 2006. Semilandmarks in three dimensions. In Modern morphometrics in physical anthropology (ed. Slice DE.), pp. 73–98. New York, NY: Kluwer Academic/Plenum Publishers. [Google Scholar]
- 30.Perez SI, Bernal V, Gonzalez PN. 2006. Differences between sliding semi-landmark methods in geometric morphometrics, with an application to human craniofacial and dental variation. J. Anat. 208, 769–784. ( 10.1111/j.1469-7580.2006.00576.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitteroecker P, Gunz P. 2009. Advances in geometric morphometrics. Evol. Biol. 36, 235–247. ( 10.1007/s11692-009-9055-x) [DOI] [Google Scholar]
- 32.Polly PD, MacLeod N. 2008. Locomotion in fossil carnivora: an application of eigensurface analysis for morphometric comparison of 3D surfaces. Palaeontol. Electron. 11, 13. [Google Scholar]
- 33.Bronstein AM, Bronstein MM, Kimmel R. 2006. Generalized multidimensional scaling: a framework for isometry-invariant partial surface matching. Proc. Natl Acad. Sci. USA 103, 1168–1172. ( 10.1073/pnas.0508601103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q, Adams B, Wicke M, Guibas L.2008. Non-rigid registration under isometric deformations. In SGP'08, Proc. of the Symposium on Geometry Processing, Copenhagen, Denmark, 2–4 July 2008, pp. 1149–1458. Aire-la-Ville, Switzerland: Eurographics Association.
- 35.Lasowski R, Tevs A, Seidel H-P, Wand M.2009. A probabilistic framework for partial intrinsic symmetries in geometric data. In IEEE Int. Conf. on Computer Vision, Kyoto, Japan, 29 September–2 October 2009, pp. 963–970. New York, NY: IEEE Publishing.
- 36.Valliant M, Glaunès J. 2005. Surface matching via currents. Lect. Notes Comp. Sci. 3565, 381–392. ( 10.1007/11505730_32) [DOI] [PubMed] [Google Scholar]
- 37.McCane B. 2013. Shape variation in outline shapes. Syst. Biol. 62, 134–146. ( 10.1093/sysbio/sys080) [DOI] [PubMed] [Google Scholar]
- 38.Laga H, Kurtek S, Srivastava A, Miklavcic SJ. 2014. Landmark-free statistical analysis of the shape of plant leaves. J. Theoret. Biol. 363, 41–52. ( 10.1016/j.jtbi.2014.07.036) [DOI] [PubMed] [Google Scholar]
- 39.Koehl P, Hass J. 2014. Automatic alignment of genus-zero surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 36, 466–478. ( 10.1109/TPAMI.2013.139) [DOI] [PubMed] [Google Scholar]
- 40.Hass J, Koehl P. 2014. How round is a protein? Exploring protein structures for globularity using conformal mapping. Front. Mol. Biosci. 1, 26 ( 10.3389/fmolb.2014.00026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bers L. 1972. Uniformization, moduli, and Kleinian groups. Bull. London Math. Soc. 4, 257–300. ( 10.1112/blms/4.3.257) [DOI] [Google Scholar]
- 42.Hass J, Koehl P.2015. A metric for genus-zero surfaces. (http://arxiv.org/abs/1507.00798. )
- 43.Fawcett T. 2006. An introduction to ROC analysis. Pattern Recogn. Lett. 27, 861–874. ( 10.1016/j.patrec.2005.10.010) [DOI] [Google Scholar]
- 44.Bowie JU, Lüthy R, Eisenberg D. 1991. A method to identify protein sequences that fold into a known three dimensional structure. Science 253, 164–170. ( 10.1126/science.1853201) [DOI] [PubMed] [Google Scholar]
- 45.Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5, 164–166. [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molec. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math. Biosci. 53, 131–147. ( 10.1016/0025-5564(81)90043-2) [DOI] [Google Scholar]
- 48.Sokal RR. 1963. The principles and practice of numerical taxonomy. Taxon 12, 190–199. ( 10.2307/1217562) [DOI] [Google Scholar]
- 49.Houle D, Govindaraju DR, Omholt S. 2010. Phenomics: the next challenge. Nat. Rev. 11, 855–866. ( 10.1038/nrg2897) [DOI] [PubMed] [Google Scholar]
- 50.Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284, 2124–2129. ( 10.1126/science.284.5423.2124) [DOI] [PubMed] [Google Scholar]
- 51.Wiens JJ. 2000. Phylogenetic analysis of morphological data. Washington, DC: Smithsonian Books. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.